Abstract
Entecavir (ETV) is a potent and selective inhibitor of hepatitis B virus (HBV) replication in vitro and in vivo that is currently in clinical trials for the treatment of chronic HBV infections. A major limitation of the current HBV antiviral therapy, lamivudine (3TC), is the emergence of drug-resistant HBV in a majority of treated patients due to specific mutations in the nucleotide binding site of HBV DNA polymerase (HBV Pol). To determine the effects of 3TC resistance mutations on inhibition by ETV triphosphate (ETV-TP), a series of in vitro studies were performed. The inhibition of wild-type and 3TC-resistant HBV Pol by ETV-TP was measured using recombinant HBV nucleocapsids, and compared to that of 3TC-TP. These enzyme inhibition studies demonstrated that ETV-TP is a highly potent inhibitor of wild-type HBV Pol and is 100- to 300-fold more potent than 3TC-TP against 3TC-resistant HBV Pol. Cell culture assays were used to gauge the potential for antiviral cross-resistance of 3TC-resistant mutants to ETV. Results demonstrated that ETV inhibited the replication of 3TC-resistant HBV, but 20- to 30-fold higher concentrations were required. To gain further perspective regarding the potential therapeutic use of ETV, its phosphorylation was examined in hepatoma cells treated with extracellular concentrations representative of drug levels in plasma in ETV-treated patients. At these concentrations, intracellular ETV-TP accumulated to levels expected to inhibit the enzyme activity of both wild-type and 3TC-resistant HBV Pol. These findings are predictive of potent antiviral activity of ETV against both wild-type and 3TC-resistant HBV.
Infection with hepatitis B virus (HBV) is a medical problem of global proportions. Despite the existence of a safe vaccine, approximately 5% of the world population is infected with HBV. Approved therapies for chronic HBV infection are treatment with alpha interferon or lamivudine (3TC). Drawbacks to treatment with alpha interferon include a low sustained response rate, undesirable side effects, the need for parenteral administration, and high cost. Therapy with 3TC is less costly and more convenient to use, but it also suffers from a low sustained response rate. Of more fundamental concern is that while initial treatment of patients with 3TC results in a rapid lowering of HBV DNA levels in the blood, its efficacy is severely compromised in most patients by the development of antiviral resistance after prolonged therapy.
The inhibition of HBV replication by nucleoside analogs results from the recognition of nucleoside analog triphosphates (TPs) by the RNA-dependent DNA polymerase of HBV (HBV Pol). Clinical resistance to 3TC results from amino acid substitutions at position 550 (methionine [M]) in the highly conserved YMDD motif of HBV Pol (31). Variants with the double mutation M550V/L526M or the single mutation M550I predominate (reviewed in reference 14). Molecular modeling studies suggest that these mutations alter the nucleotide binding site of HBV Pol to cause steric hindrance of 3TC-TP binding (1, 4). Genotypic resistance emerges in 14 to 32% of patients within the first 12 months of 3TC therapy (8, 21), increasing to 40% within 2 years of treatment (23) and 57% by year 3 (22). The development of resistance is fueled by the high rate of virus replication and the error rate of the viral polymerase. To combat this, alternative therapies that suppress HBV replication in vivo more effectively are needed. A more potent antiviral agent, for example, should suppress HBV replication and also slow the emergence of drug-resistant variants. Therapies that would also suppress the replication of 3TC-resistant HBV are needed.
Entecavir (ETV), a deoxyguanosine analog, is a potent and selective inhibitor of HBV replication; its in vitro potency is 100- to 1,000-fold greater than that of 3TC, and it has a selectivity index (concentration of drug which reduced the viable cell number by 50% [CC50]/concentration of drug which reduced viral replication by 50% [EC50]) of ∼8,000 (15, 28). Human clinical trials have demonstrated the efficacy of ETV for the treatment of chronic HBV infections at doses as low as 0.01 mg daily (7, 20). The in vivo efficacy of ETV was previously demonstrated in the woodchuck and duck models of HBV infection (13, 24). Important differences between the behavior of ETV and 3TC were observed in these models, including their roles in the development of antiviral resistance. In the woodchuck model, ETV therapy suppressed the levels of woodchuck hepatitis virus (WHV) DNA in blood by up to 8 log10 units and reduced hepatic covalently closed circular DNA levels by up to 4 log10 units. Even after 14 to 36 months of therapy with ETV, there was no emergence of drug-resistant WHV (3). These observations are in contrast to those made for 3TC therapy, which not only failed to reduce intrahepatic covalently closed circular DNA levels in the woodchuck but also led to the emergence of drug-resistant variants with mutations in WHV Pol (25). These results, while limited to the woodchuck model, clearly support the suggestion that a more potent HBV antiviral agent could slow the emergence of drug-resistant HBV in vivo. The potential for additional therapeutic benefits of ETV therapy was indicated by a reduced frequency of hepatocellular carcinoma in the woodchuck model and a prolonged life span of chronically infected animals (3). While there is limited data published on the in vitro efficacy of ETV against 3TC-resistant HBV (28), recent clinical studies have demonstrated the in vivo efficacy of ETV against 3TC-resistant HBV infections (N. Tassopoulos, S. Hadziyannis, J. Cianciara, M. Rizzetto, E. Schiff, G. Pastore, V. Rutkiewicz, N. Thomas, G. Denisky, and S. Joshi, Hepatology 34:340A, 2001). These results highlight the fact that ETV has unique properties for the inhibition of HBV replication relative to 3TC that result in excellent therapeutic benefits.
In vitro studies with HBV can provide important insights into antiviral mechanism(s) of action. Previous studies have demonstrated that ETV-TP is a competitive inhibitor of HBV Pol negative-strand synthesis and positive-strand replication, and unlike 3TC, it also inhibits the HBV priming reaction (30). A key factor contributing to the excellent potency of ETV is that HBV Pol displays a higher affinity for ETV-TP than it does for its natural substrate, dGTP (30). This report describes additional in vitro studies performed to better understand the mechanisms underlying the therapeutic efficacy of ETV against both wild-type and 3TC-resistant HBV and to relate this understanding to antiviral efficacy in vivo and the potential for antiviral resistance to ETV. A sensitive HBV Pol assay using recombinant HBV nucleocapsids was employed to measure the inhibitory activities of ETV-TP and 3TC-TP and to determine the relative binding affinities to wild-type and 3TC-resistant HBV Pol. Also, cell culture assays were used to measure the relative antiviral potency of ETV and 3TC against wild-type and 3TC-resistant HBV to gauge the potential for cross-resistance. To add an important perspective, the accumulation of ETV-TP was examined at clinically relevant exposure levels as a predictor of the potential therapeutic efficacy of ETV in wild-type and 3TC-resistant HBV infections. The results provide a level of detail that may explain the observed in vivo efficacies of ETV against both wild-type and 3TC-resistant HBV infections.
MATERIALS AND METHODS
Compounds.
ETV (formerly BMS-200475), lamivudine (3TC), and 3H-labeled ETV were chemically synthesized at Bristol-Myers Squibb.
HBV Pol constructs.
The BAC-TO-BAC Baculovirus Expression System (Invitrogen, Gaithersburg, Md.) was used to construct a panel of baculovirus recombinants expressing wild-type and mutant HBV pol genes. A SalI-to-blunted NotI fragment from pRH210 carrying the HBV pol gene (subtype ayw) flanked at the 3′ end by the DR1 and ɛ elements was cloned into the SalI-to-blunted HindIII sites of a pFastBac1 derivative (pFasBacΔAVR) in which the AvrII site in the vector was deleted, creating pFastBac-HBV-POL. Mutations were created by site-directed mutagenesis by the method of Kunkel (18), and mutagenic primers and pRH210 single-stranded DNA were used to create the mutations L526M (5′-GT-AAA-CTG-AGC-CAT-GAG-AAA-CGG-3′), M550V (5′-AA-TAC-CAC-ATC-ATC-CAC-ATA-ACT-GAA-AG-3′), M550I (5′-AA-TAC-CAC-ATC-ATC-AAT-ATA-ACT-GAA-AG-3′), V553I (5′-CC-CAA-TAC-AAT-ATC-ATC-CAT-ATA-ACT-3′), and M550V/L526M in the polymerase domain (reverse complement of mutated codon is underlined). To create pFastBac-HBV-POL derivatives having mutations L526M, M550V, M550I, V553I, or M550V/L526M, the AvrII-to-NsiI fragment was replaced with the appropriate AvrII-to-NsiI fragment from the pRH210 mutant constructs. Chimeric HBV Pol constructs having the reverse transcriptase (RT) domain from 3TC-resistant clinical HBV samples (subtype adw) were also constructed by replacing the AvrII-to-NsiI fragment of pFastBac-HBV-POL with AvrII-to-NsiI fragments having either the M550V/L526M or M550V/L526M/V553I mutations. The sequence of the clinical AvrII-to-NsiI fragment is identical to the AvrII-to-NsiI fragment (nucleotides 1587 to 2478) of the reported adw2 sequence (accession number X02763) except for a T-to-A substitution at base 1698, an A-to-T substitution at base 2045, a C-to-G substitution at base 2291, and a G-to-A substitution at base 2318. The presence of each of the mutations in the pFastBac-HBV-POL clones was verified by DNA sequence analysis. Recombinant baculoviruses and virus stocks were prepared according to the instructions of the manufacturer.
HBV genome constructs.
Plasmid pTHBV containing two head-to-tail tandem copies of the subtype ayw HBV genome (2) was used to generate a terminally redundant 1.3x HBV genome fragment. Briefly, nucleotides 1068 to 5175 (nucleotide 1 is the EcoRI site) from pTHBV were released as two fragments after digestion with NsiI/SpeI and SpeI/XbaI and recloned into the pFastBac transfer vector, yielding pBAC-HBV. To isolated recombinant baculoviruses containing the entire HBV genome, pBAC-HBV DNA was transfected into SF9 cells with BaculoGold DNA (BD Biosciences Pharmingen, San Diego, Calif.) according to the manufacturer's instructions. The recombinant baculovirus was identified by Southern dot blot hybridization to detect HBV DNA in plaque-purified baculovirus DNA. The pCMV-HBV plasmid was a generous gift from S. Goff (11). Derivatives of pBAC-HBV and pCMV-HBV having mutations responsible for 3TC resistance were created by replacing the AvrII-to-NsiI fragment of pCMV-HBV with the appropriate AvrII-to-NsiI fragments of pRH210 (see above), creating pCMV-M550I and pCMV-M550V/L526M.
Endogenous polymerase assays.
Baculovirus-derived trans HBV nucleocapsids containing wild-type or mutant polymerases were prepared as previously described (29). Briefly, SF21 cells were coinfected with baculovirus recombinants expressing HBV Pol and core proteins. Insect cell lysates were partially digested by mild protease and nuclease treatments, and recombinant HBV nucleocapsids were enriched to >80% purity via ultracentrifugation through 10% sucrose. Endogenous polymerase reactions were performed in 384-well plates (BD Biosciences Discovery Labware, New Bedford, Mass.) containing 75 mM NH4Cl, 50 mM Tris-HCl (pH 7.4), 20 mM MgCl2, 0.1% Tween 20, 100 μg of gamma globulin-free bovine albumin per ml, 100 μg of tRNA per ml, and 30 to 100 ng of recombinant HBV nucleocapsids. The plates were incubated at 37°C for 3 h. Scintillation proximity assay (SPA) beads (PVT-Protein A; Amersham Biosciences, Piscataway, N.J.) were prepared by allowing HBV core protein-specific rabbit polyclonal antibody (Dako, Carpinteria, Calif.) to bind overnight. Unbound antibody was removed, and charged SPA beads were suspended at 2.5 mg/ml in buffer A (75 mM NH4Cl, 50 mM Tris-HCl [pH 7.4], 0.1% Tween 20, 100 μg of gamma globulin-free bovine albumin per ml, 50 mM EDTA). Reactions were terminated by the addition of 200 μg of charged SPA beads. The ratio of suspended SPA bead volume to endogenous polymerase reaction volume was 3.6:1. HBV Pol-dependent incorporation of radiolabeled nucleotide was measured using a Packard TopCount counter.
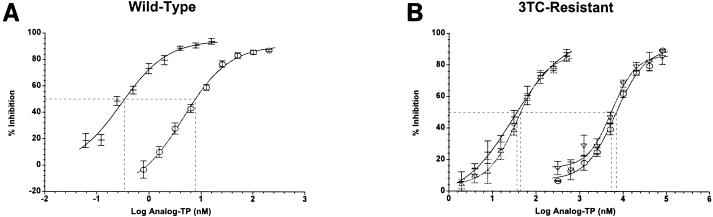
Dose-response experiments were performed with HBV nucleocapsids containing wild-type or 3TC-resistant Pol using ETV-TP (0.06 to 15.6 nM for wild-type Pol; 1.95 to 500 nM for 3TC-resistant Pol) or 3TC-TP (0.39 to 200 nM for wild-type Pol; 312 to 80,000 nM for 3TC-resistant Pol). The concentration of deoxynucleoside triphosphates (dNTPs) (Amersham Biosciences) was 1 μM (each), except in ETV-TP reaction mixtures where dGTP was replaced with 0.83 or 2.22 nM [α-33P]dGTP (NEN, Boston, Mass.) and in 3TC-TP reaction mixtures where dCTP was replaced with 0.83 or 2.22 nM [α-33P]dCTP (NEN). Dose-response curves were plotted using a four-parameter logistic equation (Fig. 1) to determine the 50% inhibitory concentration (IC50) for analog TPs.
FIG. 1.
All IC50 curves are derived from a four-parameter logistic equation. (A) Comparison of ETV-TP (−) and 3TC-TP (○) inhibition of wild-type HBV Pol. (B) Comparison of ETV-TP (−, M550V/L526M; ▵, 550I) and 3TC-TP (○, M550V/L526M; ▿, M550I) inhibition of HBV Pol YMDD mutants. Error bars represent the standard deviations from three independent experiments. The broken lines show 50% inhibition.
The Km constants for competing dNTP substrates (dGTP or dCTP) were determined with both wild-type HBV Pol (subtype ayw) and a 3TC-resistant clinical isolate RT domain (ayw/adw chimera). Initial reaction velocities for wild-type and 3TC-resistant polymerase were measured over a range of substrate concentrations from three independent experiments, and Km values were determined from direct linear plots (9). Substrate (dGTP or dCTP) concentrations ranged from 0.8 to 12.5 nM for both wild-type HBV Pol and the triple mutant HBV Pol. The concentration of noncompeting dNTPs was 1 μM, except for dTTP which was replaced with 25 nM [α-33P]dTTP (NEN). The higher concentration of radiolabel in these experiments required the removal of unincorporated counts by washing the SPA beads three times in buffer A to eliminate background. The Ki values for analog TPs were calculated from the equation Ki = IC50/(1 + [S]/Km, where [S] is the concentration of substrate) (see Table 2).
TABLE 2.
Km and Ki measurements of nucleotide substrates and analog triphosphates against recombinant HBV nucleocapsids containing wild-type or 3TC-resistant HBV Pola
| HBV Pol |
Km (nM)
|
Ki (nM)
|
Ki/Km
|
|||
|---|---|---|---|---|---|---|
| dGTP | dCTP | ETV-TP | 3TC-TP | ETV-TP | 3TC-TP | |
| Wild-type | 1.3 | 2.0 | 0.2 | 4.4 | 0.2 | 2.2 |
| M550V/L526M/ V553I | 1.6 | 2.8 | 22.1 | 6,377 | 13.8 | 2,278 |
Km and Ki values are the means from three independent experiments. Ki was calculated as follows: Ki = IC50/(1 + [S]/Km).
HBV replication in cells.
HepG2 cells (American Type Culture Collection, Manassas, Va.) were maintained at 37°C in a humidified incubator with 5% CO2 in RPMI 1640 (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS). For baculovirus-mediated gene transfer, HepG2 cells were seeded at ∼40% confluency on collagen-coated plates (BD Biosciences Discovery Labware) and then incubated overnight. The following day, cells were washed once with RPMI 1640 with 2% FBS and infected with recombinant BAC-HBV at a multiplicity of infection of 100 in RPMI 1640 with 2% FBS. Cells were infected for 3 h, the input virus was aspirated, and the cells were washed three times with growth medium. Cells were then fed with RPMI 1640 with 2% FBS with or without the indicated compound (final dimethyl sulfoxide concentration of 0.1%). Cells were incubated for 5 days, and the tissue culture supernatant was harvested for quantitation of extracellular HBV DNA. For transfection studies, plasmid DNA was transfected into HepG2 cells seeded on 24-well plates with the use of Lipofectamine (Invitrogen). Four hours after transfection, the cells were washed three times with growth medium and the cells were fed with RPMI 1640 with 2% FBS with or without the indicated compound (final dimethyl sulfoxide concentration of 0.1%). After cells were incubated for 5 to 6 days, the tissue culture supernatant was harvested for quantitation of extracellular HBV DNA.
Southern blot hybridization.
Samples of culture supernatant (150 μl) were centrifuged at 5,000 rpm in an Eppendorf model 5415C microcentrifuge, from which 100 μl was processed for blotting. Samples were combined with 1 volume of 2 N NaOH-20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), incubated for 30 min at 37°C, and applied to a Nytran membrane (Schleicher & Schuell, Keene, N.H.) in a dot blot apparatus. The membrane was washed with 4 volumes of 1 M NaCl-1 M Tris-HCl (pH 7.2) and then with 2 volumes of 20× SSC. Membranes were processed for hybridization according to the manufacturer's protocol (Gene Images Detection System; Amersham Biosciences). The hybridization probe was full-length ayw HBV DNA released by NcoI digestion of plasmid pTHBV, purified by agarose gel electrophoresis and used to generate a fluorescein-labeled probe. Membranes were removed from the dot blot apparatus, rinsed with 6× SSC for 5 min, cross-linked by UV irradiation, and then prehybridized for 30 min with Gene Block (Amersham Biosciences). Hybridization with fluorescein-labeled probe was conducted overnight in Gene Block at 60°C. Hybridized membranes were washed with 1× SSC-0.1% sodium dodecyl sulfate for 15 min at 60°C and then blocked with Liquid Block (Amersham Biosciences) in 100 mM Tris-HCl (pH 9.5) with 300 mM NaCl. Alkaline phosphatase-conjugated anti-fluorescein antibody was used for hybrid detection according to the manufacturer's protocol. Multiple film exposures were quantitated by densitometry (Molecular Dynamics SI Densitometer). A twofold serial dilution of linearized full-length HBV DNA was used as the standard for quantitation purposes.
Quantitative PCR.
Extracellular HBV DNA was quantitated directly using clarified culture supernatants (5,000 rpm for 5 min in an Eppendorf model 5415C microcentrifuge). The PCR primers and probe were designed using Primer Express software (Applied Biosystems, Foster City, Calif.). Amplification was performed in a 50-μl reaction mixture containing 2× TaqMan Universal MasterMix (Applied Biosystems), 10 μl of clarified culture supernatant, 300 pmol of forward primer (5′-GTAAACAATACCTGAACCTTTACCCC-3′; nucleotides 1126 to 1151), 300 pmol of reverse primer (5′-CGTCAGCAAACACTTGGCAC-3′; nucleotides 1194 to 1175), and 100 pmol of TaqMan probe (5′-6-carboxyfluoroscein-TTGCCCGGCAACGGCCA-3′; nucleotides 1153 to 1169). After preparation of the reaction mixtures in 96-well plates, the plates were centrifuged at 800 rpm for 1 min in a Beckman GPKR swing rotor centrifuge. Amplification and detection were performed with an ABI Prism 7700 Sequence Detection System. The PCR protocol consisted of the following: (i) a single cycle of 2 min at 50°C, followed by 10 min at 95°C; and (ii) 45 two-step cycles, with 1 cycle consisting of 15 s at 95°C and 60 s at 60°C. A 10-fold dilution series of full-length ayw linearized HBV DNA ranging from 1,000 pg to 0.5 fg was used as a DNA standard for quantitation. The limit of detection was 12.8 fg. Analysis of amplification data was done with the Sequence Detector version 1.6.3 software (Applied Biosystems).
Phosphorylation studies.
Most methods used in the phosphorylation studies were similar to those described previously (32). HepG2 cells (American Type Culture Collection) were plated at 2 × 105 to 5 × 105 cells/well in collagen-coated six-well culture dishes (9.4 cm2/well), which were then incubated overnight to allow cells to adhere. In the morning, cells were then labeled with 3H-labeled ETV (prepared at Bristol-Myers Squibb) as described in the individual experiments. Following 2 to 5 days of incubation, cells were extracted in situ with 60% methanol, and nucleotide analyses were performed as described earlier (32). Cell growth was estimated by measuring the increase in A260 of the methanol-soluble extract of washed cell monolayers. Intracellular nucleotide concentration was determined on the basis of an estimated cell volume of 10−9 ml (10).
RESULTS
Inhibition of wild-type and 3TC-resistant HBV Pol in vitro.
The potency of ETV-TP as an inhibitor of HBV DNA synthesis was examined using recombinant baculovirus-derived HBV nucleocapsids containing wild-type or 3TC-resistant HBV Pol. These recombinant nucleocapsids are competent for both the priming and reverse transcription events of HBV DNA replication, supporting the incorporation of radiolabeled nucleotides into nascent minus-strand DNA (29). In the present study, the incorporation assay was modified to include a novel immunocapture SPA detection method (Materials and Methods). In addition, reactions were done at 37°C instead of the 30°C temperature used previously. These modifications improved the assay sensitivity by increasing the reaction velocity and by reducing the background signal due to impurities in the partially purified nucleocapsid preparations.
Recombinant HBV nucleocapsids were used to measure polymerase activity in the presence or absence of ETV-TP and 3TC-TP. For each inhibitor, IC50s were derived from several independent dose-response experiments (Fig. 1A). To compare the results, IC50s were normalized to the concentration of the competing natural dNTP substrate (dGTP for ETV-TP and dCTP for 3TC-TP). Inhibition of wild-type HBV Pol (subtype ayw) by ETV-TP and 3TC-TP yielded IC50/[dNTP] ratios of 0.43 and 7.5, respectively (Table 1), comparable to values determined previously using the non-SPA detection method (30). The results indicated that ETV-TP inhibited DNA synthesis by wild-type HBV Pol with 17-fold-greater potency than 3TC-TP. Nucleocapsids containing 3TC-resistant HBV Pol were generated by site-directed mutagenesis. Two variants associated with clinical resistance to 3TC, M550V/L526M and M550I, were constructed, in addition to other single amino acid substitutions that were constructed as controls (Materials and Methods). Dose-response measurements for ETV-TP (Fig. 1B) yielded IC50/[dNTP] values of 46 and 59 for inhibition of the M550V/L526M and M550I variants, respectively (Table 1). These values indicated potent inhibition of 3TC-resistant HBV Pol, though with a significant reduction relative to wild-type HBV Pol. The M550V mutation had a minor effect on the potency of ETV-TP (∼10-fold), while the L526M and V553I single point mutations had no effect (Table 1). Dose-response measurements with 3TC-TP (Fig. 1B) yielded IC50/[dNTP] values of 6,200 and 7,600 for inhibition of the M550V/L526M and M550I variants, respectively (Table 1). These values indicated a dramatic loss of potency, up to 1,000-fold, relative to the inhibition of wild-type HBV Pol (Table 1). Nucleocapsids carrying the M550V substitution alone exhibited an intermediate level of resistance to 3TC-TP, while the L526M and V553I substitutions alone had no effect, as expected (Table 1). The findings with 3TC-TP are consistent with reports demonstrating that the M550I mutation alone confers resistance to 3TC and that the M550V mutation emerges in conjunction with the L526M mutation (1). Consequently, the intrinsic potency of ETV-TP against clinically relevant 3TC-resistant point mutants was greater than that of 3TC-TP by at least 2 orders of magnitude.
TABLE 1.
In vitro potencies of nucleoside analog TPs against recombinant HBV nucleocapsids containing wild-type or 3TC-resistant HBV Pol
| HBV Pol | IC50 of nucleoside analog/[dNTP]a
|
|
|---|---|---|
| ETV-TP | 3TC-TP | |
| Wild-type | 0.43 ± 0.06 | 7.5 ± 2.7 |
| Mutant | ||
| M550Ib | 59.2 ± 6.2 | 6,180 ± 780 |
| M550V | 4.2 ± 1.3 | 305 ± 19 |
| M550V/L526Mb | 46.1 ± 9.0 | 7,580 ± 1,920 |
| M550V/L526Mc | 51.7 ± 10.2 | 9,130 ± 3,730 |
| M550V/L526M/V553Ic | 31.3 ± 2.7 | 9,920 ± 1,770 |
| L526M | 0.67 ± 0.09 | 50.3 ± 7.6 |
| V553I | 0.54 ± 0.08 | 4.3 ± 0.3 |
Values are the means ± standard deviations from three independent experiments expressed as the ratio of IC50 of ETV-TP/[dGTP] or IC50 of 3TC-TP/[dCTP], with all concentrations given in nanomolar.
Clinically relevant 3TC-resistant point mutants in subtype ayw.
RT domain transferred from 3TC-resistant clinical isolates (ayw/adw chimeras).
To confirm that results with the YMDD point mutants were an accurate reflection of clinically resistant HBV Pol, chimeric nucleocapsids were generated in which the entire RT domain of wild-type HBV Pol was replaced with the entire RT domain from each of two 3TC-resistant clinical isolates (subtype adw). Of the two isolates, one carried the double mutation, M550V/L526M, and the other, a triple mutant, carried an additional V553I substitution that is sometimes found in association with clinically resistant M550V/L526M mutants (31). Both of these chimeras carried 25 additional amino acid substitutions in the RT domain compared to the wild-type ayw construct that are not known to be associated with 3TC resistance. In dose-response experiments, the IC50/[dNTP] ratio for 3TC-TP against the intact RT domains was increased by a factor of 1,200 to 1,350, relative to the wild type, similar to the effect of the double point mutation alone (Table 1). For ETV-TP, the two chimeric HBV Pol constructs yielded IC50/[dNTP] values of 31 and 52, respectively, also comparable to the double point mutant (Table 1). Results observed with the YMDD point mutants were, therefore, representative of HBV Pol from 3TC-resistant clinical isolates.
More detailed kinetic analysis was carried out with nucleocapsids for the wild-type HBV Pol and the triple mutant RT domain (M550V/L526M/V553I), selected as a representative 3TC-resistant HBV Pol. Using the immunocapture SPA, apparent Km values for dGTP and dCTP were determined for both Pols, and then Ki values for ETV-TP and 3TC-TP were calculated from multiple dose-response experiments, as described in Materials and Methods. For wild-type HBV Pol, the apparent Km values for dGTP and dCTP were 1.3 and 2.0 nM, respectively. The resulting Ki values for ETV-TP and 3TC-TP were 0.2 and 4.4 nM, respectively (Table 2). These results confirmed the previous finding that wild-type HBV Pol displays a binding preference for ETV-TP over dGTP (30). For the 3TC-resistant RT domain, the apparent Km values for dGTP and dCTP were 1.6 and 2.8 nM, respectively (Table 2). The resulting Ki values for ETV-TP and 3TC-TP were 22.1 nM and 6,377 nM (6.4 μM), respectively. These results indicated that YMDD mutations alter the ability of HBV Pol to bind both ETV-TP and 3TC-TP but to different degrees. Like using IC50/[dNTP] ratios to compare inhibitor potencies, Ki/Km ratios can be used to compare their relative affinities. The Ki/Km ratios for 3TC-resistant HBV Pol increased 69- and 1,035-fold for ETV-TP and 3TC-TP, respectively, relative to the wild type. This indicates a much more profound effect of the YMDD mutations on the inhibitory activity of 3TC-TP than for ETV-TP. In fact, the results show that ETV-TP was almost as potent an inhibitor of the 3TC-resistant HBV Pol (Ki/Km = 13.8) as 3TC-TP was for wild-type HBV Pol (Ki/Km = 2.2).
HBV replication in transfected HepG2 cells.
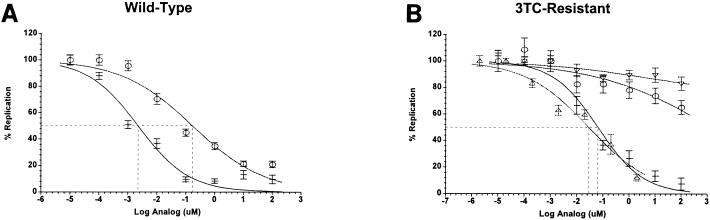
Because ETV-TP exhibited significant affinity for 3TC-resistant HBV Pol, it was of interest to examine the effects of YMDD mutations on the antiviral potency of ETV. The sensitivity of antiviral potency measurements for HBV using various cell culture assays is highly variable. For example, published EC50s for 3TC against wild-type HBV range anywhere from 5 nM (12) to >500 nM (28). A suitable application for cell-based replication assays that is less dependent on assay sensitivity is comparing the relative antiviral potencies of various HBV inhibitors (5). An important feature of cell-based replication assays is that the antiviral potency measurement is the sum of inhibitory effects on all functions of HBV Pol. In the case of ETV, for example, inhibition of the priming reaction might contribute more to the relative potency in whole-cell assays that support a full replication cycle. Two different assay formats were used to gauge the effects of 3TC resistance mutations on the antiviral activity of ETV. In the first assay format, a baculovirus vector was constructed to transfect mammalian cells with replication-competent HBV genomes, as described by Delaney and Isom (6). HBV replication was monitored in the human hepatoma cell line HepG2 by the appearance of viral DNA in virions secreted into the culture supernatant (described in Materials and Methods). In the absence of inhibitors, the time course of HBV replication was highly reproducible, and antiviral measurements were made routinely at day 6 postinfection. In dose-response experiments (Fig. 2A), the production of extracellular HBV DNA was inhibited by both ETV and 3TC with EC50s of 3 and 196 nM, respectively (Table 3). Similar EC50s were also obtained when the Huh-7 hepatoma cell line was used in place of HepG2 cells (data not shown). To assess the effects of 3TC resistance mutations on antiviral potency, a BAC-HBV construct was generated with the M550V/L526M double point mutation (BAC-M550V/L526M). The replication of this HBV variant was highly compromised in vitro, exhibiting a rate nearly 50-fold lower than that of the wild type, as measured by real-time PCR (Materials and Methods). In dose-response measurements with 3TC, an EC50 could not be obtained at concentrations up to 100 μM, indicating strong phenotypic resistance (>500-fold relative to wild-type HBV; Table 3). In clear contrast, ETV strongly inhibited the replication of the 3TC-resistant HBV mutant with an EC50 of 61 nM (Table 3). These assay results demonstrated that a 20-fold increase in ETV exposure levels was sufficient to suppress the replication of a 3TC-resistant variant of HBV. The results obtained were not related to cytotoxicity, since the concentrations of ETV used were well below its CC50 of 30 μM (15).
FIG. 2.
(A) Effect of ETV (−) or 3TC (○) on extracellular HBV production by HepG2 cells infected with wild-type BAC-HBV. HepG2 cells were infected with wild-type BAC-HBV at a multiplicity of infection of 100 for 3 h, washed, and incubated in RPMI 1640 with 5% FCS. The culture supernatant was sampled at the times indicated and analyzed for HBV DNA by DNA dot blot hybridization. (B) Effect of ETV (−, M550V/L526M; ▵, M550I) or 3TC (○, M550V/L526M; ▿, M550I) on extracellular HBV production by HepG2 cells. HepG2 cells were transfected with plasmid pCMV-M550V/L526M or pCMV-M550I HBV. Four hours after transfection, the cells were washed and fed with RPMI 1640 plus 5% FCS containing the indicated compound concentration. Culture supernatants were analyzed by DNA dot blot hybridization on day 6 posttransfection. Error bars represent the standard deviations from three independent experiments. The broken lines show 50% inhibition.
TABLE 3.
EC50s for ETV and 3TC in HepG2 cells infected with BAC-HBV derivatives or transfected with pCMV-HBV plasmids
| Virus or plasmid | EC50 (μM)a
|
3TC/ETV EC50 ratiob | Mutant/WT ETV EC50 ratioc | |
|---|---|---|---|---|
| ETV | 3TC | |||
| BAC-HBV | 0.003 | 0.196 | 65 | 1 |
| BAC-M550V/L526M | 0.061 | >100 | >1,640 | 20 |
| pCMV-HBV | 0.001 | 0.139 | 139 | 1 |
| pCMV-M550V/L526M | 0.029 | >100 | >3,450 | 29 |
| pCMV-M550I | 0.031 | >100 | >3,230 | 31 |
Values represent the average of two or three determinations.
Relative potency of ETV compared with 3TC.
EC50 of ETV in mutant constructs, relative to the EC50s in the wild type (WT).
The second assay format used plasmids with HBV genomes under transcriptional control of the human cytomegalovirus (CMV) promoter (pCMV-HBV). Use of the strong CMV promoter leads to higher levels of HBV pregenomic RNA in cells, better enabling in vitro antiviral measurements for poorly replicating HBV isolates (1, 19). For the wild-type plasmid (pCMV-HBV), EC50s for ETV and 3TC were 1 and 139 nM, respectively, close to the values measured using the baculovirus-mediated gene transfer method (Table 3). The YMDD variants pCMV-M550V/L526M and pCMV-M550I were both insensitive to 3TC, as expected (EC50 >100 μM; Fig. 2B). ETV, on the other hand, was a potent inhibitor of both mutants with EC50s of 29 and 31 nM, respectively (Fig. 2B). In these assays, a ca. 30-fold increase in ETV exposure level was sufficient to suppress the replication of 3TC-resistant HBV variants relative to the wild type (Table 3). In both assay systems described here, ETV was a more potent inhibitor of 3TC-resistant HBV replication than 3TC was of wild-type HBV replication. Taken together, the cell culture antiviral assays demonstrated that ETV is a potent inhibitor of 3TC-resistant HBV with in vitro cross-resistance of 20- to 30-fold.
Metabolic studies of ETV in HepG2 cells.
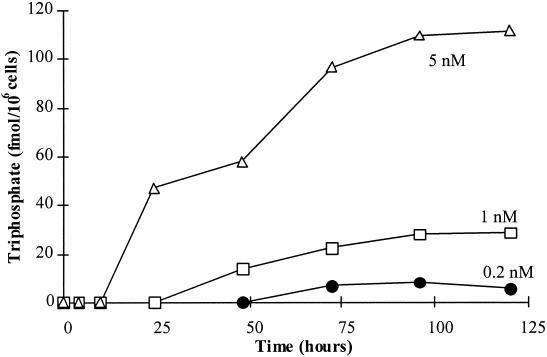
Clinical studies have demonstrated the in vivo efficacy of a once-daily dose of 0.5 mg of ETV in patients with chronic HBV infection (7) and in patients failing 3TC therapy (Tassopoulos et al., Hepatology 34:340A, 2001). The results of pharmacokinetic analyses demonstrated that the 0.5-mg dose resulted in steady-state concentrations of ETV in plasma in the low nanomolar range (average steady-state concentration of ∼2.4 nM; data not published). To gain an important perspective on the antiviral activity of ETV at clinically relevant exposure levels, experiments were undertaken to examine the accumulation of ETV-TP in human hepatoma cells treated continuously with extracellular concentrations of ETV of ≤5 nM. The phosphorylation of ETV was examined previously in cells treated with 5 to 100 nM 3H-labeled ETV (32). In these experiments, the accumulation of ETV-TP in HepG2 cells was detectable within 4 h of labeling, reached a plateau after 20 h, and was approximately linear down to the lowest concentration examined (5 nM). The intracellular half-life of ETV-TP was ∼15 h, leading to a significant accumulation of ETV-TP above the extracellular concentration. In addition, the high efficiency of ETV phosphorylation has been consistently observed in a variety of different cell types examined (32). In extending these studies to lower ETV concentrations, cells were labeled at 5, 1, and 0.2 nM. The kinetics of ETV-TP accumulation was slowed at the lower concentrations, as detectable TP did not appear for 2 to 3 days (Fig. 3). A decrease in the extracellular concentration of 3H-labeled ETV was observed at 0.2 nM, however, indicating that decreasing substrate concentration contributed to the slower kinetics. ETV phosphorylation was not detected at labeling concentrations below 0.1 to 0.2 nM. At 1 and 5 nM extracellular ETV, the levels of intracellular ETV-TP were 28 and 110 nM, respectively (Table 4). These concentrations are well above the Ki value for inhibition of wild-type HBV Pol by ETV-TP and also remain above the Ki value for inhibition of 3TC-resistant HBV Pol (Table 2). Table 4 summarizes the phosphorylation data, showing that the relative efficiency of phosphorylation was greatest at the lowest extracellular concentrations: the efficiency was 30-fold higher at 0.2 nM than it was at 25 μM. Due to limiting substrate at the lowest concentrations, the corresponding phosphorylation efficiency may actually be underestimated. Given the long intracellular half-life of ETV-TP, these findings indicate that consistent exposure to extracellular concentrations of ETV in the low nanomolar range can result in sufficient ETV-TP inside cells to inhibit not only wild-type but also 3TC-resistant HBV Pols.
FIG. 3.
The kinetics of intracellular ETV-TP accumulation was measured in HepG2 cells using 3H-labeled ETV (13.9 Ci/mmol) concentrations of 0.2, 1.0, and 5.0 nM.
TABLE 4.
Relative efficiency for ETV phosphorylation in HepG2 cells
| Extracellular ETV concn (nM) | Intracellular ETV-TP concn (nM) | Relative efficiency (ETV-TP/ETV)a |
|---|---|---|
| 0.2 | 7 | 35 |
| 1.0 | 28 | 28 |
| 5.0 | 110 | 22 |
| 25 | 270 | 11 |
| 100 | 670 | 6.7 |
| 1,000 | 6,000 | 6 |
| 10,000 | 17,000 | 1.7 |
| 25,000 | 30,000 | 1.2 |
Intracellular ETV-TP concentration/extracellular ETV concentration.
DISCUSSION
This report describes parameters influencing the antiviral activity of ETV against wild-type and 3TC-resistant HBV. To examine the intrinsic activity of ETV-TP directly, the inhibition of wild-type and 3TC-resistant HBV Pol was measured using recombinant HBV nucleocapsids formed by coexpression of the polymerase and core proteins, in which the polymerase protein is specifically primed for replication of a pseudo-pregenomic HBV RNA in an epsilon-mediated fashion (29). Assay modifications were made to further optimize the in vitro reaction conditions and to improve the sensitivity of detection using these nucleocapsids. The apparent Km and Ki values reported here for wild-type HBV Pol are about 7- to 14-fold lower than values reported previously using similar recombinant HBV nucleocapsids (30). One likely explanation for this difference is that reactions in the current study were carried out at 37°C, versus 30°C. This temperature increase would not only accelerate the enzyme reaction rate but would also increase the rate of diffusion of nucleotides into the interior of nucleocapsids where they are needed to act as substrates for HBV Pol. The resulting assay provided an optimal system to evaluate the effects of specific 3TC resistance mutations on DNA synthesis by HBV Pol. The measurements described here confirmed the excellent inhibitory potency of ETV-TP against wild-type HBV Pol and demonstrated reduced but still potent inhibition of 3TC-resistant HBV Pol. A comparison of the relative change in binding kinetics caused by YMDD mutations clearly demonstrated that the recognition of ETV-TP by HBV Pol was impacted to a lesser degree than that of 3TC-TP.
A key to understanding the potential for in vivo efficacy is to know the level of active drug that is present in HBV-infected hepatocytes. In the case of 3TC, in vitro phosphorylation measurements have been a very useful indicator of the in vivo phosphorylation profile in the peripheral blood mononuclear cells of patients infected with human immunodeficiency virus (27). In this report, we examined the phosphorylation of ETV in liver-derived cell lines exposed to clinically relevant concentrations of ETV. Human pharmacokinetic and pharmacodynamic data for both ETV and 3TC are the benchmark by which the in vitro results must be compared. The results of recent clinical trials have demonstrated the efficacy of a once-daily dose of 0.5 mg of ETV for the treatment of chronic HBV infections (7), including those that are resistant to 3TC (Tassopoulos et al., Hepatology 34:340A, 2001). Patients treated with this dose achieve an average steady-state concentration in blood of ∼2.4 nM (unpublished results). The in vitro phosphorylation data reported here (Table 4) confirm an important aspect of the activation of ETV inside cells. The results showed that the efficiency of phosphorylation to the TP form increases as the concentration of extracellular ETV is lowered. On the basis of these in vitro data, 2.4 nM ETV in blood could produce an average intracellular concentration of ETV-TP in the range of 53 to 67 nM. If so, this would be well above the Ki measured for inhibition of wild-type HBV DNA synthesis by ETV-TP, and strong inhibition of wild-type HBV replication would be expected at this exposure level. The inhibition kinetics of ETV-TP, in this respect, are consistent with the significant reduction of HBV DNA levels observed in patients treated with a 0.5-mg daily dose (7). In patients treated with a once-daily dose of 100 mg of 3TC, the average steady-state concentration in blood is estimated to be 0.85 μM (16). This could produce an average intracellular TP concentration of 0.85 to 1.7 μM, based on reports that 3TC-TP levels in hepatoma cells are one- to twofold higher than the extracellular 3TC concentration (17, 32). At this level, the concentration of 3TC-TP would also be well above its Ki for wild-type HBV Pol and consistent with the initial efficacy of 100 mg of 3TC against chronic HBV infection.
A very different picture begins to emerge when the inhibition kinetics of 3TC-resistant HBV Pol are evaluated. Given that resistance to 3TC occurs readily, it is not surprising that the inhibitory potency of 3TC-TP was reduced >1,000-fold against 3TC-resistant YMDD variants. It is notable that the Ki of 3TC-TP for the 3TC-resistant YMDD variant of HBV is above the level of intracellular 3TC-TP expected at clinically relevant exposure levels. This would be expected to result in incomplete inhibition and a consequent increase in HBV DNA replication relative to that of the wild-type HBV Pol, as occurs when resistance emerges. In contrast, the inhibition kinetics with ETV-TP clearly demonstrated potent inhibition of DNA synthesis by 3TC-resistant HBV Pol. While the potency of ETV-TP is reduced from that of the wild-type HBV Pol, the phosphorylation kinetics suggest that the intracellular concentration of ETV-TP expected in patients treated with 0.5 mg of ETV would remain above the Ki for 3TC-resistant HBV Pol (Ki = 22 nM). This leads to the expectation that clinically relevant ETV exposure levels would reduce the replication of 3TC-resistant HBV.
Another way to evaluate potential cross-resistance in vitro is to examine the complete replication cycle of HBV inside cells. It has been demonstrated that HBV Pol has a stronger preference for binding ETV-TP than its natural dGTP substrate (30) and that ETV-TP can inhibit the priming function of HBV Pol (30). Because these properties are unique relative to 3TC, it was important to determine the potential impact of YMDD mutations on ETV inhibition in an assay which involves all the replication functions of HBV Pol. For this reason, cell culture antiviral assays were used to gauge the potential for cross-resistance to ETV. Another fact to consider is that YMDD mutations have been reported to reduce the replicative fitness of HBV in cells (26), and this may have an impact on the outcome of viral replication assays which was not evident in enzyme inhibition assays. Because antiviral potency data reported in the literature for HBV vary significantly, two different cell culture systems were used to determine the effects of YMDD mutations on the antiviral potency of ETV. Similar results were obtained with either system, demonstrating that YMDD mutations conferred full resistance to 3TC but conferred no more than 20- to 30-fold cross-resistance to ETV.
The in vitro data reported here are consistent with recent clinical studies demonstrating the efficacy of ETV for the treatment of chronic HBV infections (7) and 3TC-resistant HBV infections (Tassopoulos et al., Hepatology 34:340A, 2001). The combined results indicate that the presence of YMDD mutations have a limited impact on the antiviral activity of ETV. Because of its superior potency against these variants, the development of resistance during ETV therapy may take longer to emerge and require other, or additional, mutations in HBV Pol. As a result, ETV therapy in treatment-naive patients might be expected to suppress HBV DNA levels more than 3TC therapy, due to increased potency, and for a longer duration, due to the reduced emergence of viable YMDD variants. Greater suppression of HBV DNA levels by ETV was, in fact, evident in patients treated with a 0.5-mg daily dose, which led to a drop in HBV DNA of >4 log10 units at 22 weeks, a reduction of 1 to 1.5 log10 units more than that observed for 3TC (20). The results presented here are encouraging and underscore the importance of monitoring HBV isolated from ETV-treated patients for the potential emergence of resistant HBV. They also point clearly to the development of ETV as a promising advance in the treatment of chronic HBV infections.
Acknowledgments
We gratefully acknowledge Brian Terry and Daniel Tenney for numerous informative discussions, Fiona McPhee and Jay Prendergast for help making recombinant nucleocapsids, and Amy Sheaffer for help with chemiluminescent dot blot and quantitative PCR procedures. The pCMV-HBV plasmid was kindly provided by Stephen Goff.
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K.-A. Walters, D. L. J. Tyrrell, N. F. Brown, L. D. Condreay, et al. 1998. Identification and characterization of mutations in hepatitis B resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Christman, J. K., M. Gerber, P. M. Price, C. Flordellis, J. Edelman, and G. Acs. 1982. Amplification of expression of hepatitis B surface antigen in 3T3 cells cotransfected with a dominant-acting gene and cloned viral DNA. Proc. Natl. Acad. Sci. USA 79:1815-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colonno, R. J., E. V. Genovesi, I. Medina, L. Lamb, S. Durham, M.-L. Huang, L. Corey, M. Littlejohn, S. Locarnini, B. Tennant, B. Rose, and J. M. Clark. 2001. Long-term entecavir treatment results in sustained antiviral efficacy and prolonged life span in the woodchuck model of chronic hepatitis infection. J. Infect. Dis. 184:1236-1245. [DOI] [PubMed] [Google Scholar]
- 4.Das, K., X. Xiong, H. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine and emtricitabine. J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delaney, W. E., R. Edwards, D. Colledge, T. Shaw, J. Torresi, T. G. Miller, H. Isom, C. T. Bock, M. P. Manns, C. Trautwein, and S. Locarnini. 2001. Cross-resistance testing of antihepadnaviral compounds using novel recombinant baculoviruses which encode drug-resistant strains of hepatitis B virus. Antimicrob. Agents Chemother. 45:1705-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaney, W. E., and H. C. Isom. 1998. Hepatitis B virus replication in human HepG2 cells mediate by hepatitis B virus recombinant baculovirus. Hepatology 28:1134-1146. [DOI] [PubMed] [Google Scholar]
- 7.De Man, R. A., L. M. M. Wolters, F. Nevens, D. Chua, M. Sherman, C. L. Lai, A. Gadano, Y. Lee, F. Mazzotta, N. Thomas, and D. DeHertogh. 2001. Safety and efficacy of oral entecavir given for 28 days in patients with chronic hepatitis B virus infection. Hepatology 34:578-582. [DOI] [PubMed] [Google Scholar]
- 8.Dienstag, J. L., E. R. Schiff, T. L. Wright, R. P. Perrillo, H.-W. L. Hann, Z. Goodman, L. Crowther, L. D. Condreay, M. Woessner, M. Rubin, N. A. Brown, et al. 1999. Lamivudine as initial treatment for chronic hepatitis B in the United States. N. Engl. J. Med. 341:1256-1263. [DOI] [PubMed] [Google Scholar]
- 9.Eisenthal, R., and A. Cornish-Bowden. 1974. A new graphical procedure for estimating enzyme kinetic parameters. Biochem. J. 139:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elion, G. B., P. A. Furman, J. A. Fyfe, P. DeMiranda, L. Beauchamp, and H. J. Schaeffer. 1977. Selectivity of action of an antiherpetic agent, 9-(2-hydroxyethoxymethyl)guanine. Proc. Natl. Acad. Sci. USA 74:5716-5720.202961 [Google Scholar]
- 11.Fallows, D. A., and S. P. Goff. 1995. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 69:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, L., and Y.-C. Cheng. 2000. Characterization of novel human hepatoma cell lines with stable hepatitis B virus secretion for evaluating new compounds against lamivudine- and penciclovir-resistant virus. Antimicrob. Agents Chemother. 44:3402-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genovesi, E. V., L. Lamb, I. Medina, D. Taylor, M. Seifer, S. Innaimo, R. J. Colonno, D. N. Standring, and J. M. Clark. 1998. Efficacy of the carbocyclic 2′-deoxyguanosine nucleoside BMS-200475 in the woodchuck model of hepatitis B virus infection. Antimicrob. Agents Chemother. 42:3209-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussain, M., and A. S. F. Lok. 1999. Mutations in the hepatitis B virus polymerase gene associated with antiviral treatment for hepatitis B. J. Viral Hepatitis 6:183-194. [DOI] [PubMed] [Google Scholar]
- 15.Innaimo, S. F., M. Seifer, G. S. Bisacchi, D. N. Standring, R. Zahler, and R. J. Colonno. 1997. Identification of BMS-200475 as a potent and selective inhibitor of hepatitis B virus. Antimicrob. Agents Chemother. 41:1444-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, M. A., K. H. P. Moore, G. J. Yuen, A. Bye, and G. E. Pakes. 1999. Clinical pharmacokinetics of lamivudine. Clin. Pharmacokinet. 36:41-66. [DOI] [PubMed] [Google Scholar]
- 17.Kewn, S., P. G. Hoggard, S. D. Sales, M. A. Johnson, and J. Back. 2000. The intracellular activation of lamivudine (3TC) and determination of 2′-deoxycytidine-5′-triphosphate (dCTP) pools in the presence and absence of various drugs in HepG2 cells. Br. J. Clin. Pharmacol. 50:597-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladner, S. K., T. J. Miller, and R. W. King. 1998. The M539V polymerase variant of human hepatitis B virus demonstrates resistance to 2′-deoxy-3′-thiacytidine and a reduced ability to synthesize viral DNA. Antimicrob. Agents Chemother. 42:2128-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, C., M. Rosmawati, J. Lao, H. Vlierberghe, F. Anderson, N. Thomas, and D. De Hertogh. 2001. A phase II study of entecavir vs. lamivudine in adults with chronic hepatitis B. J. Hepatol. 34:24. [Google Scholar]
- 21.Lai, C. L., R. N. Chien, N. W. Leung, T.-T. Chang, R. Guan, D.-I. Tai, K.-Y. Ng, P.-C. Wu, J. C. Dent, J. Barber, S. L. Stephenson, D. F. Gray, et al. 1998. A one year trial of lamivudine for chronic hepatitis B. N. Engl. J. Med. 339:64-68. [DOI] [PubMed] [Google Scholar]
- 22.Leung, N. W., C.-L. Lai, T.-T. Chang, R. Guan, C.-M. Lee, K.-Y. Ng, S.-G. Lim, P.-C. Wu, J. C. Dent, S. Edmundson, L. D. Condreay, and R.-N. Chien. 2001. Extended lamivudine treatment in patients with chronic hepatitis B enhances hepatitis B e antigen seroconversion rates: results after 3 years of therapy. Hepatology 33:1527-1532. [DOI] [PubMed] [Google Scholar]
- 23.Liaw, Y. F., N. W. Leung, T.-T. Chang, R. Guan, D.-I. Tai, K.-Y. Ng, R.-N. Chien, J. Dent, L. Roman, S. Edmundson, C.-L. Lai, et al. 2000. Effects of extended lamivudine therapy in Asian patients with chronic hepatitis B. Gastroenterology 119:172-180. [DOI] [PubMed] [Google Scholar]
- 24.Marion, P. L., F. H. Salazar, M. A. Winters, and R. J. Colonno. 2002. Potent efficacy observed with entecavir (BMS-200475) in duck model of hepatitis B virus replication. Antimicrob. Agents Chemother. 46:82-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason, W. S., J. Cullen, G. Moraleda, J. Saputelli, C. E. Aldrich, D. S. Miller, B. Tennant, L. Frick, D. Averett, L. D. Condreay, and A. R. Jilbert. 1998. Lamivudine therapy of WHV-infected woodchucks. Virology 24:18-32. [DOI] [PubMed] [Google Scholar]
- 26.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology 27:628-633. [DOI] [PubMed] [Google Scholar]
- 27.Moore, K. H., J. E. Barrett, S. Shaw, G. E. Pakes, R. Churchus, A. Kapoor, J. Lloyd, M. G. Barry, and D. Back. 1999. The pharmacokinetics of lamivudine phosphorylation in peripheral blood mononuclear cells from patients infected with HIV-1. AIDS 13:2239-2250. [DOI] [PubMed] [Google Scholar]
- 28.Ono, S. K., N. Kato, Y. Shiratori, J. Kato, T. Goto, R. F. Schinazi, F. J. Carrilho, and M. Omata. 2001. The polymerase L528M mutation cooperates with nucleotide binding site mutations, increasing hepatitis B virus replication and drug resistance. J. Clin. Investig. 107:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seifer, M., R. Hamatake, M. Bifano, and D. N. Standring. 1998. Generation of replication-competent hepatitis B virus nucleocapsids in insect cells. J. Virol. 72:2765-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seifer, M., R. Hamatake, R. J. Colonno, and D. N. Standring. 1998. In vitro inhibition of hepadnavirus polymerase by the triphosphates of BMS-200745 and lobucavir. Antimicrob. Agents Chemother. 42:3200-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tipples, G. A., M. M. Ma, K. P. Fischer, V. G. Bain, N. M. Kneteman, and D. L. J. Tyrrell. 1996. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology 24:714-717. [DOI] [PubMed] [Google Scholar]
- 32.Yamanaka, G., T. Wilson, S. Innaimo, G. S. Bisacchi, P. Egli, J. K. Rinehart, R. Zahler, and R. J. Colonno. 1999. Metabolic studies on BMS-200475, a new antiviral compound active against hepatitis B. Antimicrob. Agents Chemother. 43:190-193. [DOI] [PMC free article] [PubMed] [Google Scholar]