Abstract
Host defenses against the encapsulated yeast Cryptococcus neoformans involve both humoral and cell-mediated immunity. Mannoproteins (MPs) are a heterogeneous class of immunodominant glycoproteins which have been only incompletely characterized. In this study, we report on the molecular features of two novel MPs that are recognized by serum antibodies during cryptococcosis. After fractionation of extracellular cryptococcal products, MPs reacted more strongly than other components with sera from C. neoformans-infected AIDS patients. Further fractionation and Western blot analysis of MPs evidenced the presence of highly reactive bands with molecular masses of 250, 125, 115, and 84 kDa. The 115- and 84-kDa bands contained significant amounts of N-linked oligosaccharides, as shown by decreased molecular mass after peptide-N-glycosidase F treatment. N-terminal amino acid sequences of the two bands were used to search C. neoformans nucleotide databases. Homologous genomic sequences were used to synthesize DNA probes and isolate cDNA clones containing the full-length genes, which were designated MP84 and MP115. Both genes showed the presence of a serine/threonine-rich region, a potential site for heavy glycosylation. MP84 and MP115 showed homology with, respectively, polysaccharide deacetylases and carboxylesterases from other organisms. Recombinant, deglycosylated proteins expressed in Escherichia coli still reacted with sera from patients, albeit more weakly than natural MPs, indicating that at least some of the reactive epitopes were retained in the recombinant forms. In conclusion, we identified two novel MPs that are important targets of antibody responses during cryptococcosis. These data may be useful to devise alternative immunity-based strategies to control the disease.
Cryptococcus (Filobasidiella) neoformans is an encapsulated basidiomycete causing severe disease, mostly meningoencephalitis, in immunocompromised hosts, especially in AIDS patients and those subjected to immunosuppressive therapies (20, 27). Though rare, the fungus can also produce disease in individuals with intact immunity (20). Because of the limitations of currently available antifungal therapies, including their toxicity (29), much interest has focused on alternative immunity-based strategies. Both cell- and antibody-mediated mechanisms could be exploited to control cryptococcal infection (20, 23). Many studies have focused on antibody responses to glucuronoxylomannan (GXM), the main constituent of the cryptococcal capsule. GXM is essential for virulence, and some monoclonal antibodies to it have been shown to provide passive protection (9, 21).
The importance of cell-mediated responses is underscored by the frequent occurrence of cryptococcosis in patients with T-cell defects. Accordingly, much attention has been devoted to the identification of antigens that stimulate a protective cell-mediated response (17, 22, 23). Cryptococcal culture supernatants, designated CneF, have been shown to contain immunoprotective antigens (1, 17, 22, 23). A major portion of CneF proteins is represented by mannoproteins (MPs), a heterogeneous class of antigens sharing the ability to bind to concanavalin A (ConA) columns. MPs, but not other CneF components, elicited delayed-type hypersensitivity reactions in mice (24). Moreover, peripheral blood lymphocytes of patients who have recovered from cryptococcosis proliferate in response to stimulation with MPs (14).
The mechanism underlying the immunodominance of MPs likely resides in their ability to target mannose receptors on antigen-presenting cells (18, 19). We hypothesized that such a mechanism could lead to immunodominance not only in cell-mediated responses but also in humoral responses. This feature could be exploited to better characterize MPs, since historically, serologic investigations have been crucial in the identification of virulence factors and diagnostic markers. Little information is available on the molecular features of individual MPs (18, 19, 25, 28). The only MP-encoding genes that have been cloned thus far are MP98 and MP88 (12, 15), which were identified on the basis of the ability of their products to stimulate T-cell hybridomas.
In the present study, we examined various fractions obtained from MPs for their ability to react with sera from patients and experimental animals affected by cryptococcosis. A major portion of such reactivity was accounted for by a fraction containing two MPs that were cloned and expressed recombinantly. These novel antigens display features that may help gain further insights into the molecular structure of this important class of glycoproteins.
MATERIALS AND METHODS
Fungal strains.
The acapsular strain Cap 67 was kindly provided by E. Jacobson, Richmond, VA. Highly virulent C. neoformans strain H99, obtained from the American Type Culture Collection (Manassas, VA) (ATCC 208821) was used to establish a model of experimental cryptococcosis (see below). Blastomyces dermatitidis (ATCC 56123), used as a control, was also purchased from the ATCC.
Production of CneF.
The cryptococcal culture filtrate antigen (CneF) was prepared using cultures from the unencapsulated strain Cap 67. Five colonies of the Cap 67 strain were transferred from Sabouraud agar plates to 100 ml of a dialyzable chemically defined medium consisting of 2% dextrose, 0.4 mM thiamine, 1% trace elements (0.5 mg of CuSO4 · 5H2O, 200 mg of ZnSO4 · 7H2O, 3.2 mg of MnCl2 · 4H2O, 8 g of MgSO4 · 7H2O, 5.4 mg of Na2MoO4 · 2H2O, and 5.7 mg of H3BO3 per liter of endotoxin-free water), 10 g of asparagine, 0.025 g of CaCl2, and 0.4 g of K2HPO4 per liter of endotoxin-free water. Starter cultures were incubated at 30°C for 48 h under agitation and used to inoculate 200-ml aliquots of the same medium, adjusted to pH 5.0 and sterilized, in tissue culture flasks (Celbio, Pero, Italy). Supernatants were monitored daily for protein and carbohydrate content by the Bradford protein microassay method (Bio-Rad Laboratories, Milan, Italy) and the phenol-sulfuric acid method (8), respectively. After 6 days of culture, supernatants were concentrated and dialyzed using a tangential filtration system, as previously reported (1). B. dermatitidis was grown on Sabouraud broth (Difco, distributed by DID, Milan, Italy) at 37°C under agitation for 96 h, as described previously (1). Supernatants were concentrated and dialyzed by tangential filtration.
Human and murine sera.
Human sera obtained from 12 human immunodeficiency virus (HIV)-positive cryptococcosis patients were kindly provided by the National Institute for Infectious Diseases “L. Spallanzani” (Rome, Italy). Sera obtained from three laboratory workers that frequently handled C. neoformans and from five additional healthy volunteers were used as controls. For some experiments, the 12 patients' sera or the 8 control sera were pooled at an equal ratio (see Fig. 2, 3, and 6).
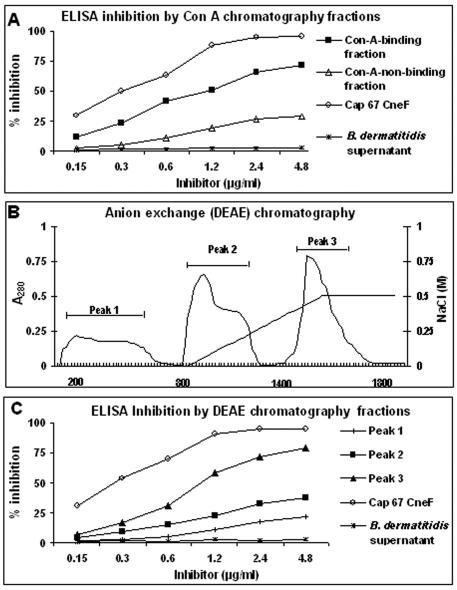
FIG. 2.
ELISA inhibition by different fractions obtained from C. neoformans CneF. Plates were coated with CneF, and pooled plasma from cryptococcosis patients was mixed with putative inhibitors before addition to the wells. Whole CneFs from Cap 67 and B. dermatitidis supernatants were used as positive and negative controls, respectively. (A) Inhibitory effects of ConA-binding or nonbinding fractions. (B) Purification of the ConA-binding fraction into three peaks by DEAE anion-exchange chromatography. Bars indicate pooled fractions obtained using a linear gradient of NaCl (right-hand axis). (C) Inhibitory effects of the different DEAE peaks. One experiment, representative of three, each performed in duplicate, is shown.
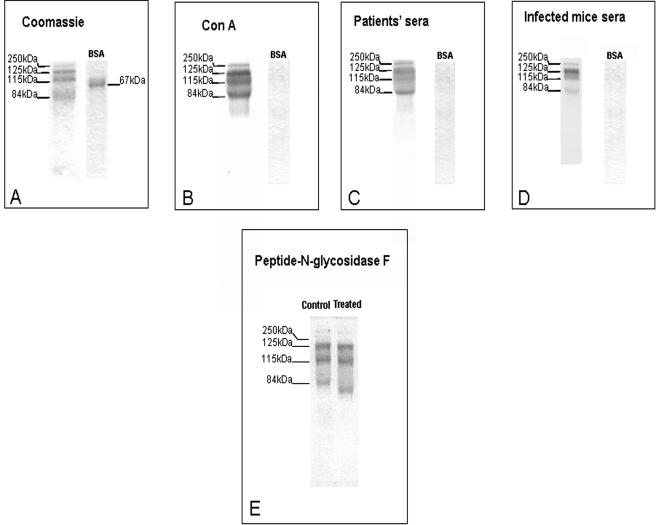
FIG. 3.
SDS-PAGE and Western blot analysis of DEAE peak 3. Material (2 μg) from peak 3 was run in 12% polyacrylamide gels and stained with Coomassie (A). Proteins resolved by electrophoresis were also transferred to nitrocellulose membranes and incubated with biotin-conjugated ConA (B) with a pool of sera from patients (C) or with a pool of sera from experimentally infected mice (D). BSA was included as a control. Panel E shows the effects of PNGase F on material from peak 3 using immunoblotting with a pool of sera from patients. To obtain a better resolution of the bands of interest, this gel was run for longer than usual (5 versus 2 h), which explains the different band pattern in this panel relative to the other panels. Pooled human and murine sera were used at a dilution of 1:100 in TTBS. Numbers on the left indicate the molecular masses in kilodaltons.
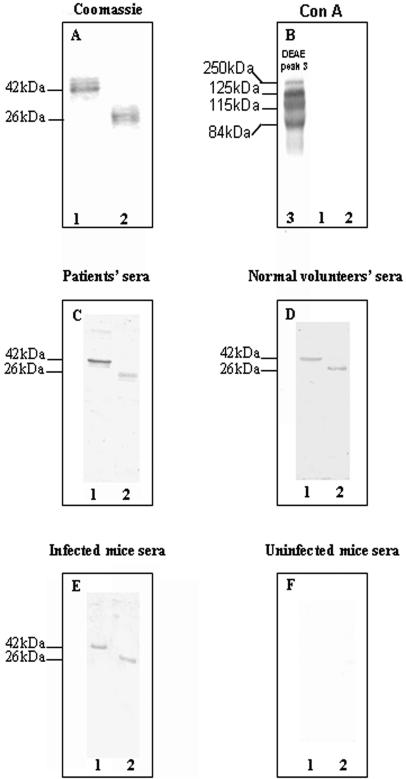
FIG. 6.
Analysis of recombinant MP84 and MP115 proteins. SDS-PAGE was followed by Coomassie staining (A), ConA blot (B), Western blot with a pool of sera from patients (C), Western blot with a pool of normal sera from volunteers (D), Western blot with a pool of sera from infected mice (E), or Western blot with a pool of sera preinfection mice (F). All pooled sera were used at a 1:100 dilution. Purified recombinant 84-kDa and 115-kDa proteins were loaded onto lanes 1 and 2, respectively. Numbers on the left indicate the molecular masses in kilodaltons.
Ten female BALB/c mice were infected intravenously with a sublethal dose of C. neoformans strain H99 (2.5 × 103 cells) as previously described (2, 3). Blood samples were obtained from the tail vein before infection and at 42 days postinfection and pooled at an equal ratio. These studies were performed in agreement with the European Union guidelines of animal care and were approved by the relevant national and local committees.
ELISA and ELISA inhibition assays.
Sera from AIDS patients infected with C. neoformans were examined for the presence of anti-CneF antibodies by enzyme-linked immunosorbent assay (ELISA). Polystyrene 96-wells plates (Nunc, distributed by Mascia Brunelli, Milan, Italy) were coated overnight at 4°C with 500 ng of antigen (protein content) diluted in 100 μl of 0.05 M sodium carbonate (pH 9.6). After washing with phosphate-buffered saline (PBS) containing 0.05% polyoxyethylene-sorbitan monolaurate (Tween 20) (PBS-Tween; Sigma, Milan, Italy), the plates were blocked with 1% bovine serum albumin (BSA) in PBS for 2 h. After washing with PBS-Tween, 100 μl of each serum sample, diluted 1:200 in 1% BSA-PBS, was added, and the plates were incubated for 1 h at 37°C. After three washes with PBS-Tween, a 1:7,500 dilution of monoclonal anti-human immunoglobulin G (IgG) alkaline phosphatase conjugate (Sigma) in PBS was added to wells and left for 1 h at 37°C, followed by the addition of p-nitrophenylphosphate (Sigma) as the substrate. Absorbance was read at 405 nm in a plate reader. Assays with sera in the absence of antigen and with antigen in the absence of sera served as controls.
Inhibition ELISA was performed as described above, with the exception that putative inhibitors were mixed with an equal volume (50 μl) of a pool of patients' sera (final serum dilution, 1:400). Inhibitors consisted of twofold dilutions of C. neoformans CneF, B. dermatitidis supernatants, or fractions thereof (see below). Percent inhibition was calculated by comparing the absorbance values of wells with inhibitors to those of wells without inhibitors.
Fractionation of CneF.
ConA-binding and nonbinding fractions were obtained from CneF by affinity chromatography. Briefly, CneF (500-mg protein content) was diluted in binding buffer (10 mM Tris-HCl containing 0.5 M NaCl, pH 7.4) and loaded onto a 2.5- by 25-cm ConA Sepharose-4B column (Amersham Biosciences, Milan, Italy), previously equilibrated with the same buffer, at a flow rate of 1.5 ml/min. The sample was recirculated twice through the column to ensure maximum binding. After washing with binding buffer until no proteins were detected, material bound to the column was eluted with a linear gradient of methyl-α-d-mannopyranoside ranging from 0 to 0.5 M. A single protein fraction was obtained under these conditions. The ConA-binding and nonbinding fractions were dialyzed against PBS and concentrated to approximately 3 mg/ml using an Amicon cell (Millipore, Vimodrone, Italy) equipped with a 10,000- Da-exclusion-limit filter. The ConA-binding fraction was further resolved by anion-exchange chromatography as described previously (1). Briefly, the ConA-reactive fraction dialyzed in binding buffer (0.01 M Tris-HCl buffer, pH 8.0) was applied on a 50-ml DEAE column (Macro-prep DEAE-Support; Bio-Rad). The column was washed until the effluent was negative for proteins and carbohydrates. Elution was performed by applying a linear gradient of NaCl (0 to 0.5 M) in 0.01 M Tris-HCl, pH 8.0, at a flow rate of 1.5 ml/min. Pooled fractions, collected according to the absorbance peaks, were dialyzed against PBS and stored at −80°C until assayed.
SDS-PAGE and Western blotting.
Analytical sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (13) on 12% polyacrylamide gels using a Mini-Protean II cell (Bio-Rad) according to the manufacturer's instructions. Protein bands were visualized with Coomassie brilliant blue R-250 (Amersham Biosciences). To perform Western blot analyses, proteins resolved by electrophoresis were transferred electrophoretically to nitrocellulose membranes (0.2-μm pore size; Amersham Biosciences) using a Minigel system (Bio-Rad) according to the manufacturer's instructions. Nitrocellulose sheets were then blocked with 2% BSA in 0.01 M Tris (pH 7.9) containing 150 mM NaCl (Tris-buffered saline) for 1 h at room temperature. After three washes with Tris-buffered saline supplemented with 0.05% Tween 20 (TTBS), membranes were incubated with ConA or sera. For ConA blots, membranes were incubated for 1 h with biotin-conjugated ConA (Sigma) diluted 1:15,000 in TTBS. For immunoblots, membranes were incubated for 1 h with 1:100 dilutions of human/murine sera. Antibody binding was detected by the addition of monoclonal anti-human IgG alkaline phosphatase conjugate or polyclonal anti-mouse IgG (γ-chain-specific) biotin conjugate (Sigma), both diluted 1:7,500 in PBS. After three washes with TTBS, streptavidin-alkaline phosphatase conjugate (Boehringer GmbH, Mannheim, Germany) was used to detect biotinylated anti-mouse IgG or ConA. Reactive bands were developed with nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate (Sigma). Precision prestained standards (Bio-Rad) were used to estimate the molecular sizes of the protein bands.
Enzymatic deglycosylation.
Asparagine-linked glycans were removed from ConA-binding proteins using peptide-N-glycosidase F (PNGase F) according to the protocols of the manufacturer (Sigma). Briefly, 12 μl (5 μg) of protein was incubated with 2 μl of PNGase F in 250 mM sodium phosphate, pH 6, for 24 h at 37°C. Protein samples incubated under the same conditions but without enzyme were used as controls.
Protein purification and amino acid sequence analysis.
Proteins from anion-exchange chromatography fractions were subjected to preparative SDS-PAGE in order to isolate single molecular species. Bands were excised, destained, and electroeluted as previously described (1). Excised bands were subjected again to SDS-PAGE and electrotransferred to an polyvinylidene difluoride membrane (Immobilon P; Millipore). The membrane-bound protein was excised, and its N-terminal amino acid sequence was determined at Midwest Analytical, Inc., St. Louis, MO, as previously described (1). Homology searches were carried out by using BLAST software of the National Center for Biotechnology Information at the National Library of Medicine (Bethesda, MD). BLAST searches were performed at both protein and DNA levels using servers available at websites of the Stanford University Technology Center C. neoformans Genome Project (http://wwwsequence.stanford.edu/group/C.neoformans/index.htlm) (16), the University of Oklahoma's Advanced Center for Genome Technology (http://www.genome.ou.edu/cneo.html), and the C. neoformans H99 Sequencing Project, Duke Center for Genome Technology (http://cneo.genetics.duke.edu/). Sequence analysis was carried out at the BCM Search Launcher server (http://searchlauncher.bcm.tmc.edu/seq-util/seq-util.html). The Signal P server (http://www.cbs.dtu.dk/services/SignalP-2.0/) and the Pfam database (http://www.sanger.ac.uk/Software/Pfam/search.shtml) were used to conduct signal peptide cleavage site and domain analyses, respectively.
Cloning and expression of the 115 - and 84- kDa proteins.
Two sense primers (5′-GGTGCTCCCGATCCCAAG-3′ and 5′-ACAATGTGGAGAATGCG-3′) coding for the N-terminal amino acids GAPDPK and NVENA of the 84- and 115-kDa proteins, respectively, were used in conjunction with an antisense universal T7 primer (1) to amplify, by PCR, DNA fragments from a λ ZAP II phage C. neoformans cDNA library prepared from strain B-3501 (Stratagene, La Jolla, CA). After labeling, these fragments were used to screen the cDNA library exactly as previously described (1). Open reading frames in positive clones were identified and cloned as described previously (1). Recombinant proteins were expressed in their mature, extracellular forms (i.e., without the signal peptide) using the plasmid PET 21b (Novagen, Inc., distributed by Inalco S.p.A., Milan, Italy) and Escherichia coli BL21(DE3) as the host strain, according to the protocols provided by the manufacturer. Protein purification was carried out as described previously (26). Briefly, after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h, six-histidine-tagged recombinant proteins were purified under denaturing conditions (6 M guanidine-HCl) from lysed cells using a BD TALON metal affinity resin according to the instructions of the manufacturer (BD Biosciences, Clontech, Palo Alto, CA). After elution with 300 mM imidazole, the recombinant proteins, partially renatured using sequentially lower concentrations of guanidine-HCl (3, 2, and 1 M), were subjected to SDS-PAGE and either stained with Coomassie or electroblotted onto nitrocellulose membranes for Western blot analysis using monoclonal anti-polyhistidine-alkaline phosphatase conjugate followed by nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate (Sigma) according to the manufacturer's instructions.
Nucleotide sequence accession numbers.
The nucleotide sequences of the MP115 and MP84 genes have been deposited in the EMBL Nucleotide Sequence Database, and they have been assigned accession numbers AJ938051 and AJ938050, respectively.
RESULTS
MPs strongly react with serum antibodies.
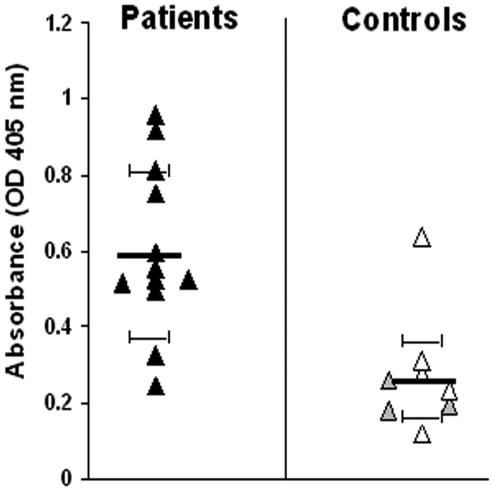
In initial experiments, all sera from AIDS patients and some sera from healthy volunteers produced positive reactions in ELISA tests in which plates were coated with CneF obtained from the unencapsulated Cap 67 strain (Fig. 1). No correlation was found between GXM levels (as defined by the latex agglutination titer) in the cryptococcosis sera and ELISA reactivity (data not shown). Next, in order to ascertain whether immune sera recognize MPs, CneF was separated by ConA affinity chromatography. ConA-binding and nonbinding fractions were collected according to absorbance peaks, concentrated, and tested in an ELISA inhibition assay. In this test, fractions were assayed at different dilutions for their ability to inhibit antibody binding to CneF-coated wells. There was no loss of inhibitory activity as a result of the fractionation procedure since total activity was accounted for by the sum of the inhibitory activities of the two fractions (Fig. 2A). The inhibition curves indicate that the ConA-binding fraction was a considerably more potent inhibitor than the nonbinding fraction, as evidenced by maximal inhibition values of 71.4% and 28.8%, respectively. As expected, CneF from Cap 67, which was used as a positive control, completely inhibited antibody binding, while no inhibition was produced by B. dermatitidis supernatant (Fig. 2A). Thus, the ConA-binding, or MP, fraction accounted for most of the antibody reactivity of CneF. Since many ill-defined positive bands were present in ConA blots (not shown), the ConA fraction was further separated by DEAE anion-exchange chromatography. Under these conditions, three peaks were obtained (Fig. 2B), which were tested in the ELISA inhibition assay. Figure 2C shows that peak 3 had significantly higher inhibitory activity than the other two peaks. Therefore, further studies concentrated on peak 3 material.
FIG. 1.
Reactivity of human sera against C. neoformans CneF determined by ELISA. Sera were obtained from 12 HIV-positive cryptococcosis patients (black triangles). Sera obtained from laboratory workers (gray triangles) and other healthy volunteers (white triangles) were used as controls. All serum samples were used at a dilution of 1:200. One experiment, representative of three, each performed in duplicate, is shown. Bars represent means ± standard deviations. OD, optical density.
Western blot analysis.
Peak 3 material was subjected to SDS-PAGE under nonreducing conditions, followed by Coomassie staining or Western blots. Under these conditions, four Coomassie-positive bands with approximate molecular masses of 250, 125, 115, and 84 kDa were detected (Fig. 3A). Western blot analysis using biotinylated ConA showed that all the bands reacted strongly with lectin (Fig. 3B). Moreover, all bands were recognized by a pool of patients' sera and by a pool of sera from experimentally infected mice (Fig. 3C and D, respectively). Figure 3D shows that infected mice tended to have lower responses against the 115-kDa band relative to the other three proteins. The reason for this phenomenon is not presently clear. To ascertain whether the ConA-reactive proteins contained mannose-rich N-linked oligosaccharides, peak 3 material was treated with PNGase F before Western blot analysis using a pool of patients' sera. This treatment did not decrease antibody reactivity but caused approximately 2-, 12-, and 23-kDa reductions in the apparent molecular masses of the 125-, 115-, and 84-kDa bands, respectively (Fig. 3E), as indicated by N-terminal amino acid sequencing of the bands before and after PNGase F treatment. This indicated the presence of N-linked oligosaccharides in each of the bands.
Cloning of the 115- and 84-kDa proteins.
In order to determine the N-terminal amino acid sequence of the immunoreactive proteins, these proteins were purified by preparative PAGE and subjected to Edman degradation. N-terminal sequences of the 250- and 125-kDa bands were identical to the Cu,Zn superoxide dismutase (SOD) gene product described previously by Hamilton and Holdom (10). No homology was found between amino acid sequences of the 115-kDa and 84-kDa bands and proteins deposited in GenBank databases. However, a search of Stanford and Oklahoma databases revealed identity with genomic and cDNA sequences, respectively. Primers, designed as detailed in Materials and Methods, were used to amplify specific fragments from a C. neoformans λ ZAP II phage cDNA library. PCR fragments were sequenced, 32P radiolabeled, and used as probes to screen the cDNA library. Positive clones containing the full-length genes were cloned and sequenced. The 84- and 115-kDa proteins were identified as the products of, respectively, open reading frames MP84 and MP115, as we named the corresponding genes.
Analysis of the deduced protein sequences.
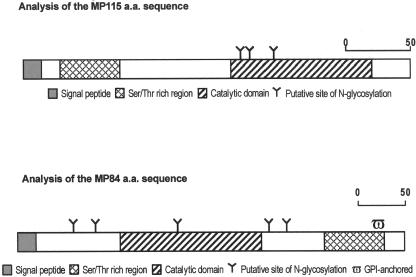
The full-length MP115 gene contained 894 bp, and the peptide sequence was predicted to be 297 amino acids in length (Fig. 4). The gene contains a putative domain of the carboxylesterase family proteins, as evidenced by a homology search in the Pfam database. Additional sequence analyses performed using Signal P and the Stanford genome database indicated a putative signal peptide (amino acids 1 to 17) and five introns of 70, 67, 55, 47, and 63 bp. There were three potential sites for N-glycosylation containing the Asn-X-Ser/Thr motif. Potential O-glycosidic linkage sites (serine/threonine) were numerous, together accounting for more than 20% of the total amino acid composition. These sites were concentrated in the N-terminal portion of the molecule (Fig. 4). A potential TATA-like box and a polyadenylation signal were present at positions −68 and +1137, respectively.
FIG. 4.
Analysis of MP115 and MP84 amino acid (a.a.) sequences. The signal sequence (shaded), catalytic domain (hatched), and Ser/Thr-rich region (cross-hatch) are shown. Sites of N-glycosylation are indicated by a Y. A putative GPI anchor site (ω) in the C-terminal portion of MP84 is also shown.
Full-length MP84 contains 1,233 bp with a deduced peptide sequence of 410 amino acids (Fig. 4). The gene contains a putative domain of polysaccharide deacetylase family proteins, as evidenced by a homology search in the Pfam database. Signal P and Stanford genome database searches revealed the presence of a putative signal peptide (amino acids 1 to 18) and five introns of 64, 63, 64, 52, and 63 bp from 5′ to 3′. There were five potential sites for N-glycosylation containing the Asn-X-Ser/Thr motif. There was a C-terminal serine/threonine-rich region that could represent a site for extensive O-glycosylation, followed by a putative glycosylphosphatidylinositol (GPI) anchor site. A potential TATA-like box and a polyadenylation signal were present at positions −54 and +1728, respectively.
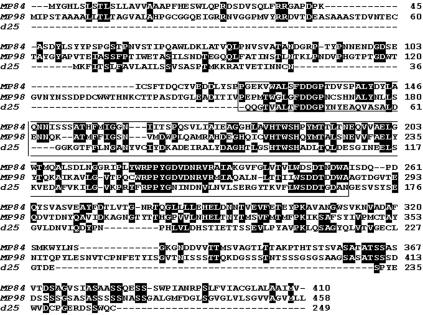
Since MP84 is the third cloned cryptococcal protein with a putative polysaccharide deacetylase domain (1, 15), we aligned similar amino acid sequences from MP84 with those of previously described MP98 and d25 putative polysaccharide deacetylases (Fig. 5). The region of homology between MP84 and MP98 was comprised between amino acids 113 and 305 of MP84 (43% identity/57% similarity). The region of homology between MP84 and the d25 was comprised between amino acids 127 and 318 of MP84 (28% identity/43% similarity).
FIG. 5.
Alignment of similar amino acid sequences from MP84, MP98, and d25. White lettering on a black background indicates identity. Amino acids within the box indicate the polysaccharide deacetylase domain.
E. coli expression of immunoreactive antigens.
PCR-generated MP115 and MP84 cDNAs were subcloned into the expression vector PET21b to yield, respectively, the pET21b-MP115 and pET21b-MP84 constructs. SDS-PAGE evidenced the presence of recombinant proteins in cell lysates obtained from the transformed E. coli strain and induced with IPTG (not shown). The two recombinant proteins were purified under denaturing conditions by His affinity chromatography as detailed in Materials and Methods. SDS-PAGE analysis of the MP115 and MP84 recombinant proteins revealed two bands of 26 and 42 kDa, respectively (Fig. 6A), which corresponded to the predicted molecular masses calculated from DNA coding sequences, including the vector-encoded peptide which contained the His tag at the C terminus. The two recombinant proteins were not recognized by ConA in Western blots, indicating the absence of protein-bound oligosaccharides capable of binding the lectin (Fig. 6 B). Moreover, Western blot analysis showed that pooled sera from patients recognized both recombinant proteins (Fig. 6C). In contrast, only a barely detectable reactivity was present using pooled sera from volunteers (Fig. 6D). Finally, antibodies reacting with the MP115 and MP84 recombinant proteins were present in the sera of experimentally infected mice (Fig. 6E), while such antibodies were not present before infection (Fig. 6F).
DISCUSSION
There is increasing evidence that antibody-mediated immunity plays an important role in host defense against cryptococcosis, and monoclonal antibodies raised against GXM have been shown to provide passive protection in experimental infection (4, 9, 21). Less is known about the occurrence and functional significance of antibody responses to antigens different from GXM (5, 6). In the present study, we showed that MPs accounted for most of the ability of secreted cryptococcal proteins to react with serum antibodies. These data are in agreement with data from previous studies (28) and support the notion that MPs are immunodominant antigens whose ability to target the immune system probably resides in their recognition by antigen-presenting cells via mannose receptors (18, 19). Despite its potential importance in immunity-based strategies to control cryptococcosis (17), this important class of antigens has been only incompletely characterized.
Thus far, only two MPs, MP98 and MP88, have been identified and cloned based on their ability to stimulate T-cell hybridomas (12, 15). In the present study, two novel MPs, MP115 and MP84, were identified by their reactivity with immune sera. As with the two previously described MPs, both MP115 and MP84 display a typical serine/threonine-rich region carrying numerous potential sites of O-glycosylation. It should be noted, however, that the serine/threonine-rich region is N terminal in MP115, while it is C terminal in MP84 and in the two previously described MPs. MP115 also differs from the latter three proteins in that it lacks a putative GPI anchor motif. Since GPI anchors are used to link proteins to the cell wall or to the cell membrane, it will be of interest to determine whether MP115 differs from the other MPs in its cellular localization.
Similar to two previously described MPs, both MP115 and MP84 have a signal peptide and several N-glycosylation sites (Fig. 4). Extensive N-glycosylation of these antigens was confirmed by marked shifts in the molecular masses of both MP115 and MP84 (12 and 23 kDa, respectively) after peptide-N-glycosidase F treatment (Fig. 3). Analysis of the sequences found outside of the Ser/Thr-rich regions provided insights into the possible functions of the novel MPs. The MP115 deduced amino acid sequence showed homology with carboxylesterases, while MP84 was homologous with polysaccharide deacetylase family proteins, including two previously described cryptococcal deacetylases (1, 15). In particular, MP84 was strongly homologous with MP98 and less homologous with d25 (Fig. 5). Thus, MP84 is the third cloned cryptococcal protein, and the second MP, with putative polysaccharide deacetylase activity.
In the present study, recombinantly expressed MP84 and MP115 retained the ability of the natural forms to react in Western blots with serum antibodies, albeit with reduced reactivity. Studies are under way to express these proteins and/or fragments thereof in a soluble, nondenaturated form and to assess the protective potential of these products, and of antibodies raised against them, in experimental cryptococcosis models.
Hamilton and Goodley have previously described an N- glycosylated 115-kDa protein recognized by immune sera (11). Since no data are available on its amino acid or nucleotide sequence, it is difficult to say if this protein is identical to protein MP115 described here. This seems unlikely, however, since antibodies raised against the protein described previously by Hamilton and Goodley were not reactive with the MP fraction (11), i.e., with the same fraction in which we detected MP115.
The MP fraction described here was also found to contain the Cu,Zn superoxide dismutase SOD1, a well-known virulence factor (7) in the form of 125- and 250-kDa species in nonreducing conditions. This is in general agreement with the data of Hamilton and Holdom, who found that SOD1 had a nonreduced molecular mass of 125 kDa (10), although the presence of SOD1 in the MP fraction has not been previously reported. Interestingly, SOD1 appeared to carry an N-linked oligosaccharide, as evidenced by a molecular weight shift after treatment with peptide-N-glycosidase F (Fig. 3). The presence of an N-linked oligosaccharide may explain the unexpected property of SOD1 to be retained by ConA columns and to react with ConA in Western blots, as documented in the present study. Collectively, our data indicate that the MP fraction may contain (i) MPs with a “classical” C-terminal serine/threonine-rich region followed by a putative GPI anchor (e.g., MP84), (ii) less typical MPs with an N-terminal serine/threonine-rich region and no GPI anchor site (e.g., MP115), and (iii) N-glycosylated proteins which perhaps should not be considered as MPs since they lack extensive O-glycosylation sites (e.g., SOD1).
In conclusion, we have identified and characterized two novel MP antigens, designated MP84 and MP115, which are highly reactive with sera from cryptococcosis patients. These data may be useful to gain further insights into the molecular structure and immunological features of MPs. Future studies will assess the potential significance of these novel MPs as markers of disease and as means to develop alternative, immunity-based strategies to control cryptococcosis.
Acknowledgments
We thank Giuseppe Ippolito, Scientific Director of the Spallanzani Hospital, for providing us with sera from AIDS patients affected by cryptococcosis. We also thank the C. neoformans cDNA Sequencing Project at the University of Oklahoma (http://www.genome.ou.edu/cneo.html), the C. neoformans H99 Sequencing Project, Duke Center for Genome Technology (http://cgt.genetics.duke.edu), and the C. neoformans Serotype D Genome Project, Stanford Genome Technology Center (http://wwwsequence.stanford.edu/group/C.neoformans/index.htlm).
This work was supported in part by the European Commission (HOSPATH Contract QLK-CT-2000-00336), by the Istituto Superiore di Sanità of Italy (Project AIDS contract no. 50 D.30), and by the Ministero dell'Istruzione, dell'Università e della Ricerca of Italy (PRIN no. 2003065895_005).
Editor: T. R. Kozel
REFERENCES
- 1.Biondo, C., C. Beninati, D. Delfino, M. Oggioni, G. Mancuso, A. Midiri, M. Bombaci, G. Tomaselli, and G. Teti. 2002. Identification and cloning of a cryptococcal deacetylase that produces protective immune responses. Infect. Immun. 70:2383-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biondo, C., C. Beninati, M. Bombaci, L. Messina, G. Mancuso, A. Midiri, R. Galbo, and G. Teti. 2003. Induction of T helper type 1 responses by a polysaccharide deacetylase from Cryptococcus neoformans. Infect. Immun. 71:5412-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biondo, C., A. Midiri, L. Messina, F. Tomasello, G. Garufi, M. R. Catania, M. Bombaci, C. Beninati, G. Teti, and G. Mancuso. 2005. MyD88 and TLR2, but not TLR4, are required for host defense against Cryptococcus neoformans. Eur. J. Immunol. 35:870-878. [DOI] [PubMed] [Google Scholar]
- 4.Casadevall, A., and L. A. Pirofski. 2004. New concepts in antibody-mediated immunity. Infect. Immun. 72:6191-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L. C., D. L. Goldman, T. L. Doering, L. Pirofski, and A. Casadevall. 1999. Antibody response to Cryptococcus neoformans proteins in rodents and humans. Infect. Immun. 67:2218-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L. C., L. A. Pirofski, and A. Casadevall. 1997. Extracellular proteins of Cryptococcus neoformans and host antibody response. Infect. Immun. 65:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, G. M., T. S. Harrison, H. C. McDade, C. P. Taborda, G. Heinrich, A. Casadevall, and J. R. Perfect. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Roberts, and F. Smith. 1956. Colorimetric method for the determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 9.Fleuridor, R., Z. Zhong, and L. Pirofski. 1998. A human IgM monoclonal antibody prolongs survival of mice with lethal cryptococcosis. J. Infect. Dis. 178:1213-1216. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton, A. J., and M. D. Holdom. 1997. Biochemical comparison of the Cu,Zn superoxide dismutases of Cryptococcus neoformans var. neoformans and Cryptococcus neoformans var. gattii. Infect. Immun. 65:488-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton, A. J., and J. Goodley. 1993. Purification of the 115-kilodalton exoantigen of Cryptococcus neoformans and its recognition by immune sera. J. Clin. Microbiol. 31:335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, C., S. H. Nong, M. K. Mansour, C. A. Specht, and S. M. Levitz. 2002. Purification and characterization of a second immunoreactive mannoprotein from Cryptococcus neoformans that stimulates T-cell responses. Infect. Immun. 70:5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Levitz, S. M., and E. A. North. 1997. Lymphoproliferation and cytokine profiles in human peripheral blood mononuclear cells stimulated by Cryptococcus neoformans. J. Med. Vet. Mycol. 35:229-236. [DOI] [PubMed] [Google Scholar]
- 15.Levitz, S. M., S. Nong, M. K. Mansour, C. Huang, and C. A. Specht. 2001. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T-cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 98:10422-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. M. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansour, M. K., L. E. Yauch, J. B. Rottman, and S. M. Levitz. 2004. Protective efficacy of antigenic fractions in mouse models of cryptococcosis. Infect. Immun. 72:1746-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansour, M. K., L. S. Schlesinger, and S. M. Levitz. 2002. Optimal T cell responses to Cryptococcus neoformans mannoprotein are dependent on recognition of conjugated carbohydrates by mannose receptors. J. Immunol. 168:2872-2879. [DOI] [PubMed] [Google Scholar]
- 19.Mansour, M. K., and S. M. Levitz. 2003. Fungal mannoproteins: the sweet path to immunodominance. ASM News 69:595-600. [Google Scholar]
- 20.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee, J., L. A. Pirofski, M. D. Scharff, and A. Casadevall. 1993. Antibody-mediated protection in mice with lethal intracerebral Cryptococcus neoformans infection. Proc. Natl. Acad. Sci. USA 90:3636-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy, J. W., F. Schafer, A. Casadevall, and A. Adesina. 1998. Antigen-induced protective and nonprotective cell-mediated immune components against Cryptococcus neoformans. Infect. Immun. 66:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy, J. W. 1988. Influence of cryptococcal antigens on cell-mediated immunity. Rev. Infect. Dis. 10:S432-435. [DOI] [PubMed] [Google Scholar]
- 24.Murphy, J. W., R. L. Mosley, R. Cherniak, G. H. Reyes, T. R. Kozel, and E. Reiss. 1988. Serological, electrophoretic, and biological properties of Cryptococcus neoformans antigens. Infect. Immun. 56:424-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitzurra, L., S. Perito, F. Baldelli, F. Bistoni, and A. Vecchiarelli. 2003. Humoral response against Cryptococcus neoformans mannoprotein antigens in HIV-infected patients. Clin. Exp. Immunol. 133:91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porath, J. 1988. High-performance immobilized-metal-ion affinity chromatography of peptides and proteins. J. Chromatogr. 443:3-11. [DOI] [PubMed] [Google Scholar]
- 27.Powderly, W. G. 1993. Cryptococcal meningitis and AIDS. Clin. Infect. Dis. 17:837-842. [DOI] [PubMed] [Google Scholar]
- 28.Reiss, E., R. Cherniak, R. Eby, and L. Kaufman. 1984. Enzyme immunoassay detection of IgM to galactoxylomannan of Cryptococcus neoformans. Diagn. Immunol. 2:109-115. [PubMed] [Google Scholar]
- 29.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]