Abstract
Legionella pneumophila, the causative agent of Legionnaires' disease, utilizes a type IVB secretion system to subvert its host cells and grow intracellularly. This type IV secretion system is composed of 25 icm (or dot) genes that probably constitute parts of a secretion complex as well as more than 30 proteins that are translocated via this system into the host cells. Three of the Icm/Dot proteins (DotD, DotC, and IcmN) contain a lipobox motif at their N terminals and are predicted to be lipoproteins. Two of these lipoproteins (DotD and DotC) were found to be essential for intracellular growth in both HL-60-derived human macrophages and in the protozoan host Acanthamoeba castellanii, while the third lipoprotein (IcmN) was found to be partially required for intracellular growth only in A. castellanii. Mutation analysis of the lipobox cysteine residue, which was shown previously to be indispensable for the lipobox function, indicated that both DotC and DotD are partially functional without this conserved residue. Cysteine mutations in both DotC and DotD or in DotC together with an icmN deletion or in DotD together with an icmN deletion were found to be additive, indicating that each of these lipoproteins performs its function independently from the others. Analysis of the transcriptional regulation of both the dotDC operon and the icmN gene revealed that both had higher levels of expression at stationary phase which were partially dependent on the LetA regulator. Our results indicate that the lipoproteins of the L. pneumophila icm (or dot) system are essential components of the secretion system and that they perform their functions independently.
Legionella pneumophila, the causative agent of Legionnaires' disease, is a facultative intracellular pathogen. In nature, this bacterium multiplies within different protozoa, and during infection in humans it multiplies inside alveolar macrophages and monocytes. The main pathogenesis system known today in L. pneumophila is the Icm/Dot type IVB secretion system (reviewed in reference [30]). This system was shown to be composed of 25 icm (or dot) genes, which probably constitute part of a secretion complex, and more than 30 effector proteins (RalF, LidA, Lep, Sid, Wip, Vip, Ylf, and others), which are probably translocated into the host cell during infection (4, 6, 8, 22, 24, 26, 37). The 25 Icm/Dot proteins can be divided into two groups according to their homologies: one group consists of 18 Icm/Dot proteins (IcmT, IcmP, IcmO, IcmM, IcmL, IcmK, IcmE, IcmG, IcmC, IcmD, IcmJ, IcmB, IcmV, IcmX, DotA, DotB, DotC, and DotD) that were found to be homologous to proteins involved in conjugation present on IncI plasmids such as R64, and the other consists of 7 proteins (IcmS, IcmR, IcmQ, IcmN, IcmF, IcmH, and IcmW) that have no homologous proteins in conjugation systems. Three of the proteins that have homologies to conjugation-related proteins (IcmB, IcmO, and DotB) contain an ATP/GTP binding motif similar to that of their homologue proteins in the type IVA secretion system (VirB4, VirD4, and VirB11, respectively). Two other proteins that belong to the first group (DotD and DotC) contain a lipobox motif which is present in proteins that are anchored to the bacterial membrane. An additional lipoprotein that belongs to the second group (IcmN) was also shown to be part of the Icm/Dot secretion system, but an insertion mutation in this gene was found previously to be dispensable for intracellular growth in HL-60-derived human macrophages and only partially required for intracellular growth in Acanthamoeba castellanii (34).
The importance of lipoproteins as building blocks of bacterial secretion systems involved in pathogenesis is well documented. In the type III secretion systems of Salmonella enterica and Shigella flexneri, InvH and MxiJ, respectively, along with VirB7 of the Agrobacterium tumefaciens type IVA secretion system, were shown to be lipoproteins as well as essential components of these systems (9, 11, 21, 28, 29).
In this report, we examined the importance of the three Icm/Dot lipoproteins (DotD, DotC, and IcmN) for intracellular growth by analyzing the requirement of their conserved lipobox motif for their functions in pathogenesis as well as by examining the functional relations between them. Our results clearly indicate that DotC and DotD are completely required for intracellular growth but also remain partially functional without a functional lipobox motif. However, all three lipoproteins were found to have additive effects on intracellular growth when examined as double mutants, indicating that they perform their functions independently from one another.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
L. pneumophila and Escherichia coli strains used in this work are listed in Table 1. Plasmids and primers used in this work are described in Table 2 and Table 3, respectively. Bacterial media, plates, and antibiotic concentrations were used as described before (32).
TABLE 1.
Bacterial strains
| Strain | Genotype and features | Reference or source |
|---|---|---|
| L. pneumophila | ||
| GS3007 | JR32 icmN::Km | 31 |
| GS-RelA | JR32 relA::Km | 43 |
| GY203 | JR32 dotD (in-frame nonpolar deletion) | This study |
| GY303 | JR32 dotC (in-frame nonpolar deletion) | This study |
| GY606 | JR32 dotDC (in-frame nonpolar deletion) | This study |
| GY806 | JR32 dotD and icmN::Km | This study |
| GY1003 | JR32 dotC and icmN::Km | This study |
| JR32 | Homogeneous salt-sensitive isolate of AM511 | 27 |
| LM1376 | JR32 rpoS::Km | 15 |
| OG2001 | JR32 letA::Km | 13 |
| E. coli | ||
| MC1022 | araD139 Δ(ara leu)7697 Δ(lacZ)M15 galU galK strA | 5 |
| SY327λpir | (lac pro) argE(Am) rif nalA recA56 pir | 10 |
TABLE 2.
Plasmids used in this study
| Plasmid | Featuresa | Reference or source |
|---|---|---|
| pGY-100 | sacB CmroriR6K | 44 |
| pMMB207αb-Km-14 | IncQ lacIq CmrmobA::kan MCS | 33 |
| pUC18 | ori ColE1 MCS Apr | 40 |
| pGS-lac-02 | Promoterless lacZ incQ Cmr | 14 |
| pGY-dotD-12 | 1kb of dotD upstream region in pUC-18 | This study |
| pGY-dotD-34 | 1kb of dotD downstream region in pUC-18 | This study |
| pGY-D-Km-01 | The dotD upstream and downstream regions with the Km cassette between them in pUC-18 | This study |
| pGY100-D-Km-01 | The insert of pGY-D-Km-01 in pGY-100 | This study |
| pGY100-D-01 | pGY100-D-Km-01 without the Km cassette | This study |
| pGY-dotC-12 | 1kb of dotC upstream region in pUC-18 | This study |
| pGY-dotC-34 | 1kb of dotC downstream region in pUC-18 | This study |
| pGY-C-Km-01 | The dotC upstream and downstream regions with the Km cassette between them in pUC-18 | This study |
| pGY100-C-Km-01 | The insert of pGY-C-Km-01 in pGY-100 | This study |
| pGY100-C-01 | pGY100-C-Km-01 without the Km cassette | This study |
| pGY-CD-Km-01 | The dotD upstream and dotC downstream regions with the Km cassette between them in pUC-18 | This study |
| pGY100-DC-Km-01 | The insert of pGY-CD-Km-01 in pGY-100 | This study |
| pGY100-DC-01 | pGY100-DC-Km-01 without the Km cassette | This study |
| pGS-Le-40-Km-1 | icmN with the Km cassette | 31 |
| pGS-BCD-01 | The dotDCB region in pUC-18 | 18 |
| pGY-dotD-02 | dotD in pMMB207αb-Km-14 | This study |
| pGY-dotDC-02 | dotDC in pMMB207αb-Km-14 | This study |
| pGY-DCB-06 | dotDCB in pMMB207αb-Km-14 | This study |
| pGY-DCB-09 | dotDCB in pMMB207αb-Km-14 | This study |
| pGY-dotC-SE-01 | Part of dotC and dotB in pUC-18 | This study |
| pGY-dotDC-SB-01 | dotD and part of dotC in pUC-18 | This study |
| pGY-D-C19S-01 | dotD C19S in pUC-18 | This study |
| pGY-C-C19A-02 | dotC C19A in pUC-18 | This study |
| pGY-D-C19S-02 | pGY-DCB-09 with dotD C19S | This study |
| pGY-C-C19A-03 | pGY-DCB-09 with dotC C19A | This study |
| pGY-DC-C19SA-01 | pGY-DCB-09 with dotD C19S and dotC C19A | This study |
| pGY-dotDlac-06 | 122 bp upstream of dotD in pUC-18 | This study |
| pGY-dotDlac-07 | 122 bp upstream of dotD in pGS-lac-02 | This study |
| pSR-N(V)-lac | 444 bp upstream of icmN in pGS-lac-02 | This study |
MCS, multiple cloning site; Ap, ampicillin.
TABLE 3.
Primers used in this study
| Primer name | Sequence (5′-3′) |
|---|---|
| dotD-2 | GTCGACACCAGCAAGAAGCGCTGAAA |
| dotD-3 | GTCGACGTCGTAGAATTGCGCTATGC |
| dotD-1 | GAATTCTCAACCAAAAACTCCGAGGC |
| dotD-4 | AAGCTTATCGAATACCAGATACTCCCTT |
| dotC-2 | GTCGACCAGGAGGGCTGATAGCAAAA |
| dotC-3 | GCGTCGACTGGCAACCTATTATAGCACC |
| dotC-1 | GAATTCGTGGCGCGTTGGGATTTGAA |
| dotC-4 | AAGCTTATCGAATACCAGATACTCCCTT |
| DotC-C19A-F | CGAGAAGAAGCGGCTACCAGGAGGGCTGAT |
| DotC-C19A-R | TGGTAGCCGCTTCTTCTCGTAATCATTACGG |
| DotD-C19S-F | TGTTCCAGCAGAACCAGCAAGAAGCGCTGAA |
| DotD-C19S-R | TTGCTGGTTCTGCTGGAACAATGAAATTTAA |
| ORF7-3 | GTCGACGCCGGTCAAGTCATTTATGG |
| dotDlac | GGATCCCCAATCTTATTGTTGTTCATTCAATG |
| icmN(V)lac | CGGGGGATCCCCAGTACGAAGTGATCTCACCTG |
| icmN-F1 | GCCGGAATTCAGGCTGCAGAGGAATTAACA |
| GSP-dotDC | CACGAAATCTGAAATGTGCTGCTT |
| RACE-DC-1 | CGCCGGATCCGCATGGAGTCACTGACTGAAACGG |
| RACE-DC-2 | CGGGGGATCCTTTAATGGTCGCATCATCACTTGG |
| GSP-icmN | TACTCTTGATCTCGCCAATGTCAA |
| RACE-N-1 | CGCCGGATCCGATCAGCAAACAGAGCGCTAGC |
| RACE-N-2 | CGGGGGATCCTTATTGTTGCATCGCATGCACC |
| dotB-GSP | AATTGTGTCGTGGCATTCGG |
| dotC-GSP | TGCTCTCCAGGCTAACCCAG |
| B-1 | GTCGCCCAATTCTGTATTGG |
| C-1 | GATGACAGGGTCTTACGCATC |
| C-3 | CTTAAAGCGGTTTCCTTAAGGG |
| D-1 | AGAATTGACTGCACGTATCGC |
| 7-1 | GTCGACGCCGGTCAAGTCATTTATGG |
Plasmid construction for complementation.
The plasmid pGS-BCD-01, which contains a 12-kb BamHI fragment that covers part of the icm (or dot) region I (dotD, dotC, and dotB), was used for the construction of four complementing plasmids. The plasmid pGS-BCD-01 was digested with BsaBI and PvuII, and a 3,535-bp fragment that contains the dotD, dotC, and dotB genes was cloned into the SmaI site of pMMB207αb-km-14 to generate pGY-7DCB-06. The plasmid pGY-7DCB-06 contains two EcoRI sites, one of which is located in the polylinker site; this plasmid was partially digested with EcoRI and self-ligated after fill-in. The resulting plasmid, pGY-7DCB-09, contains only one EcoRI site, since the polylinker site was deleted. The plasmid pGY-7DCB-06 was digested with SacI and EcoRI and self-ligated after treatment with T4 polymerase to generate pGY-dotD-02; this plasmid contains the dotD gene without the dotC and dotB genes. The plasmid pGY-7DCB-06 was digested with SphI and EcoRI and self-ligated after treatment with T4 polymerase to generate pGY-dotDC-02; this plasmid contains the dotD and dotC genes without the dotB gene.
Construction of plasmids containing mutations in the lipobox motif.
Two plasmids were generated for use as a template for the PCR mutagenesis. The plasmid pGY-7DCB-09 was digested with SacI and EcoRI, and the resulting 1,347-bp fragment was cloned into the same sites in pUC-18 to generate pGY-dotC-SE-01. The plasmid pGY-7DCB-06 was digested with SacI and BamHI, and the resulting 1,341-bp fragment was cloned into the same sites in pUC-18 to generate pGY-dotC-SB-01. These two plasmids were used as a template for a site-directed mutagenesis using the overlap extension PCR method (19). For each mutation, two primers that contain the mutation and overlap one another by 20 bp were designed (DotC-C19A-F and DotC-C19A-R; DotD-C19S-F and DotD-C19S-R; Table 3). The PCR mutagenesis includes two steps. In the first step, two PCR fragments were generated using the following primer pairs: (i) a primer located on the vector upstream from the regulatory region and one of the primers that contains the mutation, and (ii) a primer located on the vector and a second primer that contains the same mutation on the complementary strand. The resulting two fragments were gel purified and used as templates in the second step, which includes a third PCR using the two primers located on the vector. The resulting PCR products were digested with BamHI and SacI and cloned into the same sites in pGY-7DCB-09 to generate pGY-D-C19S-02, containing a C-to-S mutation in the lipobox motif of DotD, and pGY-C-C19A-02, containing a C-to-A mutation in the lipobox motif of DotC. To join the two mutations together, the plasmid pGY-7DCB-09 was digested with SacI and BamHI, the plasmid pGY-D-C19S-02 was digested with BamHI and XcmI, and the plasmid pGY-C-C19A-02 was digested with XcmI and SacI; these three fragments were ligated to generate pGY-DC-C19SA-01, which contains the mutations in both dotC and dotD.
Construction of lacZ fusions.
The promoterless lacZ vector pGS-lac-02 was used for the cloning of different fragments that originated from the regulatory region of the dotDCB operon and the icmN gene. A 122-bp fragment containing the dotD regulatory region was amplified by PCR using the 7-1 and dotDlac primers (Table 3). The resulting fragment (pGY-dotDlac-06) was cloned into pUC-18 and sequenced. pGY-dotDlac-06 was digested with BamHI and EcoRI and cloned into the pGS-lac-02 vector to generate pGY-dotDlac-07, which was used to determine the level of expression of the dotBCD operon. A 444-bp fragment containing the icmN regulatory region was amplified by PCR using the icmN(V)lac and icmN-F1 primers (Table 3). The resulting fragment was digested with BamHI and EcoRI and cloned into pGS-lac-02 to generate pSR-N(V)-lac, which was used to determine the level of expression of the icmN gene.
Plasmid construction for allelic exchange.
To generate in-frame nonpolar deletions, the allelic exchange vector pGY-100 was used (44). To generate in-frame nonpolar deletions in the L. pneumophila dotD, dotC, and dotDC genes, a 1-kb DNA fragment located on each side of the deletion planned was amplified by PCR. Each primer was planned to contain a SalI site at the place where the deletion will occur. Four fragments were amplified by use of the primers dotD-1, dotD-2, dotD-3, dotD-4, dotC-1, dotC-2, dotC-3, and dotC-4 (Table 3) and cloned into pUC-18 digested with HincII to generate pGY-dotD-12, pYG-dotD-34, pGY-dotC-12, and pGY-dotC-34. The inserts of all these plasmids were sequenced to confirm that no mutations were incorporated during the PCR. The resulting plasmids were digested with EcoRI and SalI (for plasmids that contain the fragment located upstream of the deletion) or with HindIII and SalI (for plasmids that contain the fragment located downstream of the deletion). Pairs of these fragments, including a pair that contains the dotD upstream region and the dotC downstream region, were part of a four-way ligation that contained a kanamycin (Km) resistance cassette (Pharmacia) digested with SalI and the pUC-18 vector digested with EcoRI and HindIII to generate pGY-D-Km-01, pGY-C-Km-01, and pGY-CD-Km-01. These three plasmids were digested with PvuII (this enzyme cuts on both sides of the pUC-18 polylinker), and the resulting fragments were cloned into the pGY-100 vector digested with XmnI to generate pGY100-D-Km-01, pGY100-C-Km-01, and pGY100-DC-Km-01. These three plasmids were digested with SalI and self-ligated to form pGY100-D-01, pGY100-C-01, and pGY100-DC-01, which were used for the allelic exchange as described below.
L. pneumophila allelic exchange.
The plasmids pGY100-D-01, pGY100-C-01, and pGY100-DC-01 (described above) were introduced into L. pneumophila JR32 by electroporation, grown in ACES [N-(2-acetamido)-2-aminoethamesulfonic acid] yeast extract for 5 h, and plated on ABCYE (ACES-buffered charcoal yeast extract) plates containing chloramphenicol (Cm). Transformants were patched on ABCYE plates containing Cm and then streaked on ABCYE plates containing 2% (wt/vol) sucrose (Suc) to select for cells that no longer contained vector pGY-100 sequences (Cms and Sucr). Single isolates that grow on the Suc-containing plates were patched on ABCYE plates containing Cm and on plain ABCYE plates. Cms and Sucr isolates were tested by PCR to confirm that the correct change occurred. At least six independent isolates were tested for each allelic exchange. The plasmid pGS-Le-40-Km-1 was used to generate a Km insertion in the icmN gene as described before (31).
Intracellular growth in A. castellanii.
Intracellular growth assays were performed in a way similar to that previously described (34). Amoebae (1.5 × 105) in peptone-yeast extract-glucose medium were added to wells of a 24-well microtiter dish, and the amoebae were incubated for 1 h at 37°C to let the amoebae adhere. Next, the peptone-yeast extract-glucose medium was aspirated, the wells were washed once with 0.5 ml of warm (37°C) Acanthamoeba buffer (Ac buffer), and 0.5 ml of warm Ac buffer was added to the wells. Then, L. pneumophila in Ac buffer was added to the wells at a multiplicity of infection of approximately 0.1. The plate was incubated for 30 min at 37°C, and then the Ac buffer was aspirated, the wells were washed three times with 0.5 ml of warm Ac buffer, and 0.6 ml of warm Ac buffer was added to the wells. The supernatant of each well was sampled (50 μl) at intervals of about 24 h, and CFU counts were determined by plating samples on ABCYE plates.
Intracellular growth in HL-60-derived human macrophages.
Intracellular growth assays were performed in a way similar to that previously described (34). Wells of a 24-well microtiter dish containing 6 × 106 differentiated HL-60-derived macrophages were used for infection. L. pneumophila was added to the wells at a multiplicity of infection of approximately 0.1, and the infected HL-60-derived macrophages were incubated for 1 h at 37°C under CO2 (5%). Then, the wells were washed three times, and 0.6 ml of RPMI containing 2 mM glutamine and 10% normal human serum was added to the wells. The supernatant of each well was sampled (50 μl) at intervals of about 24 h, and CFU counts were determined by plating on ABCYE plates.
Sodium sensitivity.
The sodium sensitivity assay was performed essentially as described before (42). The wild-type L. pneumophila strain JR32 and several mutants were grown for 72 h on ABCYE plates, scraped off the plate, and calibrated to an optical density at 600 nm of 4. Then, eight 10-fold serial dilutions were plated on ABCYE plates containing or lacking 100 mM NaCl. Sodium sensitivity was determined by comparing the numbers of bacteria growing on each plate.
β-Galactosidase assays.
The β-galactosidase expression levels in L. pneumophila were determined as previously described (12); the substrate for β-galactosidase hydrolysis was O-nitrophenyl-β-d-galactopyranoside.
RNA manipulations.
RNA was prepared as described before (12). To determine the transcription start sites of the dotDCB operon and the icmN gene, the 5′ rapid amplification of cDNA ends (RACE) system was used as described by the manufacturer (Invitrogen). The RACE-GSP primers (GSP-DC and GSP-icmN; Table 3) were used for generating the cDNA, these primers together with the abridged anchor primer (supplied with the kit) were used for the first PCR, and the abridged universal amplification primer (supplied with the kit) and specific primers (RACE-DC-1, RACE-DC-2, RACE-N-1, and RACE-N-2; Table 3) were used for the nested PCR. The resulting fragments from the second PCR were subsequently cloned, and about four different clones for each gene were sequenced to determine the transcription start sites of the mRNA.
To determine if the dotD, dotC, and dotB genes are located on one transcriptional unit, a reverse transcription reaction was performed with the primers (dotB-GSP and dotC-GSP) by use of avian myeloblastosis virus reverse transcriptase (Invitrogen). The cDNA product was analyzed by PCR using the primers B-1 and C-1 (Table 3) to determine whether the dotB and dotC genes are located on one transcriptional unit, and the primers C-3 and D-1 (Table 3) were used to determine whether the dotC and dotD genes are located on the same transcriptional unit. As a negative control, the primers dotDlac and 7-1 were used (Table 3).
RESULTS
Three proteins that were reported to be part of the Icm/Dot type IV secretion system were shown to contain a lipobox motif and are predicted to be lipoproteins. Two of these proteins (DotD and DotC) are homologous to proteins encoded on conjugative plasmids, and they are encoded from genes located in icm (or dot) region I. The third lipoprotein (IcmN) does not contain any homology to proteins encoded by conjugative plasmids, but it was found to contain homology to many bacterial proteins, and the gene that codes for this protein is located in the middle of icm (or dot) region II. Previously, it was shown that icmN is dispensable for intracellular growth in HL-60-derived human macrophages and only partially required for intracellular growth in amoebae; however, the phenotypes of dotC and dotD deletion mutants were not previously described. To reveal the importance of these three lipoproteins, the role of their lipobox motif, and the functional relation between them in the icm (or dot) system, they were examined as described below.
DotD and DotC are required for intracellular growth in amoebae and macrophages.
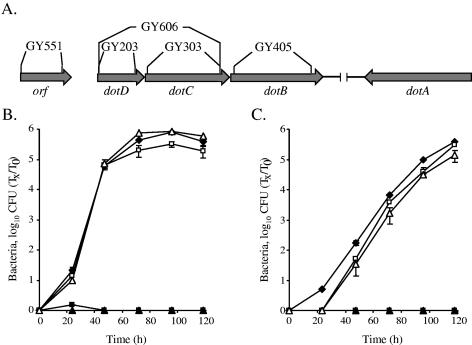
To determine the importance of the dotD and dotC genes for intracellular growth, in-frame nonpolar deletions were constructed within them. The resulting mutants (GY203 and GY303; Fig. 1A) had no changes in their growth rates in vitro (data not shown), but they were found to be completely defective for intracellular growth in the protozoan host A. castellanii (Fig. 1B) as well as in HL- 60-derived human macrophages (Fig. 1C). The intracellular growth abilities of both mutants were completely restored when a plasmid containing the dotD gene by itself (pGY-dotD-02) was introduced into the dotD mutant (GY203) and when a plasmid containing the dotDC genes (pGY-dotDC-02) was introduced into the dotC mutant (GY303) (Fig. 1B and C). These results indicated that the phenotypes observed for the dotC and dotD genes were not due to a polar effect on the downstream dotB gene but rather due to the requirement of both dotC and dotD mutants for pathogenesis. The results described here place the dotD and dotC genes together with most of the other icm (or dot) genes (icmT, icmP, icmO, icmM, icmL, icmK, icmE, icmC, icmD, icmJ, icmB, icmV, icmX, dotA, and dotB), which were shown to be completely required for intracellular growth in both HL-60-derived macrophages and amoebae (2, 3, 32-34, 36, 39); all these genes contain homologous genes on IncI plasmids and are probably part of the Icm/Dot secretion complex.
FIG. 1.
The L. pneumophila dotC and dotD genes are required for intracellular growth. (A) The icm (or dot) genomic region containing the dotDCB genes, as well as the in-frame nonpolar mutants that were constructed in these genes, is shown. Intracellular growth experiments in the protozoan host A. castellanii (B) and in HL-60-derived human macrophages (C) were performed as described in Materials and Methods. Shown are dotD mutant GY203 containing the vector pMMB207αb-Km-14 (solid boxes), the dotD gene (pGY-dotD-02) (open boxes), the dotC mutant GY303 containing the vector pMMB207αb-Km-14 (solid triangles), and the dotDC genes (pGY-dotDC-02) (open triangles). Solid diamonds represent wild-type L. pneumophila (JR32). The experiments were performed at least three times, and similar results were obtained. Tx, number of bacteria at each time point; T0, number of bacteria at time zero.
An additional phenotype that was shown to be connected to the icm (or dot) system is salt resistance (27); wild-type L. pneumophila was found to be salt sensitive, and the icm (or dot) mutants were shown to be salt resistant. When the dotD and dotC mutants were examined, they both were found to be salt resistant to the same extent as the other icm (or dot) mutants, a phenotype that was completely restored by introducing the complementing plasmids described above (data not shown).
The conserved cysteine residue of the lipobox motif of DotD and DotC is partially required for intracellular growth.
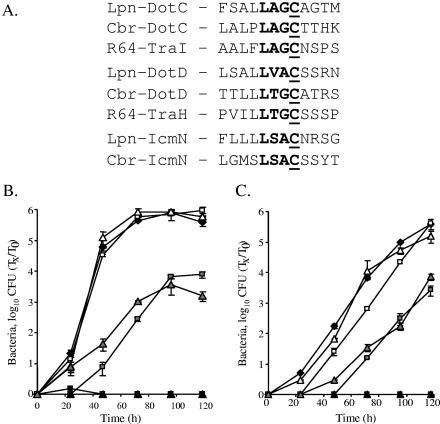
Sequence analysis of both DotD and DotC proteins revealed that they contain a conserved lipobox motif at their N-terminal regions (Fig. 2A). The lipobox motif ([LVI]-[ASTVI]-[AGS]-C; alternate amino acids for each position are shown in brackets) (23) functions in bacterial lipoproteins as a recognition signal for the lipid modification, which is made on the most conserved and essential cysteine residue (25, 38). It has been shown previously that changing this essential cysteine residue into another amino acid blocks the ability of the mutated protein to undergo the lipid modification (25, 38). To determine the importance of the lipid modification for the functions of the DotD and DotC proteins, site-directed mutagenesis was used, and the conserved cysteine residue was changed to serine (the amino acid that has the greatest structural similarity to cysteine) in each protein. When we tried to introduce the mutated DotC protein into L. pneumophila (into the dotC mutant as well as into the wild-type strain), no colonies were recovered. Further analysis of this mutant protein using a controlled expression system revealed that the mutated protein was lethal to the bacterial cells (data not shown). Due to this result, another mutation was constructed in the DotC cysteine residue, changing it to alanine, which resulted in a protein that was not lethal to the bacterial cells. The two mutant proteins constructed (DotD C19S and DotC C19A) were introduced into the dotD and dotC deletion strains, respectively, and analyzed for their abilities to complement the mutants for intracellular growth, as shown in Fig. 2B and C. When examined in A. castellanii, both mutants showed similar partial intracellular growth phenotypes (Fig. 2B). A similar result was also obtained in HL-60-derived human macrophages, but the effects of the mutations were less pronounced than the effects observed in the protozoan host. These data fit previous results obtained with several of the icm (or dot) insertion mutants showing more-severe defects for intracellular growth in A. castellanii in comparison to those in HL-60-derived human macrophages (34).
FIG. 2.
The DotD and DotC lipobox motif is partially required for intracellular growth. (A) Predicted lipobox motif in Icm/Dot proteins and their homologous proteins. The bacteria and plasmid (Lpn, L. pneumophila; Cbr, C. burnetii; R64, the IncI plasmid R64), the proteins, and the sequences of the predicted lipoboxes are indicated. The conserved amino acids ([LVI]-[ASTVI]-[AGS]-C; alternate amino acids for each position are shown in brackets) are in boldface, and the conserved cysteine residues are underlined. Intracellular growth experiments in the protozoan host A. castellanii (B) and in HL-60-derived human macrophages (C) were performed as described in Materials and Methods. Shown are the dotD mutant GY203 containing the vector pMMB207αb-Km-14 (solid boxes), the dotDCB operon (pGY-DCB-09) (open boxes), the dotDCB operon containing the mutated (C19S) dotD gene (pGY-D-C19S-01) (gray boxes), the dotC mutant GY303 containing the vector pMMB207αb-Km-14 (solid triangles), the dotDCB operon (pGY-DCB-09) (open triangles), and the dotDCB operon containing the mutated (C19A) dotC gene (pGY-C-C19A-02) (gray triangles). Solid diamonds represent wild-type L. pneumophila (JR32). The experiments were performed at least three times, and similar results were obtained. Tx, number of bacteria at each time point; T0, number of bacteria at time zero.
To determine whether the partial intracellular growth phenotypes observed with these mutants were not a result of a suppression mutation that appeared during the experiment, we used the progeny of one infection experiment for reinfection. The results obtained were very clear; the partial intracellular growth phenotypes of the mutants were the same as those observed for the original mutants used for the first infection, indicating that the partial intracellular growth phenotypes observed with the mutants were not a result of spontaneous suppression mutations.
In addition, when the amino acid located immediately downstream from the conserved cysteine residue was mutated (DotC A20D and DotD S20D), no defect for intracellular growth in either host was observed (data not shown), further indicating the importance of the conserved cysteine residue for the proper function of the protein.
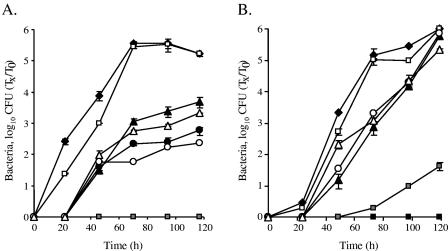
The effects of the DotD and DotC cysteine mutants are additive.
When both dotD and dotC mutants were complemented with plasmids containing the mutated cysteine residue, they had very similar intracellular growth phenotypes (Fig. 2B and C). A similar situation with regard to a partial intracellular growth phenotype was described before in the icm (or dot) system. The icmS and icmW insertion mutants had similar partial intracellular growth phenotypes (7), as did the icmH and icmF mutants (35, 42). In both of these cases, the construction of a double mutant resulted in a phenotype similar to the one obtained with each of the single mutants, indicating that both gene products perform their functions together. To examine whether a similar situation also occurs in the cases of the dotC and dotD genes, a strain containing deletions of both of these genes (GY606) was constructed (Fig. 1A). As expected from the results for the single mutations in these genes (Fig. 1B and C), the double mutant was unable to grow in A. castellanii or in HL-60-derived human macrophages (Fig. 3A and B), and it was fully complemented by a plasmid containing the wild-type dotD and dotC genes (Fig. 3A and B). Examination of plasmids containing one wild-type gene (containing the conserved cysteine residue) and one mutated gene (containing no cysteine residue in its lipobox) showed phenotypes similar to those observed for the single mutants complemented by the mutated genes (compare Fig. 2B and C and 3B and C). However, when a plasmid containing the two mutated genes (without the conserved cysteine) was introduced into the double mutant, no intracellular growth was observed in A. castellanii (Fig. 3A), and very poor growth was obtained in HL-60-derived human macrophages (Fig. 3B). These results clearly indicate that an additive effect occurs with the combined mutations, as expected from proteins that perform their functions independently. This is the first documentation of an additive effect for components of the icm (or dot) system.
FIG. 3.
The effects of the DotD and DotC cysteine mutants are additive. Intracellular growth experiments in the protozoan host A. castellanii (A) and in HL-60-derived human macrophages (B) were performed as described in Materials and Methods. Shown are the dotDC mutant GY606 containing the vector pMMB207αb-Km-14 (solid boxes), the dotDCB operon (pGY-DCB-09) (open boxes), the dotDCB operon with the mutated (C19S) dotD gene (pGY-D-C19S-01) (solid triangles), the dotDCB operon with the mutated (C19A) dotC gene (pGY-D-C19S-01) (open circles), the dotDCB operon with the mutated (C19S) dotD gene and the mutated (C19A) dotC gene (pGY-DC-C19SA-01) (gray boxes), the dotC mutant GY303 containing the dotDCB operon with the mutated (C19A) dotC gene (pGY-C-C19A-02) (solid circles), and the dotD mutant GY203 containing the dotDCB operon with the mutated (C19S) dotD gene (pGY-D-C19S-02) (open triangles). Solid diamonds represent wild-type L. pneumophila (JR32). The experiments were performed at least three times, and similar results were obtained. Tx, number of bacteria at each time point; T0, number of bacteria at time zero.
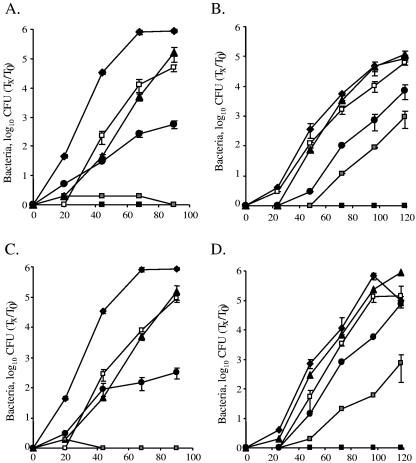
The effects of the DotD, DotC, and IcmN mutants are additive.
In addition to the fact that the DotD and DotC proteins contain a conserved lipobox motif, this motif was also found in IcmN (Fig. 2A). The corresponding gene is located in the middle of the icm (or dot) region II, and it codes for a protein that has many homologues in other bacteria but does not have a homologous protein on the IncI plasmid, on which proteins homologous to DotD and DotC (TraH and TraI, respectively) were found. An icmN insertion mutant (GS3007) was examined before for intracellular growth in HL-60-derived human macrophages and in A. castellanii, and it was found to be dispensable for intracellular growth in the human cell line and only partially required for intracellular growth in the protozoan host (34). To determine whether the minor defect in intracellular growth observed with the icmN insertion mutant results from the presence of the two other lipoproteins (DotD and DotC), double mutants were constructed containing nonpolar in-frame deletions in dotD or dotC together with an insertion mutation in icmN (GY806 and GY1003, respectively). When these double mutants were examined, they were found to be completely defective for intracellular growth, as expected from the single in-frame deletions in dotD and dotC (Fig. 4A through D). When the double mutants were complemented with a plasmid containing the dotDC genes, the intracellular growth defect in A. castellanii was similar to the one observed for the single icmN mutant, and no intracellular growth defect was observed in HL-60-derived human macrophages (an observation similar to that of the lack of the phenotype of the single icmN mutant). However, when the double deletion mutants were complemented with a plasmid containing the dotD or dotC cysteine mutants, no intracellular growth was observed in A. castellanii, and a clear additive effect was observed in HL-60-derived human macrophages (Fig. 4A through D). These results clearly indicate that the presence of two functional lipoproteins out of three (DotD and DotC, DotD and IcmN, or DotC and IcmN) is sufficient for a result of partial intracellular growth in A. castellanii, but when two of these proteins are mutated, no intracellular growth can be obtained in the protozoan host. These results clearly demonstrate that the intracellular growth phenotypes of the three lipoproteins (DotD, DotC and IcmN) are additive, and they probably perform their functions independently.
FIG. 4.
The effects of the DotD, DotC, and IcmN mutants are additive. Intracellular growth experiments in the protozoan host A. castellanii (A and C) and in HL-60-derived human macrophages (B and D) were performed as described in Materials and Methods. Shown in panels A and B are the dotD-icmN double mutant GY806 containing the vector pMMB207αb-Km-14 (solid boxes), the dotDCB operon (pGY-DCB-09) (open boxes), the dotDCB operon with the mutated (C19S) dotD gene (pGY-D-C19S-01) (gray boxes), and the dotD mutant GY203 containing the dotDCB operon with the mutated (C19S) dotD gene (pGY-D-C19S-02) (solid circles). Shown in panels C and D are dotC-icmN double mutant GY1003 containing the vector pMMB207αb-Km-14 (solid boxes), the dotDCB operon (pGY-DCB-09) (open boxes), the dotDCB operon with the mutated (C19A) dotC gene (pGY-C-C19A-01) (gray boxes), and the dotD mutant GY203 containing the dotDCB operon with the mutated (C19S) dotD gene (pGY-D-C19S-02) (solid circles). In all panels, solid diamonds represent wild-type L. pneumophila (JR32) and solid triangles represent the icmN mutant GS3007 containing the vector pMMB207αb-Km-14. The experiments were performed at least three times, and similar results were obtained. Tx, number of bacteria at each time point; T0, number of bacteria at time zero.
Analysis of the Coxiella burnetii dotDC genes.
Previously, it was found that the obligate intracellular pathogen C. burnetii contains proteins that are homologous to all the Icm/Dot proteins other than IcmR (44). In addition, it was demonstrated that some of the C. burnetii homologous genes can replace some of the icm (or dot) genes for their functions during intracellular growth, but some of them cannot replace the latter genes (41, 44). To determine whether the C. burnetii dotD and dotC genes can substitute for the corresponding L. pneumophila genes during intracellular growth, these genes were examined for complementation after they were cloned downstream to the L. pneumophila dotDCB regulatory region in a manner similar to that described before (44). Even though the C. burnetii DotD and DotC proteins are relatively homologous to the corresponding L. pneumophila proteins (39.2% identity and 45.8% similarity for DotD and 40.1% identity and 51.1% similarity for DotC), no complementation was observed (data not shown). Interestingly, similar results were also obtained previously with other C. burnetii icm genes (icmP, icmO, icmJ, icmB, and icmX), which also contain homologous genes on the conjugative plasmids (41, 44).
mRNA analysis of the dotDCB transcriptional unit and the icmN gene.
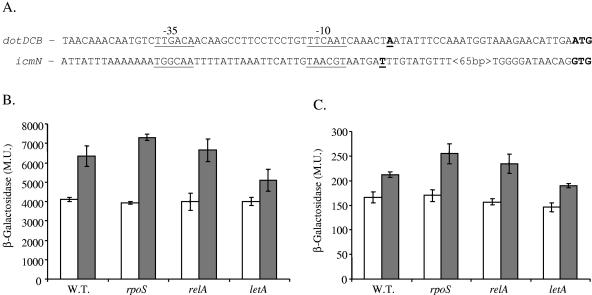
To characterize the basic regulatory elements and transcription organization of the dotD and dotC genes, reverse transcription analysis was used, and it was found that dotC, dotD, and dotB are located on the same transcriptional unit (data not shown), as was expected from the organization of the genes. In addition, using the RACE system, the transcription start sites of the dotDCB transcriptional unit and of the icmN gene were determined (Fig. 5A). The dotDCB transcription start site was found to be located 27 bp upstream from the first ATG of the dotD gene, and it is the first icm (or dot) gene in which a very clear −35 promoter element was observed (14). The icmN transcription start site was found to be located 87 bp upstream from the first ATG of the icmN gene, and it contains a relatively weak homology to the consensus promoter elements at both the −10 and the −35 promoter regions.
FIG. 5.
Analysis of dotDCB operon and icmN gene regulation. (A) Sequences of the dotDCB and icmN regulatory regions. The transcription start sites are in boldface and underlined, the initiation codons of the DotD and IcmN proteins are in boldface, and the predicted promoter sequences are underlined and indicated as −10 and −35. (B and C) β-Galactosidase activities of the dotD (B) and icmN (C) lacZ fusions in wild-type (W.T.) L. pneumophila, in the rpoS mutant (LM1376), in the relA mutant (GS-RelA) and in the letA mutant (OG2001) during exponential (white) and stationary (gray) phases. β-Galactosidase activity was measured as described in Materials and Methods. The results are the averages ± standard deviations of at least three different experiments. The β-galactosidase activities for exponential and stationary phases of both fusions and in all the strains were found to be significantly different (P > 0.0001) by the standard t test. M.U., Miller units.
The dotDCB operon and the icmN gene are expressed at higher levels at stationary phase.
To obtain additional information about the regulation of the dotDCB operon and the icmN gene, lacZ translational fusions were constructed. When examined in the wild-type strain, the dotD::lacZ fusion was found to be highly expressed, and its level of expression was about 50% higher at stationary phase (Fig. 5B). On the contrary, the icmN::lacZ fusion was expressed at low levels, and its level of expression was about 20% higher at stationary phase (Fig. 5C). To examine whether stationary phase-related regulators that were proposed to be involved in pathogenesis of L. pneumophila (1, 16, 17) are involved in the regulation of these lipoprotein-encoding genes, the levels of expression of these fusions were examined in rpoS, relA, and letA insertion mutants (Fig. 5B and C). As can be seen in Fig. 5B and C, the levels of expression of the dotD::lacZ and the icmN::lacZ fusions were unaffected by the RelA insertion mutant, were somewhat higher in the RpoS mutant at stationary phase, and were reduced in the LetA mutant at stationary phase. These results might indicate that even though the levels of expression of the dotDCB transcriptional unit and the icmN gene were found to be very different, they might be coregulated, since the effects of the three regulators examined on their expressions were minor but similar.
DISCUSSION
All the large bacterial secretion systems contain lipoproteins that probably anchor the complex to the membrane. The L. pneumophila icm (or dot) system is predicted to code for three lipoproteins, of which one (IcmN) was characterized before and found to be dispensable for intracellular growth in HL-60-derived human macrophages and only partially required for intracellular growth in the protozoan host A. castellanii. In this report, two additional lipoproteins (DotD and DotC) were characterized and found to be completely required for intracellular growth in both hosts, and the relations between these three lipoproteins were determined as well. Our results uncover three main findings about the lipoproteins in the Icm/Dot system.
First, even though the dotC and dotD deletion mutations resulted in completely null phenotypes for intracellular growth, the corresponding cysteine mutants had clear abilities to grow intracellularly, albeit not as well as the wild-type strain. In other systems where such mutations were constructed (11, 21), it was found that signal peptidase I rather than signal peptidase II (the natural processing enzyme of lipoproteins) processes the N-terminal ends of the mutated proteins, and in this way they are transferred to the periplasm. Analysis of the DotC and DotD wild-type sequences as well as of the mutated sequences using the LipoP program (20) indicated very clearly that these wild-type proteins are predicted to be recognized by signal peptidase II, while the cysteine mutants of these proteins were predicted to be recognized by signal peptidase I and expected to be cleaved at a location very similar to that for the wild-type protein. Taking this information together with the results of the intracellular growth analysis, it is most likely that the mutated DotC and DotD proteins were translocated to the periplasm and were partially able to perform their functions there, as evident from the intracellular growth results, indicating that both DotD and DotC lipoproteins have functions in the icm (or dot) system which are unrelated to their functions as lipoproteins.
Second, even though the dotC and dotD cysteine mutations resulted in similar intracellular growth phenotypes, the double mutation was found to be additive, which indicates that these two genes perform their functions independently. In addition, the Icm/Dot system was found to be partially functional for intracellular growth when all three lipoproteins were present in the bacterial cell, even if one of them did not contain a functional lipobox; however, there was no situation in which two out of the three lipoproteins were mutated and intracellular growth was observed in the protozoan host, indicating that the system can tolerate a change in one of its lipoproteins but not in two of them.
Third, the icmN gene that was characterized before had no effect on intracellular growth in HL-60-derived human macrophages and only a partial intracellular growth phenotype in A. castellanii. These results, together with the lack of homology between this protein and conjugation-related proteins, suggested that it might be that icmN, which is located in the middle of icm (or dot) region II, is not an integral part of the icm (or dot) system. However, the additive effects of the icmN insertion and the dotC and dotD cysteine mutants indicate that icmN is part of the icm (or dot) system.
In summary, as in other secretion systems, the lipoproteins of the L. pneumophila icm (or dot) system are essential components of this secretion system, and they perform their functions independently from one another.
Acknowledgments
We thank Sheli Radoshitzky for plasmid construction.
This work was supported by a grant from the “Center for the Study of Emerging Diseases” and in part by a grant (393/03) from the Israeli Science Foundation (to G.S.).
Editor: J. T. Barbieri
REFERENCES
- 1.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 2.Berger, K. H., J. J. Merriam, and R. R. Isberg. 1994. Altered intracellular targeting properties associated with mutations in the Legionella dotA gene. Mol. Microbiol. 14:809-822. [DOI] [PubMed] [Google Scholar]
- 3.Brand, B. C., A. B. Sadosky, and H. A. Shuman. 1994. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 14:797-808. [DOI] [PubMed] [Google Scholar]
- 4.Campodonico, E. M., L. Chesnel, and C. R. Roy. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 56:918-933. [DOI] [PubMed] [Google Scholar]
- 5.Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358-1361. [DOI] [PubMed] [Google Scholar]
- 7.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38:719-736. [DOI] [PubMed] [Google Scholar]
- 8.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 9.Dailey, F. E., and R. M. Macnab. 2002. Effects of lipoprotein biogenesis mutations on flagellar assembly in Salmonella. J. Bacteriol. 184:771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez, D., T. A. Dang, G. M. Spudich, X. R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 185:4908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gal-Mor, O., and G. Segal. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34:187-194. [DOI] [PubMed] [Google Scholar]
- 14.Gal-Mor, O., T. Zusman, and G. Segal. 2002. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J. Bacteriol. 184:3823-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 17.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 18.Hilbi, H., G. Segal, and H. A. Shuman. 2001. icm/dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42:603-617. [DOI] [PubMed] [Google Scholar]
- 19.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 20.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornacker, M. G., D. Faucher, and A. P. Pugsley. 1991. Outer membrane translocation of the extracellular enzyme pullulanase in Escherichia coli K12 does not require a fatty acylated N-terminal cysteine. J. Biol. Chem. 266:13842-13848. [PubMed] [Google Scholar]
- 22.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madan Babu, M., and K. Sankaran. 2002. DOLOP—database of bacterial lipoproteins. Bioinformatics 18:641-643. [DOI] [PubMed] [Google Scholar]
- 24.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 25.Narita, S., S. Matsuyama, and H. Tokuda. 2004. Lipoprotein trafficking in Escherichia coli. Arch. Microbiol. 182:1-6. [DOI] [PubMed] [Google Scholar]
- 26.Ninio, S., D. M. Zuckman-Cholon, E. D. Cambronne, and C. R. Roy. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm- mediated protein translocation. Mol. Microbiol. 55:912-926. [DOI] [PubMed] [Google Scholar]
- 27.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoenhals, G. J., and R. M. Macnab. 1996. Physiological and biochemical analyses of FlgH, a lipoprotein forming the outer membrane L ring of the flagellar basal body of Salmonella typhimurium. J. Bacteriol. 178:4200-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal, G., M. Feldman, and T. Zusman. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 29:65-81. [DOI] [PubMed] [Google Scholar]
- 31.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segal, G., and H. A. Shuman. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components on IncQ plasmid RSF1010. Mol. Microbiol. 30:197-208. [DOI] [PubMed] [Google Scholar]
- 34.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sexton, J. A., J. L. Miller, A. Yoneda, T. E. Kehl-Fie, and J. P. Vogel. 2004. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 72:5983-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sexton, J. A., J. S. Pinkner, R. Roth, J. E. Heuser, S. J. Hultgren, and J. P. Vogel. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186:1658-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shohdy, N., J. A. Efe, S. D. Emr, and H. A. Shuman. 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. USA 102:4866-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 1693:5-13. [DOI] [PubMed] [Google Scholar]
- 39.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 40.Yanish-Perron, C., J. Viera, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 41.Zamboni, D. S., S. McGrath, M. Rabinovitch, and C. R. Roy. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49:965-976. [DOI] [PubMed] [Google Scholar]
- 42.Zusman, T., M. Feldman, E. Halperin, and G. Segal. 2004. Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect. Immun. 72:3398-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zusman, T., O. Gal-Mor, and G. Segal. 2001. Characterization of a Legionella pneumophila relA insertion mutant and the role of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zusman, T., G. Yerushalmi, and G. Segal. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 71:3714-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]