Abstract
The P97 adhesin and P102 genes of Mycoplasma hyopneumoniae each have six paralogs in the genome. We tested whether these genes were expressed during infection. P102 is associated with the mycoplasma and with swine cilia. Further, most of the paralogs were transcribed in vivo in two gene transcriptional units.
Mycoplasma hyopneumoniae, the agent of enzootic pneumonia of pigs, is found throughout the world, including both third world and industrialized countries (8). In addition to the chronic disease it causes, recent evidence also suggests that M. hyopneumoniae contributes significantly to the pathogenesis of other infectious agents (10). The mechanism(s) it employs to cause disease is poorly understood, but it is thought that surface components are critically involved.
Mycoplasmas lack cell walls, and many vary their surface structure by a series of genetic events resulting in phase switching of surface lipoproteins and the presentation of a structural mosaic to the host immune system (11). Few studies have described cell surface molecules in M. hyopneumoniae (2, 5, 13), however, because the organism is genetically intractable, is a fastidious grower, and displays poor colony growth on agar surfaces. This has severely limited the experimental approaches available to study this pathogen. Genome sequencing has shown that few M. hyopneumoniae genes have the typical structure that results in phase switching (7).
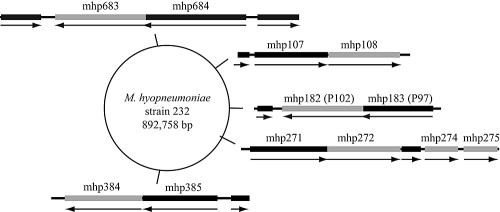
Mycoplasma hyopneumoniae binds exclusively to swine cilia, and the cilium adhesin, P97, is the most extensively characterized surface molecule (2, 3, 6, 12-14). P97 undergoes extensive posttranslational proteolytic processing, and yet the protein fragments remain associated with the mycoplasma surface (1). The gene for P97 is part of a two-gene genetic structure; the downstream or second gene codes for a 102-kDa protein designated P102 (4). The role for P97 has been clearly assigned to adherence, but the function of P102 remains unknown. Because of its close genetic linkage with P97, P102 was predicted to function in adherence either directly by interacting with host surface structures or indirectly through a supportive function for P97 activity. Recent studies have shown that both the P97 and P102 genes are members of paralogous families (1, 7), but little is known about their function or even if they are expressed. Figure 1 depicts the chromosomal location of the paralog members and their genetic organization and direction of transcription. Most of the paralogs appear to be gene fusions with nonhomologous sequences (7), which may affect their function and expression patterns. All of the P97 and P102 paralogs except for the products of mhp271, mhp385, mhp684, and mhp275 have recognizable signal peptide sequences. Further information regarding these gene sequences can be obtained at http://mycoplasma.genome.uab.edu/. While transcription and translation are tightly coupled in bacteria, there are no reports of transcription without the gene product being present in mycoplasmas. Thus, this study sought to detect mRNA transcripts of these two gene families during infection as the most direct indicator of expression for each of the paralogs.
FIG. 1.
Paralog gene structure and genome location. The gray bars indicate P102 gene paralogs, and the black bars are P97 gene paralogs. mhp182 is the P102 gene while mhp183 is the gene for P97. The arrows indicate direction of transcription. Not shown are mhp280 and mhp493.
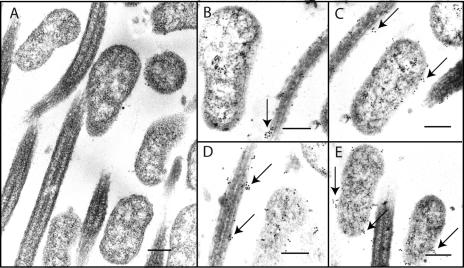
Expression of P102 in vivo. Although it is clear that P97 must be expressed in vivo to ensure colonization of the organism, it has not been determined if P102 was also expressed during disease. To examine P102 expression in vivo, immunogold labeling was performed on respiratory tissues from infected pigs. Mycoplasma-free pigs were inoculated intratracheally with M. hyopneumoniae strain 232, a derivative of strain 11, as described previously (10). At 10 or 12 days, pigs were sacrificed, tracheas were removed, and 1-cm blocks of tissue were fixed with 1% glutaraldehyde overnight, dehydrated in an acetone series, and embedded as described previously (1). Thin 80- to 90-nm sections were reacted with monospecific, polyclonal mouse anti-P102 serum raised against recombinant P102 followed by goat anti-mouse immunoglobulin G plus immunoglobulin M serum labeled with 10-nm gold particles (E. Y. Laboratories, Inc., San Mateo, Calif.) as described previously (1). Gold particles could be seen within mycoplasmas and attached to swine cilia, often in aggregates or in high concentrations (Fig. 2). Our electron microscopic data show that P102 is expressed in vivo and is secreted from the mycoplasma. Also, it appears that P102 is not directly associated with the cilium-binding R1 repeat region of P97, since that portion is found exclusively along the mycoplasma membrane (1, 12). Thus, if P102 plays an important role in virulence, it would not be through direct interaction with the cilium-binding region of P97.
FIG. 2.
Immunogold electron microscopy of mycoplasmas attached to swine cilia during infection. Infected trachea from pigs challenged with strain 232 were excised and epithelial regions prepared for electron microscopy as described. Regions containing ciliated epithelium were sectioned and stained with gold-labeled antibodies. (A) Control section stained with sera from a naïve, specific pathogen-free mouse and immunogold conjugate. (B to E) Sections stained with mouse anti-P102 antibodies followed by conjugate. Note the large numbers of gold particles (arrows). Bar, 0.5 μm.
Transcript analysis of the P97 and P102 gene paralogs.
The relationship between P97 and P102 and their paralogs was explored by determining if their genes are transcribed in vivo during disease and if gene pairs (Fig. 1) are transcribed as single mRNA molecules. To accomplish this, gene-specific reverse transcriptase PCRs (RT-PCRs8) were performed on RNA obtained from organisms isolated from M. hyopneumoniae-infected pigs. Mycoplasmas were isolated from bronchial alveolar lavage (BAL) fluids following challenge according to published protocols (10). Approximately 30 ml of BAL fluid was subjected to differential centrifugation at 4°C, first at 2,000 × g for 10 min, followed by 18,000 × g for 10 min. The final cell pellet was resuspended in 500 μl of RNAlater (Ambion, Austin, Tex.), and the cell suspension was stored at −70°C. Total RNA was isolated from BAL pellets by using an RNA isolation kit (Ambion) per the manufacturer's guidelines, including treatment with DNase I to remove contaminating genomic DNA. Control RNA from noninfected, negative control pigs was also obtained. The RNA integrity and quantity was analyzed by using an Agilent model 2100 Bioanalyzer. A first-strand cDNA synthesis reaction was performed by adding 50 to 100 ng of total RNA to 2 pmol of each gene-specific primer (Table 1) and 200 μM of deoxynucleotide triphosphates. The mixture was heated to 65°C for 5 min and then incubated on ice for 1 min. First-strand buffer, 2 mM dithiothreitol, 40 units of RNase inhibitor (Invitrogen, Carlsbad, Calif.), and 200 units of reverse transcriptase (Invitrogen) was then added to a total volume of 20 μl. The reaction mixture was incubated at 55°C for 50 min followed by 70°C for 15 min. All PCRs contained 2 U Taq DNA polymerase (New England Biolabs, Beverly, Mass.), 1 × buffer, 200 μM deoxynucleotide triphosphates, 2 mM magnesium chloride, 0.2 μM of each primer, and 2 μl of the first-strand cDNA reaction in a final volume of 50 μl. The PCR conditions were as follows: 1 cycle at 95°C for 5 min, followed by 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 30 s, with a final incubation step of 72°C for 10 min. Reaction products were analyzed on 2% agarose gels, and only fragments of the expected size were considered positive (Fig. 3). Negative control reactions for all RNA preparations included no reverse transcriptase, no primers, and no RNA (Fig. 3B, upper panel). Positive controls with 100 ng of M. hyopneumoniae strain 232 template DNA were performed with all primer pairs and are shown in Fig. 3A. To control for possible false products from swine lung RNAs in the BAL fluid preparations, RT-PCR controls were performed with total RNA isolated from whole-lung tissues harvested from mycoplasma-free control pigs.
TABLE 1.
Primers
| Primer | Function, open reading frame | Sequence (5′-3′) |
|---|---|---|
| For indicated P97 paralogs | ||
| mhp107 | ||
| 211-1020_U | Forward primer, mhp107 | GCGGATCCGATAATACTTATAATGCC |
| 211-1020_L | Reverse primer, mhp107 | ATGGATCCCTCTTTGGGAAATTGTTC |
| mhp271 | ||
| Mhp271-f | Forward primer, mhp271 | TTGGTTGGGTCTATTATTGG |
| Mhp271-r | Reverse primer, mhp271 | TAACTTTGCTAAATCATCGG |
| mhp280 | ||
| Mhp280-f | Forward primer, mhp280 | AAACAGCCGTGAAACTGC |
| Mhp280-r | Reverse primer, mhp280 | GCATTTTTGGATTCTTCTGG |
| mhp385 | ||
| 662-1020_U | Forward primer, mhp385 | ACGGATCCGATCAGCCACAGATTCAC |
| 662-1020_L | Reverse primer, mhp385 | ACGGATCCTAAAGCAAATAGTGATTCTGA |
| mhp493 | ||
| 545B-1020_U | Forward primer, mhp493B | ACGGATCCCAAAAATTAGCCACTTCC |
| 545B-1020_L | Reverse primer, mhp493B | ACGGATCCATCTATACTTCTTCAGTC |
| mhp684 | ||
| 347-1020_U | Forward primer, mhp682 | ACGGATCCAATCTTTTATCCGCTGGG |
| 347-1020_L | Reverse primer, mhp682 | ACGGATCCTGTTTGCTGATTTGAGGC |
| For indicated P102 paralogs | ||
| mhp275 | ||
| 033-1020_U | Forward primer, mhp275 | GCGGATCCATTTTTGGATGGAAACAAGAC |
| 033RT-r | Reverse primer, mhp275 | TGGGCTTACACCTTCTTTGGC |
| mhp272 | ||
| 036RT-f2 | Forward primer, mhp272 | GGCAAATGCTTTGATTTCC |
| 036RT-r2 | Reverse primer, mhp272 | GTTCTTTGCTGTGGACTTC |
| mhp108 | ||
| 210RT-f2 | Forward primer, mhp108 | GCAACAGCAAGTGGAACTGC |
| 210RT-r2 | Reverse primer, mhp108 | AAGTTGGTAAGCCGAGACTG |
| mhp683 | ||
| 348-1020_U | Forward primer, mhp683 | CGGGATCCCAAGCAAAAAATGAAAAAGAAG |
| 348RT-r | Reverse primer, mhp683 | TTTTACTTTAGCCGAAGAGGC |
| mhp384 | ||
| 663-1020_U | Forward primer, mhp384 | GCGGATCCACTAAATTATCACGAAGATCAC |
| 663RT-r | Reverse primer, mhp384 | TAGAACCATTGTAGCAGCCGG |
| For intergenic regions | ||
| P97-3′-f | Forward primer, mhp183 | GATTCAAAATCCGGTGATCC |
| P102-5′-r | Reverse primer, mhp182 | CTAGCTTGTTCTGTATTTCC |
| Mhp107-f | Forward primer, mhp107 | AAATGATGAAAATAGCCCG |
| Mhp108-r | Reverse primer, mhp108 | AGTTCCACTTGCTGTTGC |
| Mhp271-f | Forward primer, mhp271 | CCTGAAACTCCAAAAACAGAAG |
| Mhp272-r | Reverse primer, mhp272 | CAAAAGCCCGAACTAAGTCATAC |
| Mhp384-f | Forward primer, mhp384 | AGAGCATCAAAAGCGTTTAGTC |
| Mhp385-r | Reverse primer, mhp385 | TTCAGCAGCCAAGCCAGTAG |
| Mhp683-f | Forward primer, mhp683 | CCGCTTTTACTTCTTTATTAGCC |
| Mhp684-r | Reverse primer, mhp684 | GCCTCAAATCAGCAAACAAG |
| For controls | ||
| P97RT-f | Forward primer for control reaction, P97 | GGAAATTATGCCTATGAATTCG |
| P97RT-r | Reverse primer for control reaction, P97 | GTGCTCTGTTAGTTTCTAGTCC |
| P102RT-f2 | Forward primer for control reaction, pig cyclophilin | CCCGATGTGTTTTTAGATGG |
| P102RT-r2 | Reverse primer for control reaction, pig cyclophilin | CCTTTTCCTTTTCTATCGGC |
| cyclo-f | Forward primer for control reaction, pig cyclophilin | TAACCCCACCGTCTTCTT |
| cyclo-r | Reverse primer for control reaction, pig cyclophilin | TGCCATCCAACCACTCAG |
FIG. 3.
RT-PCR analysis of P97 and P102 paralog gene transcription. A. Open reading frame numbers are shown for each open reading frame examined. Left lanes indicate RNA templates from BAL samples (R); right lanes, genomic DNA template controls (D). P97, intergenic, and P102 RT-PCRs are indicated at the top of the panel. The distance between the open reading frames in base pairs (bp) of each transcriptional unit are also shown. B. Controls. (Upper panel) Lane 1, no reverse transcriptase; lane 2, no RNA; lane 3, no primers; lane 4, P97 primers with cDNA; lane 5, P97 primers with genomic DNA. (Lower panel) These products are from RT-PCRs using RNA isolated from uninfected porcine lung tissue. Lanes 1 to 11 represent RT-PCRs with specific primer pairs for open reading frames mhp275, mhp272, mhp108, mhp107, mhp271, mhp280, mhp684, mhp683, mhp493, mhp385, and mhp384, respectively. Lane 12, P97-P102 primers. Lane 13 represents the pig cyclophilin positive control reaction.
Total RNA from high-speed BAL pellets included pig RNA from host cell contamination as demonstrated by a positive cyclophilin RT-PCR (data not shown). Mycoplasma-specific primers, however, failed to react with purified pig RNA (Fig. 3B, upper panel, lanes 1 to 12), showing that pig RNA contamination would not interfere with the P97 and P102 RT-PCR analysis. Cyclophilin was used as a positive control target in pig RNA RT-PCRs to control for pig RNA quality (Fig. 3B, lower panel, lane 13). Transcripts for all of the P97 gene paralogs except for mhp280 could be demonstrated in total RNA from BAL fluid, while all of the P102 gene paralogs were transcribed (Fig. 3A). In lieu of paralog-specific antisera, this is the best evidence that these genes are expressed during disease.
Primers were also designed to bridge across the intergenic regions of the P97 and P102 gene paralogs to determine if the gene pairs produce single mRNA transcripts (Table 1). The positive reactions indicate that mhp183 (P97) and mhp182 (P102), mhp271 and mhp272, mhp384 and mhp385, and mhp683 and mhp684 are all transcribed with their partners as single mRNA molecules (Fig. 3A). We were unable to identify a transcript spanning mhp107 and mhp108, although they are separated by only 54 base pairs. This distance, however, was the largest for any of the transcripts tested (Fig. 3A) and may suggest that this distance is an upper limit for defining operon structure in M. hyopneumoniae. Two genes, mhp275 and mhp493, are single genes with no other partners. No intergenic region was tested for mhp280 because we failed to detect transcripts using gene-specific primers.
In summary, we have established by immunogold electron microscopy that P102 is expressed in vivo during disease. We also show that most of the P97 and P102 gene paralogs except for mhp280 are transcribed in vivo and, except for mhp107 and mhp108, are organized into single transcription units. Thus, these genes appear to be arranged in operons suggesting they have related functions. Cellular adhesins in other Mycoplasma species are sometimes found cotranscribed with genes for accessory proteins as is the case for Mycoplasma pneumoniae (9). Whether P102 has a function in relation to adherence has not yet been determined. Further study of the members of the P97 and P102 paralog gene families will be necessary to determine their importance to the disease process as well. At least now it is clear that they are expressed during disease.
Acknowledgments
We thank Eileen L. Thacker for assistance with obtaining lung tissues and BAL fluids from infected pigs. We also thank Jean Olsen for assistance with electron microscopy.
These studies were supported in part by funds from the Iowa Livestock Health Advisory Council and from the Healthy Livestock Initiative from the College of Veterinary Medicine, Iowa State University.
Editor: D. L. Burns
REFERENCES
- 1.Djordjevic, S. P., S. J. Cordell, M. A. Djordjevic, J. Wilton, and F. C. Minion. 2004. Proteolytic processing of the Mycoplasma hyopneumoniae cilium adhesin. Infect. Immun. 72:2791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu, T., S. Artiushin, and F. C. Minion. 1997. Cloning and functional analysis of the P97 swine cilium adhesin gene of Mycoplasma hyopneumoniae. J. Bacteriol. 179:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu, T., and F. C. Minion. 1998. Identification of the cilium binding epitope of the Mycoplasma hyopneumoniae P97 adhesin. Infect. Immun. 66: 4762-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu, T., and F. C. Minion. 1998. Molecular analysis of the P97 cilium adhesin operon of Mycoplasma hyopneumoniae. Gene 214:13-23. [DOI] [PubMed] [Google Scholar]
- 5.Kim, M. F., M. B. Heidari, S. J. Stull, M. A. McIntosh, and K. S. Wise. 1990. Identification and mapping of an immunogenic region of Mycoplasma hyopneumoniae p65 surface lipoprotein expressed in Escherichia coli from a cloned genomic fragment. Infect. Immun. 58:2637-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minion, F. C., C. Adams, and T. Hsu. 2000. R1 region of P97 mediates adherence of Mycoplasma hyopneumoniae to swine cilia. Infect. Immun. 68:3056-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minion, F. C., E. L. Lefkowitz, M. L. Madsen, B. J. Cleary, S. M. Swartzell, and G. G. Mahairas. 2004. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J. Bacteriol. 186: 7123-7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross, R. F. 1992. Mycoplasmal disease, p. 537-551. In A. D. Leman, B. E. Straw, W. L. Mengeling, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames.
- 9.Sperker, B., P. C. Hu, and R. Herrmann. 1991. Identification of gene products of the P1-operon of Mycoplasma pneumoniae. Mol. Microbiol. 5: 299-306. [DOI] [PubMed] [Google Scholar]
- 10.Thacker, E. L., P. G. Halbur, R. F. Ross, R. Thanawongnuwech, and B. J. Thacker. 1999. Mycoplasma hyopneumoniae potentiation of porcine reproductive and respiratory syndrome virus-induced pneumonia. J. Clin. Microbiol. 37:620-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yogev, D., G. F. Browning, and K. S. Wise. 2002. Genetic mechanisms of surface variation, p. 417-443. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 12.Zhang, Q., T. F. Young, and R. F. Ross. 1994. Glycolipid receptors for attachment of Mycoplasma hyopneumoniae to porcine respiratory ciliated cells. Infect. Immun. 62:4367-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, Q., T. F. Young, and R. F. Ross. 1995. Identification and characterization of a Mycoplasma hyopneumoniae adhesin. Infect. Immun. 63: 1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, Q., T. F. Young, and R. F. Ross. 1994. Microtiter plate adherence assay and receptor analogs for Mycoplasma hyopneumoniae. Infect. Immun. 62:1616-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]