Abstract
Treponema denticola is an important contributor to periodontal disease. In this study we investigated the ability of T. denticola to bind the complement regulatory proteins factor H and factor H-like protein 1 (FHL-1). The binding of these proteins has been demonstrated to facilitate evasion of the alternative complement cascade and/or to play a role in adherence and invasion. Here we demonstrate that T. denticola specifically binds FHL-1 via a 14-kDa, surface-exposed protein that we designated FhbB. Consistent with its FHL-1 binding specificity, FhbB binds only to factor H recombinant fragments spanning short consensus repeats (SCRs) 1 to 7 (H7 construct) and not to SCR constructs spanning SCRs 8 to 15 and 16 to 20. Binding of H7 to FhbB was inhibited by heparin. The specific involvement of SCR 7 in the interaction was demonstrated using an H7 mutant (H7AB) in which specific charged residues in SCR 7 were replaced by alanine. This construct lost FhbB binding ability. Analyses of the ability of FHL-1 bound to the surface of T. denticola to serve as a cofactor for factor I-mediated cleavage of C3b revealed that C3b is cleaved in an FHL-1/factor I-independent manner, perhaps by an unidentified protease. Based on the data presented here, we hypothesize that the primary function of FHL-1 binding by T. denticola might be to facilitate adherence to FHL-1 present on anchorage-dependent cells and in the extracellular matrix.
Treponema denticola, a spirochete member of the normal flora of the oral cavity, is an important contributor to the development of acute and chronic periodontal disease in humans (42, 45). It is a dominant species in periodontal lesions and at the leading front of periodontitis-associated subgingival plaque (40). Real-time PCR analyses have demonstrated that there is a strong correlation between the numbers of T. denticola cells and pocket depth in periodontitis patients (45). Adult periodontitis is the most common infection of middle-aged adults, and although it is not life threatening, its economic impact is enormous. In addition to T. denticola, other dominant species associated with periodontitis include Haemophilus actinomycetecomitans (formerly classified in the genus Actinobacillus), Porphyromonas gingivalis, and Tannerella forsythia (43). A large number of less-well-characterized species, including nearly 60 treponeme phylotypes, have also been implicated (8). There is also a strong association of T. denticola with esophageal cancers (30) and with low birth weight in preterm infants (31).
An expanding number of human pathogens, including several spirochete species belonging to the Lyme disease and relapsing fever Borrelia groups, have been shown to bind the complement regulatory protein factor H and/or factor H-like protein 1 (FHL-1) (7, 9, 11, 13, 16, 17, 25, 27-29, 33, 35, 36). In addition, recombinant Omp100 of H. actinomycetecomitans has also been demonstrated to bind either factor H or FHL-1 and to play a role in adherence and invasion of KB cells (4). Factor H and FHL-1 are comprised of a series of short consensus repeats (SCRs) consisting of approximately 50 to 60 residues (reviewed in reference 47). FHL-1 is derived from the factor H mRNA via alternative splicing and consists of the first seven SCRs of factor H plus four additional hydrophobic residues at its C terminus (12, 46). Factor H and FHL-1 regulate the alternative complement pathway by serving as a cofactors for factor I-mediated cleavage of C3b (38, 39). In addition, they inhibit the initial formation and accelerate the dissociation of the alternative pathway C3 convertase by competing with Bb for binding to C3b. In terms of bacterial pathogenesis, surface-bound factor H and FHL-1 are thought to locally increase the efficiency of C3b cleavage and thereby inhibit opsonization and phagocytosis. In addition, some pathogens may bind to extracellular matrix or cell-anchored FHL-1 as a means to facilitate adherence and intracellular localization (33).
In this report, we demonstrate that T. denticola specifically binds the complement regulatory protein FHL-1. To our knowledge, this is the first bacterial protein identified that binds specifically FHL-1 and not factor H. The FHL-1 binding protein was determined to be surface exposed and to have a molecular mass of 14 kDa. We tentatively designated this protein FhbB (FHL-1 binding protein B). Using recombinant factor H SCR constructs, we showed that the binding of FhbB to FHL-1 is inhibited by heparin and is mediated via specific charged residues in SCR 7. These analyses provide the first direct evidence for the binding of a complement regulatory protein to T. denticola and enhance our understanding of the pathogenic mechanisms of this organism. FHL-1 binding by T. denticola may represent a potential adherence mechanism and may play an important role in biofilm formation and development of periodontal disease.
MATERIALS AND METHODS
Bacterial strains and cultivation.
T. denticola American Type Culture Collection strains ATCC 35405, ATCC 33520, and ATCC 33521 were cultivated in NOS medium (ATCC medium 1357) at 37°C in an anaerobe jar with GasPak Plus (Becton Dickinson and Company, Sparks, MD). Unless otherwise indicated, all analyses were conducted using the ATCC 35405 strain. B. burgdorferi B31MI, which served as a control in some analyses, was cultivated at 33°C in complete BSK-H medium (Sigma, St. Louis, MO). Approximately 5 to 7 days was required to obtain dense cultures of all strains. Growth was monitored by dark-field microscopy. The spirochetes were recovered by centrifugation and subjected to various numbers of washes with phosphate-buffered saline (PBS) as indicated below.
Factor H and/or FHL-1 absorption assay.
T. denticola ATCC 35405 and B. burgdorferi B31MI (control) cells from 1 ml of a dense culture (∼109 cells) were recovered by centrifugation (5,000 × g, 10 min, 4°C), washed with cold PBS, resuspended in 200 μl cold PBS containing factor H/FHL-1 (0.525 mg ml−1), and incubated for 1 h at room temperature. The cells were recovered by centrifugation (3,000 × g, 5 min, room temperature), washed twice with cold PBS, solubilized in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer, electrophoresed, and immunoblotted. The membranes were then screened using the affinity ligand binding immunoblot (ALBI) assay with and without addition of factor H/FHL-1 as described below.
ALBI assays and immunoblot analyses.
Cell lysates of T. denticola were prepared by solubilization in SDS-PAGE sample buffer and boiling. The proteins were separated and analyzed by SDS-PAGE using 15% polyacrylamide precast gels (Bio-Rad). Equal loading of proteins was assessed by Coomassie blue staining. Proteins were transferred onto Immobilon P membranes (Millipore) by electroblotting as previously described (24). Factor H/FHL-1 or SCR ALBI assays were conducted as previously described previously (29) using a preparation of purified human factor H (Calbiochem) or purified recombinant SCR constructs (9). The commercial factor H preparation, as reported by the supplier, contained approximately 95% factor H and 5% FHL-1. In the ALBI assay bound factor H/FHL-1 was detected using goat anti-human factor H antiserum (1:800 dilution; Calbiochem) with horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin (IgG) antibody as the secondary antibody (1:40,000 dilution; Pierce). The goat anti-human factor H antiserum detected factor H, FHL-1, and all SCR constructs used in these analyses. The three SCR constructs analyzed spanned SCRs 1 to 7 (H7), SCRs 8 to 15, and SCRs 16 to 20. These constructs were generated as part of a previous study by Duthy et al. (9). Briefly, the SCRs of interest were PCR amplified, cloned into the yeast expression vector pPICZα (Invitrogen), expressed in Pichia pastoris, and purified by immunoaffinity chromatography.
Heparin inhibition assays.
Immunoblot strips of T. denticola ATCC 35405 cell lysates were prepared as described above. The SCR 1 to 7 construct, H7, was preincubated with or without heparin (1 μg ml−1) in 5% skim milk, PBS with 0.02% Tween 20 (PBS-T) for 30 min at room temperature before addition to the T. denticola membrane strip for 1 h at room temperature. The blot was washed with PBS-T for 30 min with three changes of buffer and then analyzed for factor H/FHL-1 binding using the ALBI assay described above.
IFAs.
For indirect immunofluorescence assays (IFAs) slides were prepared and general procedures were performed as previously described (37). To determine if FHL-1 bound to the surface of the spirochetes, cells were recovered from a dense culture of T. denticola by centrifugation (5,000 × g, 5 min), washed with PBS, pelleted by centrifugation (10,000 × g, 20 min), and resuspended in 1 ml PBS with 10% fetal goat serum (PBS-FGS). The motility and hence viability of the cells were confirmed by dark-field microscopy. Purified factor H/FHL-1 was added to the cells to a final concentration of 10 ng μl−1, and the cells were incubated at 37°C for 1 h with occasional gentle mixing. The cells were washed with PBS-FGS to remove unbound factor H/FHL-1, resuspended in 25 μl PBS-FGS, and placed on prepared slides. Bound FHL-1 was detected using goat anti-human factor H antisera (1:10 dilution; Calbiochem) and ALEXA 488-conjugated rabbit anti-goat IgG antibody (1:2,000 dilution; Molecular Probes). As a control to verify that the outer membrane was not disrupted by these procedures, a second slide was stained for the periplasmic protein FlaA. FlaA was detected by incubation with rabbit anti-FlaA (a kind gift from Chris Fenno) at a dilution of 1:10,000, followed by incubation with ALEXA 488-conjugated goat anti-rabbit IgG antibody (1:2,000 dilution). As a final control, T. denticola cells were acetone fixed and permeabilized before they were screened with the anti-FlaA antiserum as described above. Images were captured using a MagnaFire camera and software (Olympus).
C3b cleavage assay.
The ability of factor H/FHL-1 bound to T. denticola ATCC 35405 to cleave C3b was assessed using a C3b cleavage assay described previously (23). In brief, cultures of T. denticola were pelleted, washed with ice-cold PBS, resuspended in cold PBS with 10 mM MgCl2, and incubated with 0 or 50 ng of purified human factor H/FHL-1 for 1 h at 37°C. The cells were washed with PBS to remove unbound factor H/FHL-1. Factor I (150 ng; Calbiochem) and C3b (250 ng; Calbiochem) were then added, and the mixture was incubated for 2 h at 37°C. The samples were fractionated by SDS-PAGE, immunoblotted, and screened with anti-human C3b antiserum (1:800 dilution; Accurate). The immunoblot methods used have been described previously (25). As additional controls for the reaction conditions employed, various combinations of purified factor H/FHL-1 (150 ng), factor I (150 ng), and/or C3b (250 ng) were incubated at 37°C in PBS for 2 h. In addition, T. denticola and B. burgdorferi were incubated as described above with factor I, factor H/FHL-1, or both omitted from the reaction. The reaction products were analyzed by SDS-PAGE and Coomassie blue staining.
RESULTS
Assessment of factor H and/or FHL-1 binding to T. denticola.
To determine if T. denticola can bind factor H, FHL-1, or both, an absorption assay was performed. T. denticola ATCC 35405 was incubated with a purified factor H/FHL-1 preparation. According to the supplier, the preparation contained approximately 95% factor H and 5% FHL-1. After washing to remove unbound factor H/FHL-1, the cells were solubilized, and the resulting cell lysates were fractionated by SDS-PAGE and immunoblotted. Screening of the blots with anti-factor H/FHL-1 antiserum demonstrated that FHL-1 and an unidentified ∼37-kDa protein were absorbed by T. denticola (Fig. 1A). The 37-kDa protein may have been FHR-1 or, alternatively, a degradation product of FHL-1 generated by one of the many proteases produced by T. denticola (5, 6, 21). Since reagents specific for FHR-1 are not available, the identity of this protein could not be determined. Verification that the anti-factor H/FHL-1 antiserum was specific and was not binding to treponeme proteins came from the demonstration that T. denticola that was incubated without factor H/FHL-1 did not react with the antiserum. It is important to note that in spite of the higher concentration of factor H in the assay mixture, no detectable binding of factor H was observed. FHL-1 was significantly concentrated by absorption, as shown by the fact that it was only weakly detected in the control lane of the purified factor H/FHL-1 preparation. In contrast to the results observed with T. denticola, B. burgdorferi absorbed predominantly factor H, indicating that there was a striking difference in the specificity of binding to different members of the factor H family by these spirochetes.
FIG. 1.
Detection of an FHL-1 binding protein in T. denticola ATCC 35405. To determine if factor H or FHL-1 or both can bind to T. denticola, absorption assays were performed. Whole cells were incubated with purified factor H/FHL-1, recovered by centrifugation, washed to remove unbound ligand, solubilized, and immunoblotted, and bound factor H/FHL-1 was detected by immunoblotting using anti-factor H/FHL-1 antiserum (A). All methods are described in the text. B. burgdorferi B31MI and purified factor H/FHL-1 served as positive controls for absorption and immunoblot detection, respectively. The abilities of different T. denticola strains (indicated above the lanes) to bind FHL-1 and the molecular mass of the binding protein were determined using the ALBI assay (B). In brief, cell lysates of each strain were fractionated by SDS-PAGE and immunoblotted. The blots were incubated with purified factor H/FHL-1 and then screened with anti-factor H/FHL-1 antiserum. B. burgdorferi B31MI, which produces multiple factor H binding proteins (BBA68 and OspE), served as the positive control. The positions of molecular mass markers are indicated on the right.
To determine the molecular mass of the FHL-1 binding protein of T. denticola ATCC 35405 and to determine if it is produced by other strains, cell lysates of multiple strains were immunoblotted and screened using the factor H/FHL-1 ALBI assay. Equal loading of protein in all lanes was determined by Coomassie blue staining (data not shown). A protein with a molecular mass of approximately 14 kDa was found to bind FHL-1. This protein was detected in all T. denticola strains, although there was some variation in the amount produced. The protein was designated FhbB for FHL-1 binding protein B (Fig. 1B). This designation reflects the function and is consistent with the nomenclature used for the functionally related FhbA protein of the relapsing fever Borrelia (18, 19). Homologs of known factor H binding proteins produced by other spirochetes or bacterial species were not detected in the T. denticola ATCC 35405 genome sequence. However, this is not surprising since there is little or no sequence homology among the factor H binding proteins that have been identified to date. Instead, conservation is primarily at the structural level, with coiled-coil motifs appearing to be key critical structural motifs (22, 24). B. burgdorferi B31MI, which produces multiple factor H binding proteins (1, 16, 22, 25, 29, 44), served as the positive control and readily bound factor H.
Demonstration that FhbB of T. denticola ATCC 35405 binds to SCR 7 and that this interaction is mediated by charged residues within SCR7.
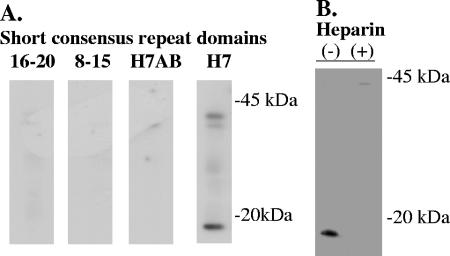
To further assess the specificity of the interaction of FhbB with FHL-1 and to determine if SCRs 8 to 20 (which are not part of FHL-1) have the potential to bind to FhbB, recombinant SCR constructs spanning SCRs 1 to 7 (H7), 8 to 15, and 16 to 20 were employed. Only H7 was found to bind to FhbB (Fig. 2A). This provided further support for the unique specificity of the interaction of FhbB with FHL-1 but not factor H. Weak binding of the H7 construct to one or more proteins with molecular masses of approximately 42 kDa was also observed. These proteins may have been additional FHL-1 binding proteins, possibly trimers of FhbB, or simply may have represented nonspecific background.
FIG. 2.
Demonstration that SCRs 1 to 7 bind to the FhbB protein of T. denticola ATCC 35405 and that binding is dependent on charged residues within SCR 7 and inhibited by heparin. To determine which SCRs of factor H/FHL-1 can bind to T. denticola, recombinant SCR constructs were used to screen immunoblot strips using the ALBI assay format (A). The SCR constructs used to screen the blots are indicated above the blots. The effects of charged residues within SCR 7 on binding to T. denticola were assessed using a site-directed mutant of the SCR 1 to 7 construct, H7AB. The influence of heparin on SCR1 to 7 binding was determined by preincubating the H7 construct with (+) or without (−) heparin before immunoblot strips of T. denticola lysate were screened using the ALBI assay (B).
SCR 7 has three regions with basic residues designated sites A (R369 and K370), B (R386 and K387), and C (K392) (13). To determine if charged residues within specific domains of SCR 7 are involved in binding to FhbB, a site-directed mutant of the H7 construct designated H7AB that was previously generated and described by Giannakis et al. (13) was used in the binding assay. In the H7AB mutant, the charged residues within sites A and B are replaced with alanine. It has been demonstrated that H7AB cannot bind heparin (13). H7AB did not bind to any of the T. denticola FHl-1 binding proteins, indicating that specific charged residues within site A and/or site B are necessary for binding.
Inhibition of FHL-1 binding to FhbB by heparin.
Since the charged residues altered in H7AB also mediate heparin binding, we determined whether heparin can inhibit the binding of the H7 construct to FhbB. Immunoblot strips of T. denticola lysate were screened with H7 that had been preincubated with or without heparin, and binding was assessed using the ALBI approach. Heparin completely inhibited the binding of H7 to FhbB (Fig. 2B). The inhibition of FHL-1 binding to FhbB by heparin, which has been shown to bind to SCR 7, provides additional support for the hypothesis that FhbB binds directly to SCR 7.
Demonstration that FhbB of T. denticola is surface exposed.
To indirectly demonstrate that FhbB of T. denticola is surface exposed, the ability of intact cells to bind FHL-1 to their surfaces was assessed using indirect immunofluorescence (Fig. 3). Unfixed cells readily bound FHL-1, indicating that the FHL-1 binding protein is surface exposed. As a control, cells were also screened with antiserum to the FlaA protein, which is an inner membrane-anchored protein that resides primarily in the periplasm. No binding to FlaA was observed in unfixed cells, but binding was observed in acetone-fixed (permeabilized) cells.
FIG. 3.
Demonstration of FHL-1 binding to the surface of T. denticola ATCC 35405 determined by IFAs. IFAs were performed as described in the text using unfixed and acetone-fixed cells (panels A and B, respectively). The antiserum used in the IFA is indicated below each image where appropriate. The images in panels A and B were obtained with a ×100 (oil immersion) objective. The dark-field image is presented for each preparation for reference to indicate the location of the spirochetes.
Analysis of the nature of the interaction of FhbB with the T. denticola cell surface.
The association of FhbB with the spirochete cell surface was also assessed following successive washes of actively growing cells with PBS. Cells washed from zero to four times with PBS were lysed and immunoblotted, and FHL-1 binding was assessed using the ALBI assay (Fig. 4). With each successive wash, detectable FHL-1 binding to the resulting T. denticola cell lysates decreased. In contrast, when identical samples were screened with anti-FlaA antiserum, no decrease in FlaA detection by immunoblotting was observed. The effect of washing on detection of FHL-1 binding suggests that FhbB is loosely associated with the outer membrane.
FIG. 4.

Demonstration that the FHL-1 binding protein of T. denticola ATCC 35405 is loosely associated with the cell surface. To assess the nature of the interaction of FhbB with the T. denticola cell surface, T. denticola cells were subjected to one, two, three, or four washes with PBS after recovery from culture media. Two blots were generated; one was screened for FlaA protein by immunoblotting (A), and the other was screened for FHL-1 binding using the ALBI assay (B). B. burgdorferi (Bb) B31MI served as the positive control in the ALBI assay. The positions of molecular mass standards are indicated on the right.
Demonstration of FHL-1-independent cleavage of C3b by T. denticola.
To determine whether FHL-1 bound to the surface of T. denticola is capable of serving as a cofactor in factor I-mediated cleavage of C3b, an in vitro cleavage assay was employed (23) (Fig. 5). Cleavage that resulted in the expected C3b digestion products was observed in the control reactions, in which factor I and C3b were incubated with the purified factor H/FHL-1 preparation. In addition, factor H/FHL-1-dependent cleavage of C3b by factor I, which has previously been demonstrated, was also observed (1, 3, 16, 20, 23). When FHL-1 was bound to T. denticola and then factor I and C3b were added, significant digestion of C3b was observed. However, the pattern of digestion products was inconsistent with the expected specific cleavage at serine residues of C3b, and when C3b was incubated with T. denticola alone, significant cleavage was also observed. C3b cleavage was also observed with culture supernatant alone. These analyses suggest that T. denticola produces a factor I-independent protease with C3b cleavage activity. Furthermore, the data suggest that the primary biological function of T. denticola-bound FHL-1 may not be in immune evasion but perhaps in adherence.
FIG. 5.
Demonstration of FHL-1-independent C3b cleavage by T. denticola ATCC 35405. T. denticola ATCC 35405 and B. burgdorferi B31MI were incubated with or without factor H/FHL-1 (fH/FHL-1), as indicated above the lanes. Unbound ligand was removed by washing with PBS. The cells were then incubated with or without factor I (fI) and/or C3b as indicated above the lanes, analyzed by SDS-PAGE, immunoblotted, and screened with anti-C3b antiserum. Additional controls in which various combinations of purified C3b, factor I, and/or factor H/FHL-1 were combined and treated as described above were also included. All methods are described in the text.
DISCUSSION
Periodontal disease is caused by a complex polymicrobial population of endogenous bacteria and several host-determined factors. The bacteria associated with periodontal disease act in a synergistic fashion to establish a productive population in the subgingiva that tenaciously adheres and invades host cells and tissues. The host response is characterized by significant inflammation, a humoral response, and complement activation within the periodontal pocket (41). In spite of this robust host response, the bacteria survive and thrive. In this study we sought to determine whether T. denticola exploits the binding of the complement regulatory protein factor H and/or FHL-1 in immune evasion, adherence, or tissue penetration. Numerous pathogens circumvent the alternate complement pathway and subsequent opsonophagocytosis by binding factor H and/or FHL-1 (4, 7, 9, 11, 13, 16, 17, 25, 27-29, 33, 35, 36). Through a variety of mechanisms, these proteins contribute to the downregulation of C3b production and locally decrease the levels of the opsonin C3b. Additionally, and potentially of great importance in T. denticola pathogenesis, the binding of FHL-1 by proteins exposed on the surface of group A streptococci have also been shown to facilitate adherence and invasion (33). In light of the ability of T. denticola to adhere to and persist within periodontal lesions we sought to determine if T. denticola is capable of binding factor H and/or FHL-1.
As a first step in assessing the binding of complement regulatory proteins to T. denticola a whole-cell absorption assay was performed. Viable cells were incubated with a purified factor H/FHL-1 preparation that consisted of 95% factor H and 5% FHL-1 (as determined by the supplier). It has not been determined if other members of the factor H family, such as the FHR proteins, also copurify with factor H. After incubation of T. denticola with factor H/FHL-1, the cells were washed and then analyzed by SDS-PAGE and immunoblotting. Screening of the immunoblot with the anti-factor H/FHL-1 antiserum demonstrated that the molecular masses of the proteins bound by T. denticola were approximately 43 and 37 kDa. It is striking that in spite of its higher concentration, no factor H (150 kDa) was absorbed by T. denticola. In contrast, factor H, but not FHL-1, was readily absorbed by B. burgdorferi. The absorption of 37- and 43-kDa proteins suggests that FhbB not only binds to FHL-1 but may also bind to other members of the factor H family of proteins, possibly FHR-1. FHR-1 exists as two forms, FHR-1α (37 kDa) and FHR-1β (43 kDa), and the difference in mass is due to differences in carbohydrate modification (10). However, it is equally plausible that the smaller bands observed are simply FHL-1 degradation products. Based on the intensity of the signal observed when the blot was screened with anti-factor H/FHL-1 antiserum, it is evident that the stoichiometry of FHL-1 binding to T. denticola is relatively high, as the cells significantly concentrate this protein. It is noteworthy that the control for these analyses, B. burgdorferi B31MI, bound predominantly, if not exclusively, factor H. Alitalo and colleagues reported that the B. burgdorferi IA strain bound both factor H and FHL-1 (2). However, other isolates in the report of these workers bound only factor H, and based on this finding and the data presented here it is appears that FHL-1 binding may vary between Lyme disease spirochete strains. Since factor H and FHL-1 can have distinct functional roles, it is likely that Borrelia and T. denticola exploit the binding of complement regulatory proteins for different purposes.
The number, mass, and distribution among isolates of the T. denticola FHL-1 binding proteins were determined using the ALBI assay. An FHL-1 binding protein with a molecular mass of 14 kDa was detected in all T. denticola strains tested and was designated FhbB (for FHL-1 binding protein B). A functionally related protein, FhbA, has been identified in the relapsing fever spirochetes (18), and hence the designation FhbB is intended to be consistent with bacterial protein nomenclature guidelines, to reflect functional activity, and to provide consistency in the nomenclature of factor H/FHL-1 binding proteins.
The data presented above strongly suggest that FhbB specifically binds to FHL-1 but not factor H. To verify this, recombinant proteins consisting of SCRs 1 to 7, 8 to 15, and 16 to 20 were tested for the ability to bind to FhbB. Only H7 bound to T. denticola. As described above, FHL-1 is comprised of the first seven SCRs of factor H plus four additional C-terminal residues. The FhbA protein of Borrelia hermsii (19) and the group A streptococcal M and Fba proteins also have been reported to bind specifically to SCRs 1 to 7 (13, 32). Similar to results obtained for the M and Fba proteins, heparin inhibited the binding of H7 to T. denticola. Previous studies identified interaction sites for heparin within factor H SCRs 7, 12, 13, 14, and 20 (47). Hence, the ability of heparin to inhibit FHL-1 binding to FhbB is consistent with the localization of both FhbB and heparin interaction sites within SCR 7 of FHL-1.
Previous analyses of the molecular interaction between factor H and the M protein of group A streptococcus using an SCR 1 to 7 mutant designated H7AB demonstrated that charged residues at specific sites in SCR 7 are directly or indirectly involved in binding (13). In the H7AB mutant, specific charged residues in SCR 7 were replaced by alanine. The H7AB mutant was found to be incapable of binding to the M protein. In this study, we demonstrated that H7AB also could not bind to FhbB. This finding further confirms the specific involvement of SCR 7 in binding and the importance of positively charged residues in the FHL-1/FhbB interaction.
For FHL-1 binding to FhbB to be of biological significance, FhbB would need to be exposed on the surface of intact cells. To assess this, several different approaches were employed. First, intact cells were subjected to treatment with proteinase K, which should degrade surface proteins but not internal proteins. This approach has been successfully employed to demonstrate the surface exposure of several spirochete proteins, including FhbA (factor H binding protein A) of B. hermsii ((18). While treatment with proteinase K led to complete elimination of FHL-1 binding by T. denticola (data not shown), some digestion of the inner membrane-anchored, periplasmic FlaA protein was also observed. This suggests that the integrity of the outer membrane was compromised and that the outer membrane of T. denticola is labile and fragile. The fragility of the outer membrane and the apparent loose association of FhbB with the cell surface were confirmed by analysis of the FHL-1 binding ability of T. denticola cells that had been subjected to increasing numbers of gentle washes with PBS. With each successive wash, the amount of FHL-1 bound by the cell lysate proteins derived from the washed cells decreased significantly, indicating that FhbB is easily removed by washing. This observation suggests that FhbB is loosely associated with the cell surface. Surface exposure and outer membrane localization of FhbB were further confirmed by IFA analyses of unfixed cells. Direct binding of FHL-1 to the cell surface was readily demonstrated, and since FhbB is the dominant, if not only, FHL-1 binding protein produced by T. denticola, it can be concluded that FhbB is at least partially surface exposed.
The potential of T. denticola surface-bound FHL-1 to participate as a cofactor in the factor I-mediated cleavage of C3b was also assessed in this study. Factor I is a serine protease that cleaves C3b at its serine residues, yielding a specific cleavage pattern (14, 26). The C3b cleavage assay employed here was previously employed by McDowell and colleagues to demonstrate the complement regulatory activity of factor H bound to the relapsing fever spirochetes B. hermsii and Borrelia parkeri and the Lyme disease spirochete, B. burgdorferi (23). When factor H/FHL-1 was incubated with T. denticola and then tested for its ability to cleave C3b in the presence of factor I, the resulting cleavage patterns were atypical and suggested that cleavage was occurring at non-Ser residues. C3b cleavage was also observed when C3b was incubated with T. denticola and factor H/FHL-1 without factor I, demonstrating that C3b cleavage by T. denticola is not FHL-1 and factor I dependent. Instead, C3b may be targeted by one or more of the proteases produced by T. denticola (34). While this observation does not rule out a role for surface-bound FHL-1 in C3b cleavage, it suggests that the main biological function of FHL-1 binding by T. denticola may not be related to complement circumvention.
In summary, T. denticola produces a novel protein, FhbB, that specifically binds FHL-1 and not factor H. FHL-1 has been demonstrated to be present on anchorage-dependent cell types, where it binds to integrin receptors, and to interact with the extracellular matrix through SCR 4 (15). Our hypothesis is that T. denticola binds to host cell surface-bound FHL-1 via its FhbB protein and that this interaction facilitates adherence, biofilm formation, and possibly tissue penetration. Additional studies are now under way to further characterize FhbB and to define its relative contribution to the molecular pathogenesis of T. denticola and periodontal disease.
Editor: D. L. Burns
REFERENCES
- 1.Alitalo, A., T. Meri, T. Chen, H. Lankinen, Z.-Z. Cheng, T. Sakari Jokiranta, I. J. T. Seppala, P. Lahdenne, P. S. Hefty, D. R. Akins, and S. Meri. 2004. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 172:6195-6201. [DOI] [PubMed] [Google Scholar]
- 2.Alitalo, A., T. Meri, L. Rämä, T. S. Jokiranta, T. Heikkilä, I. J. T. Seppala, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alitalo, A., M. T., H. Lankinen, I. Seppala, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 4.Asakawa, R., H. Komatsuzawa, T. Kawai, S. Yamada, R. B. Goncalves, S. Izumi, T. Fujiwara, Y. Nakano, H. Shiba, M. A. Taubman, H. Kurihara, and M. Sugai. 2003. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 50:1125-1139. [DOI] [PubMed] [Google Scholar]
- 5.Bian, X.-L., H.-T. Wang, Y. Ning, S. Y. Lee, and J. C. Fenno. 2005. Mutagenesis of a novel gene in the prcA-prtP protease locus affects expression of Treponema denticola membrane complexes. Infect. Immun. 73:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correia, F. F., A. R. Plummer, R. P. Ellen, C. Wyss, S. K. Boches, J. L. Galvin, B. J. Paster, and F. E. Dewhirst. 2003. Two paralogous families of a two-gene subtilisin operon are widely distributed in oral treponemes. J. Bacteriol. 185:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dave, S., S. Carmicle, S. Hammerschmidt, M. K. Pangburn, and L. S. McDaniel. 2004. Dual roles of PspC, a surface protein of Streptococcus pneumoniae, in binding human secretory IgA and factor H. J. Immunol. 173:471-477. [DOI] [PubMed] [Google Scholar]
- 8.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 9.Duthy, T. G., R. J. Ormsby, E. Giannakis, A. D. Ogunniyi, U. H. Stroeher, J. C. Paton, and D. L. Gordon. 2002. The human complement regulator factor H binds pneumococcal surface protein PspC via short consensus repeats 13 to 15. Infect. Immun. 70:5604-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estaller, C., W. Schwaeble, M. Dierich, and E. H. Weiss. 1991. Human complement factor H: two factor H proteins are derived from alternatively spliced transcripts. Eur. J. Immunol. 21:799-802. [DOI] [PubMed] [Google Scholar]
- 11.Fischetti, V. A., R. D. Horstmann, and V. Pancholi. 1995. Location of the complement factor H binding site on streptococcal M6 protein. Infect. Immun. 63:149-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friese, M. A., J. Hellwage, T. S. Jokiranta, S. Meri, H. H. Peter, H. Eibel, and P. F. Zipfel. 1999. FHL-1/reconection and factor H: two human complement regulators which are encoded by the same gene are differentially expressed and regulated. Mol. Immunol. 36:809-818. [DOI] [PubMed] [Google Scholar]
- 13.Giannakis, E., T. S. Jokiranta, D. A. Male, S. Ranganathan, R. J. Ormsby, V. A. Fischetti, C. Mold, and D. L. Gordon. 2003. A common site within factor H SCR7 responsible for binding heparin, C-reactive protein and streptococcal M protein. Eur. J. Immunol. 33:962-969. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, D. L., J. Rice, J. J. Finlay-Jones, P. J. McDonald, and M. K. Hostetter. 1988. Analysis of C3 deposition and degradation on bacterial surfaces after opsonization. J. Infect. Dis. 157:697-704. [DOI] [PubMed] [Google Scholar]
- 15.Hellwage, J., S. Kuhn, and P. F. Zipfel. 1997. The human complement regulatory factor-H-like protein 1, which represents a truncated form of factor H, displays cell-attachment activity. Biochem. J. 326:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppala, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 17.Horstmann, R. D., H. J. Sievertsen, J. Knobloch, and V. A. Fischetti. 1988. Antiphagocytic acitivity of streptoccocal M protein: selective binding of complement control protein factor H. Proc. Natl. Acad. Sci. USA 85:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovis, K., J. V. McDowell, L. Griffin, and R. T. Marconi. 2004. Identification and characterization of a linear plasmid encoded factor H-binding protein (FhbA) of the relapsing fever spirochete, Borrelia hermsii. J. Bacteriol. 186:2612-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hovis, K. M., T. Sadlon, R. Gauri, D. L. Gordon, and R. T. Marconi. Submitted for publication.
- 20.Kraiczy, P., J. Hellwage, C. Skerka, M. Kirschfink, V. Brade, P. F. Zipfel, and R. Wallich. 2003. Immune evasion of Borrelia burgdorferi: mapping of a complement inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 33:697-707. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. Y., X. L. Bian, G. W. Wong, P. M. Hannam, B. C. McBride, and J. C. Fenno. 2002. Cleavage of Treponema denticola PrcA polypeptide to yield protease complex-associated proteins Prca1 and Prca2 is dependent on PrtP. J. Bacteriol. 184:3864-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDowell, J. V., M. E. Harlin, E. Rogers, and R. T. Marconi. 2005. Putative coiled-coil structural elements of the BBA68 protein of the Lyme disease spirochetes are required for formation of its factor H binding site. J. Bacteriol. 187:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDowell, J. V., E. Tran, D. Hamilton, J. Wolfgang, K. Miller, and R. T. Marconi. 2003. Analysis of the ability of spirochete species associated with relapsing fever, avian borreliosis, and epizootic bovine abortion to bind factor H and cleave C3b. J. Clin. Microbiol. 41:3905-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDowell, J. V., J. Wolfgang, L. Senty, C. M. Sundy, M. J. Noto, and R. T. Marconi. 2004. Demonstration of the involvement of outer surface protein E coiled-coil structural domains and higher order structural elements in the binding of infection-induced antibody and the complement-regulatory protein, factor H. J. Immunol. 173:7471-7480. [DOI] [PubMed] [Google Scholar]
- 25.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 71:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medicus, R., J. Melamed, and M. A. Arnarout. 1983. Role of human factor I and C3b receptor in the cleavage of surface-bound C3bi molecules. Eur. J. Immunol. 13:465-470. [DOI] [PubMed] [Google Scholar]
- 27.Meri, T., A. Hartmann, D. Lenk, R. Eck, R. Wurzner, J. Hellwage, S. Meri, and P. F. Zipfel. 2002. The yeast Candida albicans binds complement regulators factor H and FHL-1. Infect. Immun. 70:5185-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meri, T., T. S. Jokiranta, J. Hellwage, A. Bialonski, P. F. Zipfel, and S. Meri. 2002. Onchocerca volvulus microfilariae avoid complement and attach by direct binding of factor H. J. Infect. Dis. 185:1786-1793. [DOI] [PubMed] [Google Scholar]
- 29.Metts, S., J. V. McDowell, M. Theisen, P. R. Hansen, and R. T. Marconi. 2003. Analysis of the OspE determinants involved in the binding of factor H and OspE targeting antibodies elicited during infection in mice. Infect. Immun. 71:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narikiyo, M., C. Tanabe, Y. Yamada, H. Igaki, Y. Tachimori, H. Kato, M. Muto, Montensano, H. Sakamoto, Y. Nakajima, and H. Sasaki. 2004. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 95:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Offenbacher, S., H. L. Jared, P. G. O'Reilly, S. R. Wells, G. E. Salvi, H. P. Lawrence, S. S. Socransky, and J. D. Beck. 1998. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann. Periodontol. 3:233-250. [DOI] [PubMed] [Google Scholar]
- 32.Pandiripally, V., E. Gregory, and D. Cue. 2002. Acquisition of regulators of complement activation by Streptococcus pyogenes serotype M1. Infect. Immun. 70:6206-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandiripally, V., L. Wei, C. Skerka, P. F. Zipfel, and D. Cue. 2003. Recruitment of complement factor H-like protein 1 promotes intracellular invasion by group A streptococci. Infect. Immun. 71:7119-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontology 24:153-192. [DOI] [PubMed] [Google Scholar]
- 35.Ram, S., D. P. McQuillen, S. Gulati, C. Elkins, M. K. Pangburn, and P. A. Rice. 1998. Binding of complement factor H to loop 5 of porin protein 1A: a molecular mechanism of serum resistance of nonsialyated Neisseria gonorrhoeae. J. Exp. Med. 188:671-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ram, S., A. K. Sharma, and S. D. Simpson. 1998. A novel sialic acid binding site on factor H mediates serum resistance of non-sialylated Neisseria gonorrhoeae. J. Exp. Med. 187:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, D., M. Caimano, J. McDowell, M. Theisen, A. Holm, E. Orff, D. Nelson, S. Wikel, J. Radolf, and R. Marconi. 2002. Environmental regulation and differential expression of members of the Bdr protein family of Borrelia burgdorferi. Infect. Immun. 70:7033-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruddy, S., and K. F. Austen. 1969. C3 inactivator of man. I. Hemolytic measurement by the inactivation of cell-bound C3. J. Immunol. 102:533-543. [PubMed] [Google Scholar]
- 39.Ruddy, S., and K. F. Austen. 1971. C3b inactivator of man. II. Fragments produced by C3b inactivator cleavage of cell-bound or fluid phase C3b. J. Immunol. 107:742-750. [PubMed] [Google Scholar]
- 40.Saglie, R., M. G. Newman, F. A. Carranza, and G. L. Pattison. 1982. Bacterial invasion of the gingiva in advanced periodontitis in humans. J. Periodontol. 53:217-222. [DOI] [PubMed] [Google Scholar]
- 41.Schenkein, H. A. 1991. The role of complement in periodontal diseases. Crit. Rev. Oral Biol. Med. 2:65-81. [DOI] [PubMed] [Google Scholar]
- 42.Simonson, L. G., C. H. Goodman, J. J. Bial, and H. E. Morton. 1988. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect. Immun. 56:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Socransky, S., A. Haffajee, M. Cugini, C. Smith, and R. J. Kent. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 44.Wallich, R., J. Pattathu, V. Kitiratschky, C. Brenner, P. F. Zipfel, V. Brade, M. M. Simon, and P. Kraiczy. 2005. Identification and functional characterization of complement regulator acquiring surface protein 1 of the Lyme disease spirochetes Borrelia afzelii and Borrelia garinii. Infect. Immun. 73:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida, A., M. Kawada, N. Suzuki, Y. Nakano, T. Oho, T. Saito, and Y. Yamashita. 2004. TaqMan real-time polymerase chain reaction assay for the correlation of Treponema denticola numbers with the severity of periodontal disease. Oral Microbiol. Immunol. 19:196-200. [DOI] [PubMed] [Google Scholar]
- 46.Zipfel, P. F., and C. Skerka. 1999. FHL-1/reconectin: a human complement and immune regulator with cell-adhesive function. Immunol. Today 20:135-141. [DOI] [PubMed] [Google Scholar]
- 47.Zipfel, P. F., C. Skerka, J. Hellwage, S. T. Jokiranta, S. Meri, V. Brade, P. Kraiczy, M. Noris, and G. Remuzzi. 2002. Structure-function studies of the complement system. Biochem. Soc. Trans. 30:971-978. [DOI] [PubMed] [Google Scholar]