Abstract
Candida albicans is the most common fungal pathogen of humans. The recent discovery of sexuality in this organism has led to the demonstration of a mating type locus which is usually heterozygous, although some isolates are homozygous. Tetraploids can be formed between homozygotes of the opposite mating type. However, the role of the mating process and tetraploid formation in virulence has not been investigated. We describe here experiments using a murine model of disseminated candidiasis which demonstrate that in three strains, including CAI-4, the most commonly used strain background, tetraploids are less virulent than diploids and can undergo changes in ploidy during infection. In contrast to reports with other strains, we find that MTL homozygotes are almost as virulent as the heterozygotes. These results show that the level of ploidy in Candida albicans can affect virulence, but the mating type configuration does not necessarily do so.
Candida albicans exists as a commensal in the gastrointestinal tract in 50 to 75% of humans and is the most common human fungal pathogen, causing both mucosal and bloodstream infections. The attributed mortality for disseminated infections is 30 to 50% (22), with immunocompromised patients being particularly susceptible to this organism. Intensive analysis over the last 10 years has yielded evidence that pathogenesis in this fungus is multifactorial.
C. albicans is diploid as usually isolated, and, with one exception, WO-2, a strain with several translocations (32), no stable haploids or aneuploids with fewer than two copies of each chromosomal homologue have been reported, although many laboratory strains can spontaneously become trisomic for chromosome 1 (7), and reversible trisomy for chromosome 2 seems to be frequent (37). C. albicans was thought to be asexual until Hull and Johnson demonstrated the existence of a mating type locus (MTL, mating type-like) which is similar to the MAT locus in Saccharomyces cerevisiae and exists as two alleles, MTLa and MTLα, in most C. albicans strains (16). Subsequently C. albicans laboratory strains were engineered to be homozygous or hemizygous at the MTL locus and shown to be capable of mating to form tetraploids, either in the laboratory or after infection in a mouse (17, 31).
This process was connected to the well-studied white-opaque phenotypic transition when it was shown that white-opaque switching required strains homozygous at the MTL locus and that the opaque forms were manyfold more effective at mating than the white forms (28, 33). Tetraploids formed by mating have been shown to return to the diploid state under some conditions, although the process seems to involve random and sequential chromosome loss leading eventually to diploidy rather than classical meiosis (4). Mating is not restricted to laboratory constructs, since MTL-homozygous strains exist among clinical isolates (25, 28) and have been shown to be capable of cell fusion, mating, and formation of tetraploids (5, 25, 27). The isolation of mating products from an experimental infection and a recent report demonstrating that cell fusion can occur on the skin in an animal model suggest that the mating process can occur in association with the host, at least in experimental models of infection (17, 24). The relatively frequent occurrence of strains with the potential to mate (3 to 10%, depending on the study) and their demonstrated ability to mate in vivo suggest that homozygosity at the MTL locus and mating could play a significant role in the virulence of this fungus, even though evidence suggests that it reproduces most frequently, although not exclusively, in a clonal manner (2, 14, 36).
Despite the evidence suggesting the existence of mating in vivo, there are no reports of tetraploids among clinical isolates. One possible explanation is that there has been no careful survey of clinical isolates for ploidy. Indeed, only flow-cytometric analysis would provide definitive evidence for tetraploidy, but experiments designed to look at haplotypes might give some indication if, for example, three or four haplotypes were found in a single strain. However, analyses of heterozygosities have reported only two alleles for any given strain (11, 30, 38).
A second possible explanation is that tetraploids are formed in vivo but are rapidly lost, because they are intrinsically less able to survive the host defenses (and may be outcompeted by the parental diploids) or because they rapidly return to the diploid state via meiosis or random chromosome loss (4). Inoculation of mice with tetraploid strains in the presence or absence of diploids would provide a way of determining the fate of tetraploids in vivo in at least one experimental model. In this paper we report that such experiments suggest that tetraploids are less virulent than isogenic diploids, and both competition by diploids and chromosome loss may contribute to the rapid loss of tetraploids in the mouse model of disseminated disease.
While the role of mating and the MTL loci in the virulence of C. albicans is unknown, mating plays an important role in virulence in other pathogenic fungi. In the yeast Cryptococcus neoformans, strains with the MATα configuration at the mating type locus have been shown to be more virulent in one serotype (23). In similar studies of C. albicans, Lockhart et al. have shown that, in several strains which generate MTL homozygotes at a high rate, largely due to nondisjunction (41), the homozygotes are very much diminished in virulence, and MTLa/α strains outcompete the corresponding MTL homozygotes (29). A reconstructed heterozygote, in which the MTLα2 gene was replaced ectopically in an MTLa homozygote, regained partial virulence.
These studies, however, used strains which are atypical, in that they seem to undergo nondisjunction at a very high rate, and furthermore they are prototrophs and thus difficult to manipulate in the laboratory. Finally, they are uncharacterized with respect to their karyotypes and genomic sequence except for a few genes. In order to take advantage of the growing number of techniques and large amount of genomic information presently available concerning C. albicans, we asked about the role of mating type in virulence, using strains that are genomically and/or genetically characterized in order to determine whether a wide variety of strains, including the one best characterized genetically, show the severe impairment of virulence observed by Lockhart et al. (29). The availability of close-to-isogenic MTL homozygotes and heterozygotes has enabled us to answer this question. We show that in two separate isolates MTL homozygotes are diminished in virulence only slightly, if at all, compared to the heterozygotes.
MATERIALS AND METHODS
Strains and strain construction.
The strains used in this work are listed in Table 1.
TABLE 1.
Genotypes of Candida albicans strains used in this study
| Strain | Genotype | MTL genotype | Source and/or reference |
|---|---|---|---|
| SC5314 | Wild typea | a/α | 13 |
| 3116b | Wild type | a/a | Sorbose selection from SC5314 (this work) |
| 3117b | Wild type | α/α | Sorbose selection from SC5314 (this work) |
| 4448b | Wild type | a/a | Sorbose selection from SC5314 (this work) |
| 4449b | Wild type | α/α | Sorbose selection from SC5314 (this work) |
| 4450 | Wild type | a/a/α/α | Mating product of 4448 and 4449 (this work) |
| CAI4.URA | URA3/ura3 | a/α | CAI-4 (8) |
| 3142 (CHY444) | ura3/ura3 MTLa/mtlα1::hisG mtlα2::hisG | a/Δα | 17 |
| 3153 | ura3/ura3 ade2::hisG/ade2::hisG mtla1a2::hisG/MTLα | α/Δa | Derived from CHY477 (33) (this work) |
| 3153.URA | URA3/ura3 ade2::hisG/ade2::hisG mtla1a2::hisG/MTLα | α/Δa | Derived from 3153 (this work) |
| 3832 | URA3/ura3/ura3/ura3 ADE2/ADE2/ade2/ade2 MTLa/mtlα1::hisG mtlα2::hisG MTLα/mtla1a2::hisG | a/α/Δa/Δα | Mating product of 3142 (35) and 3153.URA |
| 3834 | Same as 3832 | a/α/Δa/Δα | Mating product of 3142 (35) and 3153.URA |
| 3609 | ura3/ura3/ura3/ura3 ADE2/ADE2/ade2::hisG ade2::URA3 MTLa/mtlα1::hisG mtlα2::hisG MTLα/mtla1a2::hisG | a/α/Δa/Δα | Mating product of 3142 (35) and CHY477 (33) |
| 1035 | Wild type | a/α | FC18 (40) |
| 3567b | Wild type | a/a | Sorbose selection from FC18 (this work) |
| 3568b | Wild type | α/α | Sorbose selection from FC18 (this work) |
| 3316b | arg/arg ilv/ilv | a/a | Sorbose selection from A505 (20) (this work) |
| 3320b | trp1/trp1 lys2/lys2 | α/α | Sorbose selection from A658 (21) (this work) |
| 3148 | arg/arg/ARG/ARG ilv/ilv/ILV/ILV trp1/trp1/TRP1/TRP1 lys2/lys2/LYS2/LYS2 | a/a/α/α | 3316 × 3320 (31) |
| AF27 | ura3/ura3 his1::hisG/his1::hisG gal1::HIS1/gal1::URA3 | a/α | 9 |
Wild type refers to an absence of auxotrophic markers and known gene disruptions.
Strains constructed by growth on sorbose of parent strain.
Strains 3116, 3117, 4448, and 4449 were isolated from SC5314 after growth on sorbose as described previously (31). Strains 3567 and 3568 were similarly derived from 1035. Strain 4450 was derived from strains 4448 and 4449 by mixing the cultures, isolating a mating pair, and then analyzing the progeny to verify that they were tetraploid. Thus, this strain is an isogenic tetraploid of SC5314.
Restoring auxotrophies.
In order to avoid positional effects related to the integration of the URA3 gene, we restored one copy of the native URA3 locus for 3153 (derived from CAI-4). Strain CHY477, containing URA3 inserted into one allele of the ade2 locus and hisG in the other allele, was rendered Ura− by plating onto synthetic complete medium containing 0.1% 5-fluoroorotic acid (Zymo Research) and uridine (80 μg/ml). The resulting strain was 3153. To prepare the transformation construct, a 3.9-kb NheI/PstI fragment encompassing URA3 and the adjacent IRO1 gene was cloned from a genomic DNA library from C. albicans SC5314. The URA3-IRO1 fragment was subcloned into XbaI/PstI-digested pBSK (Stratagene) to yield pBSK-URA3. Finally, NotI/PstI digestion was used to release the URA3-IRO1 fragment from pBSK-URA3. This fragment was used to transform the Ura− strains using the lithium acetate method (6). Integration of the URA3-IRO1 fragment into the correct locus was confirmed by PCR and resulted in a reconstruction of the original URA3-IRO1 locus. The URA3 gene was then replaced as above to yield 3153.URA. Strain CAI-4.URA was restored to uridine prototrophy in the same way.
Construction of tetraploids.
In order to test the effect of ploidy on virulence, we generated three sets of tetraploids closely related to the diploid strains used in the virulence tests. All tetraploids were constructed by mating as described by Magee and Magee (31). Strains 3832 and 3834 were isolates of a mating between strains 3153.URA and 3142, and 3609 was a mating product of 3142 and CHY477. Strain 3148 was a mating product of strains 3316 and 3320. The parents were derived from FC18 by UV mutagenesis (20, 21) and rendered homozygous at the MTL locus by growth on sorbose.
Growth and preparation of strains.
All strains were maintained in YEPD (10 g yeast extract, 10 g peptone, and 20 g glucose per liter)-glycerol (50%) or YEPD-dimethyl sulfoxide (7%) suspensions at −80°C. Strains were grown in vitro at 30°C. Media used were YEPD and Min (6.7 g yeast nitrogen base without amino acids and 20 g of glucose per liter). Supplements (20 mg/liter) were added as needed. Sorbose medium was Min without glucose and with 2% sorbose (Fluka 85541). Gal− strains were selected on Min minus glucose plus 3% glycerol plus 0.1% 2-deoxygalactose (2-dG; Aldrich; 04407) (9). All plates contained 15 g/liter agar (Sigma; A1296).
Murine model of disseminated candidiasis.
C. albicans strains were passaged overnight at room temperature in YEPD three times prior to infection in order to insure a highly active log-phase culture. Cells were harvested, washed, counted, and resuspended at a density of 106/ml in nonpyrogenic phosphate-buffered saline (Irvine Scientific, Irvine, CA). Male BALB/c mice weighing 20 to 23 g (NCI) were injected through the lateral tail vein with 0.5 ml containing approximately 5 × 105 blastospores. The infectious inocula were verified by counting the CFU plated on YEPD agar from appropriately diluted samples. Mouse survival was monitored three times daily, and moribund mice were euthanized. For tissue fungal burden experiments, mice were sacrificed at selected time periods postinfection and their kidneys were removed by sterile dissection, weighed, homogenized, diluted with saline, and quantitatively cultured on YEPD at 30°C. The plates were counted after 3 days. Values were expressed as CFU per gram of tissue. Cells from these plates were analyzed as described below for their ploidy. All procedures involving mice were approved by the institutional animal use and care committee, according to the National Institutes of Health guidelines for animal housing and care.
Statistical analysis.
Kaplan-Meier curves were pairwise compared with the nonparametric log rank test. Median survival times were compared by using the nonparametric Wilcoxon rank sum test for multiple comparisons. Comparisons with P values of <0.05 were considered significant.
Analysis of tetraploid cells recovered from moribund mice.
In order to determine whether there was a reduction in ploidy during passage through mice, we used two complementary strategies to analyze cells recovered from the kidneys of mice infected with tetraploids.
Replica plating on 2-dG.
Kidney homogenates from mice were plated on YEPD at several dilutions in order to get ∼100 to 200 colonies/plate, and the resulting plates replicated to 2-dG medium (9). The colonies which were 2-dG resistant in the mouse isolates were taken to be derived from the inoculated diploids.
Flow-cytometric analysis.
Colonies were picked from the YEPD plates containing the kidney homogenates, restreaked on YEPD, stained with Sytox green, and analyzed for DNA content by flow-cytometric analysis as previously described (31). Some colonies were picked from the 2-dG plates and similarly analyzed.
Determination of viability in stationary phase.
SC5314 and 4450 were inoculated from overnight cultures into YEPD and shaken at 30°C. At intervals, samples were taken, their cell counts were determined with a hemocytometer, and the samples were diluted and plated for CFU.
RESULTS
Tetraploids are less virulent than diploids.
Mating and tetraploid formation can occur between clinical isolates which are homozygous and complementary at the MTL locus (25, 27). However, there has not been a report of the isolation of a tetraploid C. albicans outside the laboratory. Possible reasons for this failure include a strong selection against tetraploids in vivo or a reduction to diploidy within the host, either via meiosis or by random chromosome loss. We examined the virulence of tetraploids in the mouse model to see whether the tetraploids are hypovirulent and unable to compete with diploid cells in the host.
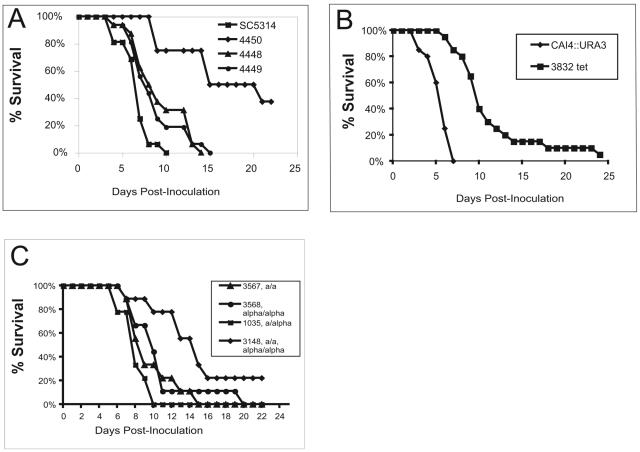
Figure 1A shows the results of infecting mice with 4450, a tetraploid mating product of strains 4448 and 4449. This strain exhibited reduced virulence compared with SC5314 and its MTL-homozygous derivatives (P = 0.001 for SC5314); median survival was 14 days, and, after 21 days, 40% of the mice were still alive. We therefore examined three other tetraploids; two (3832 and 3834) were separate isolates of a cross of strain 3142 (MTLa/mtlα1::hisG mtlα2::hisG ura3::imm434/ura3::imm434) and 3153.URA (MTLα/l mtla1a2::hisG URA3/ura3::imm434 ade2::hisG/ade2::hisG). One parent carries deletions of MTLa1, MTLa2, and both alleles of URA3; the other carries deletions of MTLα1 and MTLα2, both alleles of ADE2, and one allele of URA3. The other URA3 allele of 3153.URA is intact through gene replacement. Both are derived from the parent strain CAI-4. Thus, while not isogenic to CAI-4, both parents are closely related to this strain. Tetraploid 3609 was a mating product of 3142 and CHY477 (33). It is similar to 3832 in that it lacks two intact ADE2 alleles and three intact URA3 alleles, but in 3609 a URA3 allele is inserted into the disrupted ade2 locus.
FIG. 1.
Survival curves of mice infected with C. albicans strains differing in MTL configuration or ploidy. The mice were infected as described in Materials and Methods. A. SC5314 (MTLa/α), 4448 (MTLa/a), 4449 (MTLα/α), and 4450 (MTLa/a/α/α). The data from two separate experiments involving the same strains were combined. B. CAI-4.URA (MTLa/α) and 3832 (MTLa/a/α/α). The data from two separate experiments involving 3832 were combined. C. 1035 (MTLa/α), 3567 (MTLa/a), 3568 (MTLα/α), and 3148 (MTLa/a/α/α).
Figure 1B shows that mice infected with 3832 survive significantly longer than those infected with the parent diploids. The time to 50% mortality with 3832 is about 9 to 10 days, and 10% of the mice survived for the duration of the experiment, while for 3834 survival was somewhat longer. We have no explanation for the fact that these sister strains differed in virulence, but the difference was significantly less than the difference between any tetraploid and any diploid. For strain 3609, 50% of the mice died between 8 and 11 days (data not shown) and all died after 18 days. The differences from diploids are highly significant, with a P value of 0.0002 for 3832 compared to the repaired CAI-4 strain. Strain 3832 was among the fastest-growing C. albicans strains used in the infection experiments, so that its reduced virulence must be related to something other than growth rate in rich medium. The reduced virulence of tetraploids was not strain specific, since tetraploid 3148, constructed from two auxotrophic derivatives of FC18, was also less virulent than the parental diploid, median survival being 13 to 14 days versus 7 to 8 days (P = 0.007 by log rank test) (Fig. 1C), and 20% of the mice survived. The two auxotrophic parents of this last diploid were also tested for virulence; one, 3320 (Trp− Lys−), was avirulent, while the other, 3316 (Arg− Ilv−), was less virulent than the prototrophic diploid parent but more virulent than the prototrophic tetraploid. Thus, every tetraploid tested was less virulent than any prototrophic diploid, whether from the same or a different strain, and the virulence of the tetraploids was not dependent on whether they were constructed of auxotrophs or prototrophs. Table 2 summarizes the virulence of tetraploids compared to that of SC5314.
TABLE 2.
Comparison of virulence of tetraploid strains
| Strain | Strain background | Relative virulence (days to 50% mortality) |
|---|---|---|
| SC5314 (control) | SC5314 | 6-6.5 |
| 4450 | SC5314 | 15 |
| 3832 | CAI-4 | 9-10 |
| 3834 | CAI-4 | 14-15 |
| 3609 | CAI-4 | 8-11 |
| 3148 | FC18 | ≥12 |
Competition between tetraploids and diploids in infected mice.
In order to determine whether tetraploids were intrinsically less virulent than diploids, we used strain AF27, which was derived from CAI-4 by disruption of both copies of the GAL1 locus on chromosome 1 and replacement of URA3 at the GAL1 locus. This strain has been shown to be equal in virulence to SC5314 (9). Its advantage is that it can be selected on medium containing 2-dG. We coinfected mouse strains with inocula containing equal amounts of AF27 and either of two tetraploids, 3832 or 4450, sacrificed the mice at various time points, excised the kidneys and homogenized them, and then plated the homogenate on YEPD. The colonies were then replica plated to 2-dG to determine the fraction of diploids and tetraploids. Table 3 shows that, for the AF27-3832 combination, the number of diploids was roughly equivalent to the number of tetraploids after 2 days, but by 4 days the diploids constituted between 70 and 88% of the cells isolated from the mouse kidneys. The coinfection with AF27 and 4450 gave similar results; in this case the diploids predominated after 2 days, and by 4 days constituted more than 90% of the cells recovered from the kidneys (Table 3). By 6 days, the tetraploids were in a very small minority or in some cases absent.
TABLE 3.
Selection in vivo of diploids in a mixed infectionc
| Strains | Day | Mouse | No. of:
|
Fraction diploid | |
|---|---|---|---|---|---|
| YEPD colonies | 2-dG colonies | ||||
| AF27 and 3832 | |||||
| 1 | 1a | 129 | 68 | 0.52 | |
| 2a | 62 | 32 | 0.52 | ||
| 3a | 88 | 34 | 0.39 | ||
| 2 | 1a | 13 | 9 | 0.69 | |
| 2a | 36 | 16 | 0.44 | ||
| 3a | 19 | 11 | 0.57 | ||
| 4 | 1b | 135 | 119 | 0.88 | |
| 2a | 208 | 151 | 0.73 | ||
| 3a | 54 | 45 | 0.83 | ||
| AF27 and 4450 | |||||
| 0 | Inoculum | 86, 85 | 47, 44 | 0.46 | |
| 2 | 1a | 222 | 200 | 0.90 | |
| 2a | 144 | 118 | 0.82 | ||
| 4 | 3b | 34 | 31 | 0.91 | |
| 4b | 40 | 38 | 0.95 | ||
| 6 | 5a | 214 | 212 | 0.99 | |
| 6b | 100 | 100 | 1.0 | ||
| 7 | 7a | 78 | 71 | 0.91 | |
| 7b | 208 | 183 | 0.88 | ||
| 8b | 84 | 84 | 1.0 | ||
| 8 | 9b | 8 | 6 | 0.75 | |
| 9a | 203 | 203 | 1.0 | ||
| 10b | 423 | 423 | 1.0 | ||
1:10 dilution.
1:100 dilution.
A 2-deoxygalactose-resistant (Gal−) diploid, AF27, was mixed with either of two tetraploids in a 1:1 ratio and injected into mice. At the times indicated, mice were sacrificed, their kidneys were excised and homogenized, and the homogenate was plated on YEPD. The cells were grown for several days on YEPD and then replica plated to Min plus 2-deoxygalactose. The number of 2-dG-sensitive colonies is taken as the number of colonies derived from the tetraploid inoculum.
Recovery of tetraploids from infected mice.
The differential virulence of tetraploids during infection could be attributable to any of several causes. The tetraploids could be cleared more rapidly than the diploids by the mouse. Alternatively, the tetraploids could lose chromosomes either randomly or by a hitherto-undetected meiotic cycle. Such aneuploid cells or meiotic products could be diminished in virulence.
If differential clearance is the explanation, the 2-deoxygalactose-sensitive cells recovered from the mice would be expected to be largely tetraploid. However, if the cells undergo frequent changes in ploidy in the mouse, one would expect the sensitive cells to be diploid or at least to vary significantly from tetraploid. We therefore analyzed the colonies recovered from the kidneys of the moribund mice infected with 4450 for their ploidy. Several random colonies were selected (both from YEPD and from 2-dG), and the DNA content was determined by flow-cytometric analysis to look for evidence of reduction in ploidy.
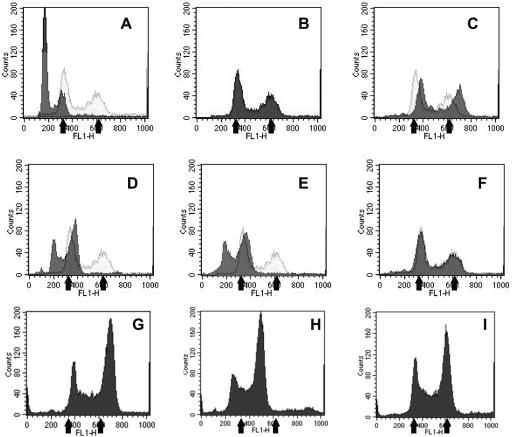
Figure 2 shows examples of strains with altered karyotypes taken from the competition experiment. Figure 2A shows the distribution of G1 and G2 cells from the diploid AF27 (dark trace) and Fig. 2B gives the G1 and G2 cells from the tetraploid 4450; the tetraploid distribution (light trace) is superimposed on the diploid in Fig. 2A and on the cells in Fig. 2C, D, E, and F. Figure 2D shows diploid cells from a 2-dG-resistant colony taken at 6 days. Figure 2C, E, and F show 2-dG-sensitive cells; the cells in Fig. 2C have a ploidy greater than 4×, those in Fig. 2E are approximately diploid, and Fig. 2F shows a tetraploid. Table 4 shows the results of the analysis of 17 recovered strains as well as the parents. There are several important results. First, as expected the resistant strains remained diploid throughout the entire experiment. Second, up until 48 h, all the sensitive strains tested were tetraploid. Third, by 96 h the tetraploids were beginning to show variations in ploidy (MC13 to MC18 and MC23). Finally, there was a tetraploid strain, MC26, which had DNA content greater than 4N. Thus, the DNA content of the tetraploids begins to vary as the infection progresses. In single-infection experiments with tetraploids, the ploidy also varies, but cells with 4N DNA content are found as late as 10 days into infection. Figure 2G to I show flow-cytometric data from an infection with 3834, a sister strain of 3832. This strain is one of the least-virulent tetraploids we have tested (Table 2). Figure 2G shows a colony containing cells with ploidy greater than 4N, while the isolate in Fig. 2H is intermediate between 2N and 4N and the strain in Fig. 2I is close to tetraploid. These results suggest that the reduced virulence of tetraploids in the mouse model of disseminated disease is due to a variety of factors, one of which is probably production of aneuploid cells.
FIG. 2.
Flow-cytometric analysis of cells isolated from the diploid-tetraploid competition and from single-tetraploid infection. The panels show the DNA content of cells from colonies isolated from mouse kidneys. The fluorescence peak for G1 diploid cells is at 160 units and for G2 cells is at 320 units. For tetraploids the corresponding peaks are at 350 and 620 units (black arrows). A to F. Colonies from the competition experiment. A. AF27 preinoculation (MC3). B. 4450 preinoculation (MC4). C. 2-dG-sensitive cells (6 days, MC26). D. 2-dG-resistant cells (8 days, MC28). E. 2-dG-sensitive cells (8 days, MC29). F. 2-dG-sensitive cells (8 days, MC12). The preinoculum 4450 trace is superimposed on panels A and C to F. The MC numbers refer to strains in Table 4. G to I. Colonies from a single infection with tetraploid 3834.
TABLE 4.
DNA content of strains recovered from mixed infectiona
| Strain | Sensitivity to 2-dG | Day | Mouse | Ploidy |
|---|---|---|---|---|
| MC2 | Sensitive | 0 | 4× | |
| MC3 | Resistant | 0 | 2× | |
| MC4 | Sensitive | 0 | 4× | |
| MC5 | Sensitive | 2 | M1 | 4× |
| MC6 | Sensitive | 2 | M1 | 4× |
| MC7 | Sensitive | 2 | M1 | 4× |
| MC8 | Sensitive | 2 | M1 | 4× |
| MC9 | Resistant | 2 | M1 | 2× |
| MC10 | Resistant | 2 | M1 | 2× |
| MC11 | Resistant | 2 | M1 | 2× |
| MC12 | Resistant | 2 | M1 | 2× |
| MC13 | Sensitive | 4 | M2 | 2× |
| MC14 | Sensitive | 4 | M2 | ∼4× |
| MC15 | Resistant | 4 | M2 | 2× |
| MC16 | Resistant | 4 | M2 | 2× |
| MC17 | Sensitive | 4 | M3 | <4× |
| MC18 | Sensitive | 4 | M3 | <4× |
| MC19 | Resistant | 4 | M3 | 2× |
| MC20 | Resistant | 4 | M3 | 2× |
| MC21 | Sensitive | 4 | M4 | 4× |
| MC22 | Sensitive | 4 | M4 | 4× |
| MC23 | Sensitive | 4 | M4 | <4× |
| MC24 | Resistant | 6 | M5 | 2× |
| MC25 | Resistant | 6 | M5 | 2× |
| MC26 | Sensitive | 6 | M5 | >4× |
| MC27 | Resistant | 6 | M5 | 2× |
| MC28 | Resistant | 8 | M6 | 2× |
| MC29 | Sensitive | 8 | M6 | 2× |
| MC30 | Sensitive | 8 | M6 | 4× |
| MC31 | Resistant | 8 | M6 | 2× |
Colonies were picked from YEPD or 2-dG medium and restreaked on YEPD. They were then assayed for DNA content by flow cytometry as described in Materials and Methods.
Tetraploids and diploids from C. albicans die at equal rates in stationary phase.
One possible reason for the failure of tetraploids to compete in the mouse infections would be a tendency to die in stationary phase more rapidly than diploids, as shown in Saccharomyces cerevisiae (1). We tested this possibility by monitoring the viability of tetraploid 4450 and its parent, diploid SC5314, over 4 weeks. Table 5 shows that the viability of the diploid and the tetraploid declined at the same rate. By the end of the experiment, the viability of SC5314 had declined 98.5%, while that of 4450 had declined 98.7%.
TABLE 5.
Viability of diploids and tetraploids in stationary phase
| Viable cells/ml of strain:
|
||
|---|---|---|
| Day | SC5314 | 4450 |
| 4 | 2.05 × 108 | 1.59 × 108 |
| 7 | 5.55 × 107 | 7.10 × 107 |
| 11 | 2.80 × 106 | 2.15 × 106 |
| 14 | 6.69 × 106 | 3.34 × 106 |
| 16 | 4.27 × 106 | 1.42 × 106 |
| 18 | 4.72 × 106 | 2.57 × 106 |
| 29 | 3.05 × 106 | 2.04 × 106 |
Correlation of MTL genotype with virulence.
In order to determine whether the configuration at the MTL locus affected virulence, we analyzed MTL homozygotes constructed from SC5314, the strain used to determine the C. albicans genome sequence, by plating the MTLa/α heterozygote on sorbose-containing plates. Cells which are monosomic for chromosome 5, the site of the MTL locus, are able to grow on these plates (Sou+ [18]) but undergo a reduplication event when grown on rich medium, leading to euploid strains completely homozygous for chromosome 5. The two homologues are lost with approximately equal frequency, so among the Sou+ strains are homozygotes for either mating type (26, 31).
To determine the effects of MTL homozygosity on virulence, these homozygous strains were used to infect mice. The inocula varied from 3.2 × 105 to 5 × 105 cells per strain. Figure 1A shows the combined results of two experiments using 16 mice in total. Strain 4448, the MTLa/MTLa strain derived from SC5314, and 4449, the corresponding MTLα/MTLα strain, appeared to be less virulent than the MTLa/MTLα strain, SC5314 (P = 0.009 for 4448 and P = 0.034 for 4449), and the two homozygotes were equivalent to one another (P = 0.7). In a parallel experiment using a second, independent set of MTL homozygotes, 3116 (a/a) and 3117 (α/α), 3116 was not different from SC5314 (P = 0.47) but 3117 was less virulent (P = 0.002) (data not shown).
To broaden the number of strains examined, we used MTL homozygotes of an unrelated strain, FC18. Figure 1C shows that once again the MTLα/MTLα cells were less virulent than the MTLa/MTLα cells (P = 0.03). Although the MTLa/MTLa homozygotes were slightly less virulent than the parent in this experiment, the difference was not statistically significant (P = 0.15). Thus, in two genetic backgrounds, strains lacking the MTLa locus (MTLα homozygotes) were somewhat less virulent than the heterozygote (although still capable of lethal infections at the standard dose), suggesting that there may be genes associated with virulence which are upregulated by the product of the MTLα1 or MTLα2 gene or downregulated by the MTLα genes (39). The behavior of the strains lacking the MTLα allele was variable. SC5314-derived homozygotes were diminished in virulence like the MTLα homozygotes while FC18-derived a/a homozygotes were equal in virulence to the parent. However, in all cases MTL homozygotes were significantly virulent.
DISCUSSION
Since the discovery of the MTL locus in Candida albicans (16) and the subsequent demonstration that mating is possible in this pathogenic yeast (17, 31), the question of the biological role of mating and especially its role in virulence has been an intriguing one. MATα is more virulent than MATa in serotype D C. neoformans (23), but in serotype A the levels of virulence of the two genotypes are equivalent (34). Since mating in C. albicans involves loss of some or all heterozygosity on chromosome 5, a switch in cell type from white to opaque, and a mating product which is tetraploid, it might be expected to have a significant effect on host-fungus interactions. We therefore investigated the virulence of various forms involved in mating. The results presented here demonstrate that tetraploidy has an effect on virulence in Candida albicans, while in two commonly used laboratory strains, homozygosity at the MTL locus has little effect.
We have shown that a number of independently isolated tetraploids from several strain backgrounds are less virulent than their diploid parents. This is true whether they are tested individually or in competition experiments. The difference is unrelated to growth rate, and it does not seem to be due to differential loss of viability in stationary phase. Although the tetraploid cells were rapidly displaced in competition experiments, they were nevertheless able to cause lethal infections when inoculated alone. In several experiments, the difference in the time to 50% mortality for isogenic or nearly isogenic diploid/tetraploid pairs was about 6 to 8 days, suggesting that tetraploids are less able to survive and multiply in the animal, despite the fact that they grow well in vitro.
What might be the mechanism for this diminished virulence? One possibility raised by the results of Lockhart et al. is that the process of becoming homozygous at the MTL locus attenuates a cell and that mating does not restore virulence. However, in our hands the parents of the tetraploids we analyzed were as virulent or almost as virulent as the heterozygous diploids from which they were derived. The tetraploids grow as fast as or faster than the diploids in vitro. However, growth in vitro is a test of the ability to grow at a rapid rate, while infection requires the ability to withstand host defense factors, to grow under difficult conditions, and to adapt to particular niches. All of these characteristics require particular gene expression patterns, and it may be that in C. albicans, as in S. cerevisiae, gene expression is affected by ploidy. A study of the effect of ploidy on gene regulation in S. cerevisiae found that 10 genes were ploidy induced and 7 were ploidy repressed (12). One of the repressed genes, FLO11, is involved in invasive growth, a process related to filamentation in Candida; however, an orthologue of this gene has not been identified in the C. albicans genome. As discussed by Galitsky et al., two possible mechanisms for ploidy-regulated gene expression are sensing gene dosage and sensing total DNA content (12). If expression of genes related to virulence is similarly regulated in C. albicans, the diminished virulence might be explained. Experiments are ongoing to look at gene expression in tetraploids of C. albicans compared to isogenic diploids to determine whether there are significant differences dependent upon ploidy and whether these differences involve virulence factors. If so, it will be important to determine how the affected genes are regulated.
Downregulation of some genes for some virulence factors in tetraploids may explain part of the virulence discrepancy, but it seems likely that an important factor may be the fact that tetraploids lose chromosomes under stress (4, 15). Indeed, chromosome changes, such as apparent loss of a homologue followed by reduplication of the remaining one, can occur during infection even in diploids during infection (10). Since we found that many colonies derived from tetraploid cells in the mixed-infection experiment had altered ploidy, the stress of infection likely causes significant karyotypic changes in tetraploids. Aneuploids began to appear in the mixed infections after about 4 days, and the fraction increased as the infection continued. Aneuploidy has been shown in vitro to lead to slower growth (3, 19) and diminished virulence (7). If slow-growing aneuploids are produced in large numbers from tetraploids in vivo, they could effectively diminish the inoculum. A population shift to predominately aneuploid cells over the course of the infection in the mouse would explain the fact that in five of six independent experiments, from 10 to 40% of the mice survived infection with an inoculum of tetraploids. However, no tetraploid was avirulent. The cells with ploidy higher than 4× pose a special puzzle. It seems highly unlikely that chromosome loss leads to MTL homozygotes with ploidy greater than 2 which then mate. The cells with very high DNA content are most likely due to the same sort of nondisjunction events which lead to aneuploids with intermediate DNA content. The karyotypes of all the cells with altered DNA content are under investigation. The fact that cells of ploidy intermediate between diploid and tetraploid were isolated from infected mice argues that, as in the in vitro experiments of Bennett et al. (4), reduction in ploidy does not occur via classical meiosis.
We determined the virulence of a total of three pairs of MTL homozygotes derived from two strains: SC5314 and FC18. In contrast to previous results (29), we found that the strains we tested that were homozygous at the MTL locus are all somewhat virulent, with the exception of one Trp− Lys− auxotroph (data not shown). For example, in Fig. 1A, the two homozygotes differ in virulence from the parental heterozygote by only 1 to 2 days in the time to 50% survival of the infected mice. In replicate experiments, the 50% mortality time difference varied from 0 to about 3 days. In strains derived from FC18, the difference was 1 to 2 days, and the a/a strain was statistically equivalent in virulence to the parent, while the α/α strain was slightly less virulent. All the prototrophic MTL homozygotes were more virulent than any tetraploid.
Several differences may explain the divergence between these results and those of Lockhart et al., who found that MTL homozygotes derived from three different clinical isolates, all of which have a high frequency of nondisjunction for chromosome 5, were avirulent or only slightly virulent and that restoring the α2 gene to an a/a homozygote partially restored virulence (29). Among the possible explanations is that nondisjunction of one chromosome may often be accompanied by nondisjunction of others, leading to progeny which are genetically dissimilar to the parents. This could lead to genotypes of much lower virulence. It seems unlikely that the tendency to nondisjoin itself is a factor, since the parents were all virulent. This possibility does not explain the complete avirulence of one homozygote which arose by mitotic recombination or the restoration of virulence by α2. A second possible explanation is that the homozygotes may have reduced growth rates. It is also possible that there are fundamental differences in the regulatory network controlled by the MTLa/α loci between the group of strains used by Lockhart et al. and the three used here. For example, if the strains used by Lockhart et al. are able to switch to the less-virulent opaque cell type at higher temperatures, the discrepancy would be resolved, since addition of the α2 gene would prevent switching and restore virulence. A final possibility may be the difference in mice and the conditions used to infect them; we used BALB/c mice and a several-step growth preparation for our inocula, while Lockhart et al. used ND4 mice and a single-step growth protocol for inoculation. Whatever the explanation for the previously reported complete avirulence associated with MTL homozygosity, it does not occur in the experiments reported here.
The observation that tetraploids are less virulent than diploids may explain the failure to isolate tetraploids from clinical samples. However, since a comprehensive survey does not appear to have been conducted, we cannot say that higher-ploidy cells do not exist in vivo; we can only point out that they have not been observed. Our results suggest that mating does not play a significant role in virulence, since the mating products are rapidly selected against, at least in the mouse model of disseminated disease. However, if mating products are formed in vivo, the selection against them will rapidly lead to a return to the diploid state. This in itself could serve as a means to genetic diversity.
It seems clear that ploidy can play a role in the pathogenic potential of C. albicans, at least in the murine model of hematogenously disseminated disease. A correlation of virulence with mating type is not unprecedented among human fungal pathogens; mating type is important in the virulence of Cryptococcus neoformans var. neoformans (23) and not important in C. neoformans var. grubii (34). However, our results show that the particular configuration of the mating type does not seem to be important in the virulence in C. albicans in the most frequently used strain background. Thus, any experiments which involve MTL homozygotes in virulence must take into account the possible strain-specific virulence differences associated with MTL homozygosity.
Acknowledgments
A.S.I. is a Burroughs Wellcome Fund New Investigator in Pathogenic Mycology. D.C.S. was supported by a Canadian Institutes of Health Clinician Scientist Award, and Career Award in the Biomedical Sciences from the Burroughs Welcome Fund. This work was supported by grant number R01AI16567 from the National Institute of Allergy and Infectious Disease to P.T.M.
We acknowledge the assistance of the Flow Cytometry Core Facility of the University of Minnesota Cancer Center, a comprehensive cancer center designated by the National Cancer Institute, supported in part by P30 CA77598.
Editor: T. R. Kozel
REFERENCES
- 1.Andalis, A. A., Z. Storchova, C. Styles, T. Galitski, D. Pellman, and G. R. Fink. 2004. Defects arising from whole-genome duplications in Saccharomyces cerevisiae. Genetics 167:1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. B., C. Wickens, M. Khan, L. E. Cowen, N. Federspiel, T. Jones, and L. M. Kohn. 2001. Infrequent genetic exchange and recombination in the mitochondrial genome of Candida albicans. J. Bacteriol. 183:865-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, R. C., and K. Gull. 1992. Isolation, characterization, and genetic analysis of monosomic, aneuploid mutants of Candida albicans. Mol. Microbiol. 6:171-177. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, R. J., and A. D. Johnson. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, R. J., M. G. Miller, P. R. Chua, M. E. Maxon, and A. D. Johnson. 2005. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol. Microbiol. 55:1046-1059. [DOI] [PubMed] [Google Scholar]
- 6.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277: 105-109. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., B. B. Magee, D. Dawson, P. T. Magee, and C. A. Kumamoto. 2004. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol. Microbiol. 51:551-565. [DOI] [PubMed] [Google Scholar]
- 8.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forche, A., G. May, J. Beckerman, S. Kauffman, J. Becker, and P. T. Magee. 2003. A system for studying genetic changes in Candida albicans during infection. Fungal Genet. Biol. 39:38-50. [DOI] [PubMed] [Google Scholar]
- 10.Forche, A., G. May, and P. T. Magee. 2005. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during infection. Eukaryot. Cell 4:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forche, A., G. Schonian, Y. Grayser, R. Vilgalys, and T. G. Mitchell. 1999. Assessment of genetic relatedness of vaginal isolates of Candida albicans from Africa. Fungal Genet. Biol. 28:107-125. [DOI] [PubMed] [Google Scholar]
- 12.Galitski, T., A. J. Saldanha, C. A. Styles, E. S. Lander, and G. R. Fink. 1999. Ploidy regulation of gene expression. Science 285:251-254. [DOI] [PubMed] [Google Scholar]
- 13.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 14.Graser, Y., M. Volovsek, J. Arrington, G. Schonian, W. Presber, T. G. Mitchell, and R. Vilgalys. 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93:12473-12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilton, C., D. Markie, B. Corner, E. Rikkerink, and R. Poulter. 1985. Heat shock induces chromosome loss in the yeast Candida albicans. Mol. Gen. Genet. 200:162-168. [DOI] [PubMed] [Google Scholar]
- 16.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 17.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289: 307-310. [DOI] [PubMed] [Google Scholar]
- 18.Janbon, G., F. Sherman, and E. Rustchenko. 1999. Appearance and properties of L-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics 153:653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95:5150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakar, S. N., and P. T. Magee. 1982. Genetic analysis of Candida albicans: identification of different isoleucine-valine, methionine, and arginine alleles by complementation. J. Bacteriol. 151:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kakar, S. N., R. M. Partridge, and P. T. Magee. 1983. A genetic analysis of Candida albicans: isolation of a wide variety of auxotrophs and demonstration of linkage and complementation. Genetics 104:241-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kullberg, B. J., and S. G. Filler. 2002. Candidemia, p. 327-340. In R. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 23.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lachke, S. A., S. R. Lockhart, K. J. Daniels, and D. R. Soll. 2003. Skin facilitates Candida albicans mating. Infect. Immun. 71:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legrand, M., P. Lephart, A. Forche, F. M. Mueller, T. Walsh, P. T. Magee, and B. B. Magee. 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52:1451-1462. [DOI] [PubMed] [Google Scholar]
- 26.Lephart, P. R., H. Chibana, and P. T. Magee. 2005. Effect of the major repeat sequence on chromosome loss in Candida albicans. Eukaryot. Cell 4:733-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lockhart, S. R., K. J. Daniels, R. Zhao, D. Wessels, and D. R. Soll. 2003. Cell biology of mating in Candida albicans. Eukaryot. Cell 2:49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockhart, S. R., C. Pujol, K. J. Daniels, M. G. Miller, A. D. Johnson, M. A. Pfaller, and D. R. Soll. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockhart, S. R., W. Wu, J. B. Radke, R. Zhao, and D. R. Soll. 2005. Increased virulence and competitive advantage of a/α over a/a or α/α offspring conserves the mating system of Candida albicans. Genetics 169:1883-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lott, T. J., B. P. Holloway, D. A. Logan, R. Fundyga, and J. Arnold. 1999. Towards understanding the evolution of the human commensal yeast Candida albicans. Microbiology 145:1137-1143. [DOI] [PubMed] [Google Scholar]
- 31.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTL a and MTL α strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 32.Magee, B. B., and P. T. Magee. 1997. WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology 143:289-295. [DOI] [PubMed] [Google Scholar]
- 33.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panwar, S. L., M. Legrand, D. Dignard, M. Whiteway, and P. T. Magee. 2003. MFα1, the gene encoding the α mating pheromone of Candida albicans. Eukaryot. Cell 2:1350-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pujol, C., J. Reynes, F. Renaud, M. Raymond, M. Tibayrenc, F. J. Ayala, F. Janbon, M. Mallie, and J. M. Bastide. 1993. The yeast Candida albicans hasa clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc. Natl. Acad. Sci. USA 90:9456-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selmecki, A., S. Bergmann, and J. Berman. 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55:1553-1565. [DOI] [PubMed] [Google Scholar]
- 38.Tavanti, A., N. A. Gow, S. Senesi, M. C. Maiden, and F. C. Odds. 2003. Optimization and validation of multilocus sequence typing for Candida albicans. J. Clin. Microbiol. 41:3765-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsong, A. E., M. G. Miller, R. M. Raisner, and A. D. Johnson. 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115:389-399. [DOI] [PubMed] [Google Scholar]
- 40.Whelan, W. L., R. M. Partridge, and P. T. Magee. 1980. Heterozygosity and segregation in Candida albicans. Mol. Gen. Genet. 180:107-113. [DOI] [PubMed] [Google Scholar]
- 41.Wu, W., C. Pujol, S. R. Lockhart, and D. R. Soll. 2005. Chromosome loss followed by duplication is the major mechanism of spontaneous mating-type locus homozygosis in Candida albicans. Genetics 169:1311-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]