Abstract
The merozoite surface antigen 2 (MSA-2) proteins of Babesia bovis are members of the variable merozoite surface antigen (VMSA) family that have been implicated in erythrocyte invasion and are important targets for antibody-mediated blocking of invasion. Extensive sequence variation in another VMSA member, MSA-1, has been shown in all vaccine breakthrough isolates. To test the hypothesis that the msa-2 genes of vaccine breakthrough isolates would also encode a diverse set of proteins, the complete msa-2 locus was characterized from 12 Australian B. bovis strains and isolates, including two vaccine strains and eight vaccine breakthrough isolates, and compared to the loci in previously and newly characterized American strains. In contrast to American strains, the msa-2 loci of all Australian strains and isolates examined contain, in addition to msa-2c, only a solitary gene (designated msa-2a/b) closely related to American strain msa-2a and msa-2b. Nevertheless, the proteins encoded by these genes are quite diverse both between and within geographic regions and harbor evidence of genetic exchange among other VMSA family members, including msa-1. Moreover, all but one of the Australian breakthrough isolate MSA-2a/b proteins is markedly different from the vaccine strain from which immune escape occurred, consistent with their role in strain-specific protective immunity. The densest distribution of polymorphisms occurs in a hypervariable region (HVR) within the carboxy third of the molecule that is highly proline rich. Variation in length and content of the HVR is primarily attributable to differences in the order and number of degenerate nucleotide repeats encoding three motifs of unknown function.
Proteins found on the cell surface of apicomplexan hemoparasitic merozoites, such as Babesia and Plasmodium species, are important for erythrocyte invasion and, consequently, host immunity. How these proteins function and the mechanisms utilized to avoid immune recognition are critical to developing strategies for vaccination. The variable merozoite surface antigen (VMSA) family of Babesia bovis, an important cause of bovine babesiosis, encodes immunodominant, glycosylphosphatidylinositol (GPI)-anchored, membrane surface proteins coexpressed in both merozoites and sporozoites (16, 17). The family is defined by conserved amino- and carboxy-terminal regions, encoding leader and anchor signals, respectively, and a characteristic central hydrophilic structure (6, 10, 26). Members of the family include merozoite surface antigen 1 (MSA-1), MSA-2, and the distantly related BabR (6). Unlike msa-1, which is present as a single-copy gene (26), the msa-2 locus of the biological clone Mo7 is organized into four tandemly arranged and expressed genes (two msa-2a genes, msa-2b, and msa-2c) (10). Because the proteins encoded by the two msa-2a genes and msa-2b have a high degree of identity to each other (78 to 90%) with distinctly conserved regions, a shared function is postulated. The potential role that the MSA-2a and -2b proteins have in erythrocyte invasion is illustrated by the ability of MSA-2a and -2b monospecific antisera to significantly inhibit sporozoite and merozoite invasion of erythrocytes (16), consistent with their coexpression on the merozoite surface.
Although attenuated vaccines generally provide protection against bovine babesiosis, disease outbreaks involving heterologous isolates occur (4, 5). These heterologous field isolates, termed vaccine breakthrough isolates, are genetically distinct from the vaccine strain used to immunize animals (13). However, the composite genetic and antigenic changes that result in vaccine breakthrough are not defined. LeRoith et al. recently showed that significant MSA-1 variation was present in every vaccine breakthrough isolate examined (12). Based on studies using American strains (defined by geographic origins), MSA-2 has been postulated to be less divergent despite apparent intragenic sites of genetic exchange (10). However, because of their familial relationship, we hypothesized that MSA-2 would demonstrate similar sequence divergence when examined in isolates of B. bovis able to escape vaccine-induced immunity. Further, this divergence was postulated to arise in part from genetic exchange among VMSA family members. To test these hypotheses, we characterized the complete msa-2 locus from 12 Australian strains and isolates, including two vaccine strains and eight vaccine breakthrough isolates, and compared the msa-2 genes to the two previously characterized loci (10) as well as one new American isolate. The results identify a unique structure of the msa-2 locus in Australian strains and isolates and confirm the hypothesis that significant sequence variation occurs in breakthrough isolates. While genetic exchange does appear to contribute to sequence diversity, unique degenerate nucleotide repeats encode much of the variation in a proline-rich hypervariable region (HVR) occupying the carboxy third of MSA-2a/MSA-2b proteins.
MATERIALS AND METHODS
Parasites and DNA.
The origins of all strains and isolates used, along with relevant references, are shown in Table 1. Strains are defined as well-characterized, passaged or cultured stocks of B. bovis and include the T and K vaccine strains and the T2Bo, L, and S strains. Isolates are those parasite stocks isolated during Australian B. bovis outbreaks. The biological clone Mo7, the T2Bo strain, and the L strain were maintained in microaerophilic stationary-phase blood cell cultures as described elsewhere (8). Genomic DNA was isolated by sodium dodecyl sulfate lysis-proteinase K digestion, followed by phenol-chloroform extractions and a standard ethanol precipitation (24); for the Australian S strain, DNA was isolated from stabilates by guanidine hydrochloride extraction utilized in the QIAamp DNA Blood Midi kit (QIAGEN, Valencia, CA).
TABLE 1.
Strains and isolates used in msa-2 locus and gene analyses
| Strain | Isolation yr | Origin | Comment | Reference(s) |
|---|---|---|---|---|
| American strains | ||||
| Mo7 | 1983 | Mexico | Biological clone obtained by limiting dilution of a field isolate | 23 |
| R1A | 1990 | Argentina | Vaccine strain attenuated by rapid passage in splenectomized calves | 2 |
| T2Bo | 1978 | South Texas | Virulent strain passaged 2× through splenectomized calves | 19 |
| Australian strains/isolates | ||||
| L | 1965 | New South Wales | Virulent strain isolated from field-infected ticks passaged 2× in splenectomized calves | 15 |
| S | 1969 | Queensland | Virulent field strain passaged 3× in splenectomized calves | 15 |
| K | 1975 | Queensland | Vaccine strain attenuated by rapid passage in splenectomized calves | 4, 5 |
| T | 1987 | Queensland | Vaccine strain attenuated by rapid passage in splenectomized calves | 4, 5 |
| F3, F28, F35, F40 | 1989-1990 | Queensland | Breakthrough field isolates from cattle vaccinated with K vaccine strain | 13 |
| G06, G36, G51, G52 | 1991-1993 | Queensland | Breakthrough field isolates from cattle vaccinated with T vaccine strain | 13 |
Probe synthesis and Southern blotting.
T2Bo strain and L strain msa-2a/ b-specific, amino-modified, digoxigenin-labeled, oligonucleotide probes (Integrated DNA Technologies, Coralville, IA) were designed from the leader sequence of msa-2b and msa-2a/b sequences, respectively (sequences are shown in Table 2). For blotting experiments, approximately 2 micrograms of genomic DNA was digested overnight with HindIII/EcoRI (L strain) or PstI (T2Bo strain); HindIII/EcoRI cuts just outside the L msa-2a/b gene, and PstI cuts upstream of msa-2a1 (the most 5′ msa-2a/b gene) and 176 bp from the 3′ end of msa-2b (the most 3′ msa-2a/b gene) in T2Bo. Digested DNA was electrophoresed on an 0.8% agarose gel and blotted overnight to positively charged nylon membranes (Roche Applied Science, Indianapolis, IN). The membranes were hybridized in DIG Easy Hyb (Roche Applied Science) with digoxigenin-labeled probes overnight at 42°C. The membranes were washed in 0.5% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate at 42°C for 30 min. Chemiluminescence detection of the probe was carried out as directed (Roche Applied Science).
TABLE 2.
Oligonucleotides used for the amplification of msa-2 loci and msa-2 genes and Southern blot experiments
| Primer | Application(s) | Sequence (5′-3′) |
|---|---|---|
| msa-2c-F | Locus PCR, PCR | ATGGTGTCTTTTAACATAATAAC |
| B42/44-R | PCR | AAAATGCAGAGAGAACG |
| B42/44-R-f | PCR | CTCTCTGCTACTTCGTTCTCTCTGC |
| msa-2-F1 | PCR | ATGATCGGGAAAATCTTC |
| msa-2-F1-r | PCR | CTGTGATTCCTCAGAAGCAGACAC |
| msa-2-F1-k | PCR | ATGATCGGCAAAATCTTT |
| aus-IR-F | PCR | GTGTAGATTTCTACCATTGCATTCGC |
| orf-b-F | Locus PCR, PCR | TATGGCGGGTTATGGATAACTCC |
| orf-b-F3 | Locus PCR | CTACTGGCAAATGACGATTTCGTG |
| orf-b-R | PCR | CGGGCGGTATTTTATGTATTTCCG |
| L-msa2-p | Southern blots | ATGATCGGCAAAATCTTC |
| T2Bo-msa2-p | Southern blots | ATGATCGGGAAAATCTTC |
Amplification and sequencing of msa-2 loci from B. bovis isolates and strains.
The msa-2 locus (extending from the start codon of msa-2c to orfB) of S, L, T, and T2Bo strains and the G series, F3, F35, and F40 isolates was amplified from genomic DNA employing temperature gradient PCR. The locus of T2Bo, S, L, T, F35, G36, and G51 was amplified with primer set msa-2c-F and orf-b-F, and the locus of G06, G52, F3, and F40 was amplified with primer set msa-2c-F and orf-b-F3. The final concentrations in reaction mixtures were 200 μM deoxynucleoside triphosphates, 2 mM MgSO4, 500 nM primers, 2% dimethyl sulfoxide, and 1 U Platinum Taq DNA High Fidelity polymerase (Invitrogen). Cycling, following a 5-min 95°C hot start, was carried out as follows: denaturation for 30 s at 95°C, annealing for 30 s ranging from 50°C to 60°C, and extension for 8 min at 68°C for 35 cycles, concluding with a 10-min extension at 72°C. Appropriately sized amplicons were cloned into pCR-4-TOPO. The T2Bo msa-2 amplicon, after agarose gel isolation, was cloned into pCR-XL-TOPO (Invitrogen, Carlsbad, CA). Positive clones were analyzed by PCR or restriction enzyme digestion.
The cloned loci, except for T2Bo, were sequenced by subcloning four overlapping PCR amplicons that spanned the original insert. The msa-2c and -2a/b genes were amplified from plasmids containing complete loci using the primer sets described below and cloned into pCR-4-TOPO. The intergenic region between msa-2c and -2a/b and the intergenic region between msa-2a/b and orfB were amplified from plasmid DNA using primer sets B42/44-R-f with msa-2-F1-r and B42/44-R-f with orf-b-F3, respectively. Miniprep DNA was sequenced using ABI chemistries and analyzed on an ABI3100 genetic analyzer (Applied Biosystems, Foster City, CA). The four subclone sequences were compiled into a single contiguous sequence using ContigExpress (Invitrogen). The T2Bo msa-2 locus sequence was derived from shotgun sequencing of a BAC clone (1K11) generated from a genomic DNA library of the T2Bo strain (www.vetmed.wsu.edu/research_vmp/babesia-bovis), followed by verification via subcloning and sequencing.
Amplification and sequencing of msa-2 genes from B. bovis isolates and strains.
msa-2a and -2b of the T2Bo strain and msa-2a/b of strains or isolates L, S, F28, F35, T (T-1 msa-2a/b), and G06 were amplified from genomic DNA using primer set msa-2-F1 and B42/44-R with Taq DNA polymerase (Invitrogen). msa-2a/b of the K strain was amplified from genomic DNA using primer set msa-2-F1-k and B42/44-R. msa-2c of the K, S, L, T, and T2Bo strains and the F and G isolates was amplified with primer set msa-2c-F and B42/44-R. Additional msa-2a/b genes were amplified from cloned msa-2 loci (see above). msa-2a/b of isolates G36 and F35 was amplified from locus clones with primer set msa-2-F1 and B42/44-R. An msa-2a/b gene from the T strain (sequence T-2 msa-2a/b) and msa-2a/b from isolates G06, G51, G52, F3, and F40 were amplified from locus clones with primer set aus-IR-F and B42/44-R (primer sequences shown in Table 2). Amplicons were cloned into pCR-4-TOPO (Invitrogen). Screening for positive clones, end sequencing, and analyses were done as described above. Nine additional T msa-2a/b clones—amplified with forward primer msa-1-F1, aus-IR-F, or msa-1-F1-k and reverse primer B42/44—were sequenced and nearly identical to either T-1 or T-2 msa-2a/b. Similarly, nine additional K msa-2a/b clones—amplified with forward primer msa-1-F1-k or aus-IR-F and reverse primer B42/44—were sequenced and nearly identical to K msa-2a/b.
The protein secondary structure of MSA-2a/b was analyzed by SSpro (www.igb.uci.edu/tools/scratch/) and nnPredict (www.cmpharm.ucsf.edu/∼nomi/nnpredict.html).
GenBank sequence accession numbers.
GenBank accession numbers of the previously published genomic DNA sequences from which protein sequences were derived are as follows: Mo7 msa-2 locus, AY052538; R1A msa-2a1, AY052539; R1A msa-2a2, AY052540; R1A msa-2b, AY052541; and R1A msa-2c, AY052542.
Nucleotide sequence accession numbers.
GenBank accession numbers of the new genomic DNA sequences from which protein sequences were derived are as follows: F28 msa-2a/b, DQ173946; F3 msa-2a/b, DQ173947; F35 msa-2a/b, DQ173948; F40 msa-2a/b, DQ173949; G06 msa-2a/b, DQ173950; G36 msa-2a/b, DQ173951; G51 msa-2a/b, DQ173952; G52 msa-2a/b, DQ173953; K vaccine strain msa-2a/b, DQ173954; L strain msa-2a/b, DQ173955; S strain msa-2a/b, DQ173956; T vaccine strain msa-2a/b T-1, DQ173957; T vaccine strain msa-2a/b T-2, DQ173958; T2Bo strain msa-2a1, DQ173959; T2Bo strain msa-2a2, DQ173960; T2Bo strain msa-2b, DQ173961; F28 msa-2c, DQ173962; F3 msa-2c, DQ173963; F35 msa-2c, DQ173964; F40 msa-2c, DQ173965; G06 msa-2c, DQ173966; G36 msa-2c, DQ173967; G51 msa-2c, DQ173968; G52 msa-2c, DQ173969; K vaccine strain msa-2c, DQ173970; L strain msa-2c, DQ173971; S strain msa-2c, DQ173972; T vaccine strain msa-2c, DQ173973; T2Bo strain msa-2c, DQ173974; F3 msa-2 locus, DQ173975; F35 msa-2 locus, DQ173976; F40 msa-2 locus, DQ173977; G06 msa-2 locus, DQ173978; G36 msa-2 locus, DQ173979; G51 msa-2 locus, DQ173980; G52 msa-2 locus, DQ173981; L strain msa-2 locus, DQ173982; S strain msa-2 locus, DQ173983; T vaccine strain msa-2 locus, DQ173984; T2Bo strain msa-2 locus, DQ173985.
RESULTS
Characterization of the msa-2 locus.
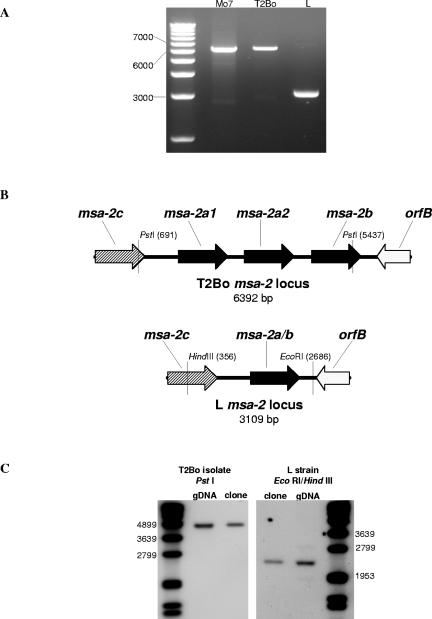
The msa-2 locus of the B. bovis biological clone Mo7 has four tandemly arranged msa-2 genes separated by identical intergenic regions (10). To determine whether this architecture is preserved across other American and Australian strains and isolates, amplification of the entire locus from T2Bo, a virulent strain from Texas, and L, a virulent Australian strain, was performed utilizing PCR (Fig. 1A). Products of approximately 6.3 kb were generated from Mo7 (the positive control) and T2Bo, which correlates well with the expected size based upon the published Mo7 sequence and the sequence of a BAC clone that contains the T2Bo msa-2 locus. Amplification of the L strain, in contrast, resulted in an amplicon of approximately 3.2 kb, a length which allows for only two msa-2 genes. To confirm the apparent reduction in msa-2 genes, long-distance PCRs were cloned, subcloned, and sequenced. The results are shown in a schematic of the loci in Fig. 1B. While T2Bo has a locus organization similar to that of Mo7, L has only one additional msa-2 gene that lies between msa-2c and orfB. This Australian L strain msa-2 gene is similar in length, position within the locus, and sequence to both msa-2a and -2b from previously characterized loci (10). Because of the evolutionary relationship between this gene and both msa-2a and -2b genes of Mo7 and R1A (10), we have designated this gene msa-2a/b. For the purposes of genetic comparisons in this study, we will refer to the group of all msa-2a, -2b, and -2a/b sequences as msa-2a/b genes and the predicted proteins that they encode as MSA-2a/b proteins.
FIG. 1.
The Australian virulent B. bovis L strain contains a single msa-2a/b gene. (A) Ethidium bromide-stained agarose gel of the msa-2 locus amplified from biological clone Mo7 (lane 2), Texas T2Bo strain (lane 3), and Australian virulent L strain (lane 4) with a 1.0-kb marker in lane 1. (B) Schematic of the msa-2 locus from T2Bo, based on the sequence of BAC clone 1K11 (see Materials and Methods), and L, based on the nucleotide sequence of the amplified msa-2 locus. (C) Southern blot of T2Bo genomic DNA and a T2Bo msa-2 locus clone digested with PstI and probed with digoxigenin-labeled T2Bo msa-2a/b-specific oligonucleotide (left panel) and L strain genomic DNA and L msa-2 locus clone digested with EcoRI/HindIII and probed with digoxigenin-labeled L msa-2a/b-specific oligonucleotide (right panel). Probe detection was by chemiluminescence. Size markers are in the far left and far right lanes, and the relevant markers are designated in base pairs to the left and right of the panels.
Amplifications over long distances have been reported to generate in vitro recombinants (27). To verify that the organization of the msa-2 loci had been maintained throughout the amplification process, Southern blots of T2Bo and L genomic and cloned DNA digested with PstI (T2Bo) or EcoRI/HindIII (L) and probed with msa-2a/b-specific oligonucleotides were examined (Fig. 1C). The digestions resulted in the predicted 4,700-bp fragment from T2Bo and 2,300-bp fragment from L genomic and cloned DNA. Based on these results, long-distance PCR appeared to faithfully amplify the msa-2 locus from genomic DNA.
The msa-2 loci from the Australian T vaccine strain, vaccine breakthrough isolates of the F and G series (F3, F35, F40, G06, G36, G51, and G52), and the virulent S strain were amplified from genomic DNA and produced, similarly to the L strain, a 3.2-kb amplicon. All loci have an organization identical to that of the L strain, with one msa-2c gene at the 5′ end of the locus and one additional msa-2a/b gene located 3′ of msa-2c.
Analysis of MSA-2a/b protein sequences.
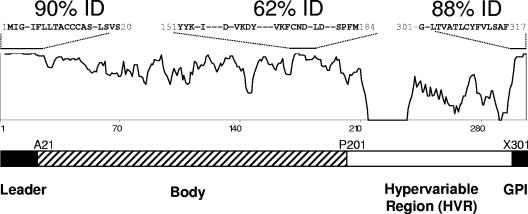
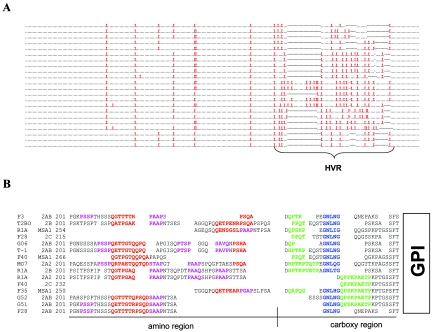
msa-2a/b genes were sequenced from the virulent Australian L and S strains, the Australian K and T vaccine strains, several Australian vaccine breakthrough isolates (F and G), and the T2Bo strain (Table 1). These sequences along with the msa-2a/b genes from the South American R1A vaccine strain and the Mo7 lab strain were aligned and analyzed. The MSA-2a/b molecule can be split into four different regions: the leader sequence (M1 to S20), the body (A21 to X200), the HVR (P201 to X300), and the GPI anchor signal sequence (X301 to F317) (Fig. 2). The leader and GPI anchor signal sequences, both of which are predicted to be absent from the surface-expressed protein, are the only highly conserved regions of the protein, with 90% and 88% overall identity among all sequences, respectively. The body of the molecule is less conserved but has several small islands of complete identity. Notable is a 34-residue block (Y151 to M184, 62% identity) that contains one of the most hydrophilic regions of the molecule (10). The block has three stretches of absolute conservation: YYK, VKFCND, and SPFM, as identified in previously examined American strains (10). Additional short regions of relative conservation are present in the amino half of the body and include a 16-residue stretch (T32 to T48) with 59% identity. However, the conservation of 129FNAFLNDNP137, found among all R1A and Mo7 MSA-2a/b sequences (10), is not maintained in the Australian strains and isolates. In contrast to the body of the molecule, the HVR is a highly divergent region with few conserved residues and is analyzed in depth below.
FIG. 2.
The MSA-2a/b proteins are diverse and have a hypervariable region in the carboxy half of the molecule. An alignment similarity plot of all examined MSA-2a/b proteins is shown. Regions of relative high identity are indicated by an overlying bar with sequence information and composite percent identity provided. Dashes in sequences indicate nonconserved residues. A schematic of MSA-2a/b molecular organization based on sequence variation and known function is shown below. Protein residues noted at the junction of sequence segments are initiating residues.
Comparison of MSA-2a/b molecules from vaccine strains to their respective breakthrough isolates.
To determine whether the level of diversity throughout the entire MSA-2a/b molecule is associated with vaccine breakthrough, the MSA-2a/b molecules of two Australian vaccine strains, T and K, were compared to four each of their respective breakthrough isolates. Sequence alignments of these comparisons are shown in Fig. 3A and B. The deduced amino acid sequences of all msa-2a/b T vaccine breakthrough isolates are different from the encoded sequence of T-1 msa-2a/b. Breakthrough MSA-2a/b amino acid identity to the T-1 sequence ranges from 61% to 85% (G06 and G52) with overall pairwise identities among all G outbreak isolates that range from 64% to 79%. Thus, the disparity between T-1 and each outbreak isolate is no greater than the disparity among all isolates. Polymorphisms among the sequences span the entire mature molecule with the highest number of changes in the HVR, sparing the leader and the GPI anchor signal sequences. The body of the molecule, in contrast, has several large regions with far fewer changes separated by a few small stretches of complete conservation. The longest of these conserved runs include an eight-residue hydrophilic stretch near the amino end of the molecule, 41ANYIKFLT48, and a 10-residue stretch, 169LVKFCNDELD178, near the carboxy end. A second T vaccine strain sequence, T-2 MSA-2a/b, was also found. The G outbreak isolates are comparably divergent to this vaccine sequence with the exception of G52, which has 99% identity to T-2.
FIG. 3.
Vaccine breakthrough isolate MSA-2a/b protein sequences differ from their associated vaccine strains. (A) MSA-2a/b sequence alignment of the Australian T vaccine strain compared to the vaccine breakthrough G isolates (G52, G51, G36, and G06). (B) MSA-2a/b sequence alignment of the Australian K vaccine strain compared to the vaccine breakthrough F isolates (F3, F28, F35, and F40). The underlined sequences indicate stretches of complete conservation. (C) MSA-2a/b amino-terminal region alignment of T vaccine strain T-1, vaccine breakthrough isolate G06, and Mo7 2a1. Boxes highlight regions of high identity between G06 and Mo7 2a1. Spaces denote identity, and dashes indicate gaps. Alignments were constructed by CLUSTALW.
Similar to the relationship between the T vaccine breakthrough isolates and the T vaccine strain, all K vaccine breakthrough isolate MSA-2a/b molecules are different from the K vaccine strain (Fig. 3B). As a group, pairwise disparity among F breakthrough isolates ranges from 67% to 100%. Compared to the vaccine strain, for which only one sequence was identified, F breakthrough identities range from 59% to 71% (F28 and F35/F40). The leader and GPI anchor signal sequences are well conserved with comparable changes throughout the body and HVR as described for the T vaccine breakthrough isolates. The only completely conserved sequence in the body of the molecule is a 10-residue stretch, 169LVKFCNDELD178, near the carboxy end.
Comparisons between vaccine strain and respective vaccine breakthrough isolate MSA-2a/b molecules also demonstrated defined stretches of polymorphism. Similar stretches of sequence can be found in other Australian and American strains. This is exemplified in Fig. 3C, which shows a multiple sequence alignment for T-1, G06, and Mo7. While G06 MSA-2a/b has high identity to T-1 MSA-2a/b (84.9%), two segments of G06 (denoted in boxes in Fig. 3C) have nearly complete identity to Mo7 MSA-2a1. Similar examples have been observed among the Mo7 and R1A MSA-2a/b molecules. As has been previously proposed, these regions appear to be sites of genetic exchange (10).
Analysis of the hypervariable region.
Comparison between vaccine strains and their respective breakthrough isolates indicates that the HVR contains the greatest amount of diversity in the molecule and that it varies in length. To determine the basis for this, we examined the amino acid and nucleotide sequences of the HVR from 22 MSA-2a/b proteins. The HVRs of MSA-2a/b proteins are extremely hydrophilic (data not shown) and are defined at the amino end by a completely conserved P201 and at the carboxy end by the predicted GPI anchor signal sequence cleavage site of SFT (Fig. 2). The length of this region varies widely from 34 (F28 MSA-2a/b) to 84 (Mo7 MSA-2a1) amino acids.
The HVR is rich in proline residues, especially in comparison to the remainder of the molecule (Fig. 4A). Sixteen percent of residues in the combined pool of HVR codons (443 codons total) are proline. They are irregularly spaced and have an intervening sequence length ranging from 0 to 13 residues. To understand why this part of the molecule is proline rich, manual alignment of representative HVRs, including the carboxy region of two msa-1- and two msa-2c-encoded proteins, was performed (Fig. 4B). These manual alignments identify three major, semiconserved, proline-containing motifs. The first motif of 11 residues (red sequence in Fig. 4B) has a consensus sequence of QGTTGTQ-[PQ]-SQD and is present at least once, in whole or in part, in all HVRs at a well-conserved position near the amino end of the HVR. The second motif of four residues (purple sequence in Fig. 4B) has a consensus sequence of PAAP and follows the first motif with a variable number of intervening amino acids. The third motif of nine residues (green sequence in Fig. 4B) has a consensus sequence of QPTKPAETP and is limited to the carboxy half of the HVR. It is present in whole or truncated form in all but one of the sequences and in all but one MSA-2a/b sequence is associated with a semiconserved nonrepeated segment with a consensus sequence of GNLNG (blue sequence in Fig. 4B).
FIG. 4.
The HVR of MSA-2a/b contains a series of three, proline-rich, repeating motifs. (A) Amino acid alignment of all 22 MSA-2a/b proteins examined. A period replaces all nonproline residues, a red “X” replaces all proline residues, and dashes indicate gaps in sequences. (B) Manual amino acid alignment of 10 representative MSA-2a/b HVRs along with the 3′ region upstream of the GPI anchor signal sequence of F35, F40, and R1A MSA-1 and F40 and F28 MSA-2c. Alignments are based on both amino acid and nucleotide sequences. Motifs are highlighted by color. Sequences of the first motif are red, sequences of the second motif are purple, sequences of the third motif are green, and the common GNLNG sequence is blue. Residues that are not assigned to a motif are in black letters. Blank spaces indicate gaps. A black bisected underline splits the HVR into amino and carboxy regions as labeled.
To determine the codon basis of the proline-containing motifs, representative nucleotide sequences of variable lengths from the 5′ half of the msa-2a/b HVRs were aligned. The alignment demonstrates that nearly all nucleotide sequences in the 5′ half of the HVRs can be mapped to the consensus sequence shown in Fig. 5A.
FIG. 5.
The amino-terminal region of the HVR is encoded by a series of degenerate nucleotide repeats. (A) Nucleotide sequence alignment of representative segments from the amino-terminal region (Fig. 4B) of HVRs. Base pair substitutions in reference to the consensus sequence are italicized; base pairs identical with the consensus sequence are red. The encoded amino acid sequences of these segments, with their relative positions in the 5′ region of the HVR, are aligned in the left half of the figure. The amino acid alignment is partitioned into three segments and numbered as indicated at the top of the alignment. “Md” indicates sequences derived from the middle portion of the amino half of the HVR region. Spaces denote gaps. (B) Sequences of the of representative HVRs with transformation of the amino acid sequence fragments of the 5′ region with the segment number to which their nucleotide sequences mapped.
The architecture of the HVR was delineated by dividing the full-length consensus sequence into three segments, labeled 1, 2, and 3. Amino acid sequence segments of the HVRs were then replaced with the segment number to which they mapped. The product of this transformation applied to representative MSA-2a/b HVRs (Fig. 5B) illustrates that this region is largely composed of a chain of degenerate repeats that varies in number and order. For example, the HVR of F3 MSA-2a/b is one of the shortest regions, partially attributable to the presence of only four degenerate repeat segments with the order 1, 2, 2, 3. In contrast, Mo7 MSA-2a1 has one of the longest HVRs, which is due to the presence of at least 10 degenerate repeat segments with the order 1, 2, 3, 2, 2, 3, 2, 1, 2, 3. The repeat region also contains a few residues that are not clearly assigned to any one repeat segment because of their short length.
Comparison of the MSA-2a/b HVR to other VMSA family members.
All VMSA family members share a well-conserved GPI anchor signal sequence. In contrast, this carboxy-terminal signal sequence is preceded by an HVR in MSA-2a/b that has very few overall similarities to other family members. There are several examples, however, where segments of the HVR have high identity to a corresponding sequence in other VMSA family members (Fig. 6).
FIG. 6.
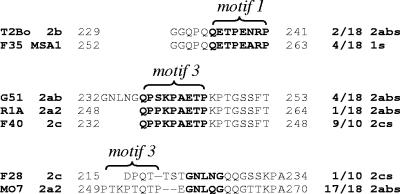
The HVR has sequence similarities to other VMSA family members. Three separate amino acid alignments are shown. Sequences shared among VMSA family members are boldfaced. The bracketed segments highlight motifs 1 and 3 as specified in the text (Fig. 4). Dashes indicate sequence gaps. The fractions listed at the right of the sequences are the frequency with which the representative sequence segments are present in the indicated VMSA family members that were examined.
One example is between T2Bo and Mo7 MSA-2b and the Australian MSA-1 proteins. The middles of the HVRs of these two proteins have a full-length repeated and degenerate first motif (Fig. 4B) that is most closely related to a similar sequence in a similar position in the carboxy region of four unique Australian MSA-1 proteins, of which F35 MSA-1 is representative. These sequence fragments, including five residues upstream, are 92% identical. Over that same stretch of sequence, the next closest sequence relation is to G51 MSA-2a/b (31% identity).
A second example is the sequence similarity and motif associations of R1A MSA-2a2 to MSA-2c. The third motif of R1A MSA-2a2, unlike all other MSA-2a/b proteins, is not associated with the GNLNG sequence (Fig. 4B). However, this arrangement of the third motif without the adjoining sequence is representative of nearly all MSA-2c proteins examined (9/ 10). Moreover, the sequence of R1A MSA-2a/b is identical over this stretch to F40 MSA-2c, including the eight remaining downstream residues of the HVR.
Lastly, the region just upstream of the GPI anchor cleavage site of F28 MSA-2c, unlike all other MSA-2c proteins, contains the GNLNG sequence present in the HVR of most MSA-2a/b proteins. This region, along with seven residues downstream, has 77% identity to Mo7 MSA-2a2 and shares four residues upstream with the truncated third motif in T2Bo MSA-2b (PPQT) (Fig. 4B). Collectively, these data suggest that genetic exchange may have occurred among all members of the VMSA family, including msa-2a/b, msa-2c, and msa-1 genes.
Analysis of MSA-2c.
Analysis of the deduced amino acid sequences of msa-2c from all Australian and American strains is consistent with previous findings that these proteins are more highly conserved than MSA-2a/b across the entire molecule (10, 28). Nearly all proteins are 265 residues, and they collectively have 75% identity, with pairwise comparisons ranging from 100 to 85% identity. In contrast to the highly polymorphic MSA-2a/b proteins, the vaccine breakthrough MSA-2c proteins have 92% to 100% identity to the MSA-2c of their respective vaccine strain. F28 MSA-2c, however, is an exception to this group. The protein is 254 residues long as a result of three short deleted segments, with pairwise comparisons ranging from 53 to 55% identity to MSA-2c of all other strains and isolates. Interestingly, however, F28 MSA-2c also is closely related to BabR 0.8 (50% overall identity), another VMSA family member not present in the msa-2 locus (10), with 73% identity to the first 180 amino acid residues. All other MSA-2c proteins are more distantly related to BabR, with overall identities that range between 39% and 40%.
DISCUSSION
B. bovis vaccine breakthrough isolates are genetically and antigenically distinct from vaccine strains (12, 13). These differences are, in part, due to the MSA-1 proteins, which in these breakthrough isolates have recently been shown to encode strain-specific epitopes not shared with vaccine strains (12). Comparison of MSA-2a/b proteins reveals that, like MSA-1, nearly all MSA-2a/b proteins from Australian B. bovis vaccine breakthrough isolates examined are significantly different from the MSA-2a/b proteins of the vaccine strains used to immunize these animals. These differences could reflect allelic prevalence in the field population working independently of immune selection. In addition, the occurrence of outbreaks in vaccinated herds is clearly a multifactorial event that may not always correlate with laboratory challenge (5). However, the observations that these genes encode exceptionally polymorphic, surface-expressed proteins and that monospecific sera directed toward them can block erythrocyte invasion (16) are all consistent with immunologic selection for change within the breakthrough isolate MSA-2a/b proteins. Many of the changes between the vaccine strain and the breakthrough isolates are in hydrophilic regions of the body and in the repeat region within the HVR, both of which are predicted to be highly antigenic (data not shown).
The only region of the predicted mature molecule that is semiconserved among vaccine strains and their breakthrough isolates, as well as in MSA-2a/b proteins of all other strains, is in the central region of the molecule containing the YYK motif. A similar motif (YFK), also in the central region of the molecule, is completely conserved in the MSA-1 proteins examined from Australia and America (12), suggesting that this part of the molecule in both MSA-1 and MSA-2a/b may have the same function. The tripeptide along with two semiconserved downstream residues, YYK-[KN]-[EH], is reminiscent of the binding motif (YY-[LIV]-[DN]-H) contained within WW modules, well-described motifs that bind proline-rich and phosphorylated targets (14). The regions containing these two motifs, intriguingly, are predicted to have similar secondary structures (data not shown).
The HVR is one of the most hydrophilic regions of the molecule and, interestingly, is proline rich as a result of three, frequently repeated, proline-containing, semiconserved motifs. HVR-specific monoclonal antibodies (11) bind to the surface of live merozoites (20), and monospecific antiserum directed against MSA-2a/b proteins blocks erythrocyte attachment and invasion (16). Taken together, these findings imply that the HVR may be involved with the initial interactions between merozoites and the erythrocyte surface during invasion. Several well-described Plasmodium surface proteins involved in cell invasion and the procyclins of Trypanosoma brucei also contain proline-rich regions (PRRs). Plasmodium vivax and Plasmodium knowlesi Duffy receptor proteins have central PRRs (9) that have recently been shown to contain the elements responsible for binding Duffy antigens (25), allowing invasion only in Duffy antigen-positive individuals. Duffy blood group antigens have also been described for cattle, and the ability of B. bovis to infect erythrocytes from different species of cattle positively correlates with the prevalence of Duffy antigen expression in different species (18). The central repeat region II of the circumsporozoite protein (CSP) of several Plasmodium spp. also constitutes a PRR (7, 22) that appears to function as a linker between two binding domains implicated in hepatocellular adhesion. The procyclins of T. brucei are GPI-anchored surface proteins expressed during the life cycle in the tsetse fly that, similar to the MSA-2a/b proteins, have PRRs abutting the GPI anchor signal sequence (29). Acosta-Serrano et al. postulate that the function of this region could be to protect the parasite during its passage through the midgut (1).
How and when MSA-2a/b proteins undergo variation are not clear. The amino acid changes within the body of the molecule range from numerous scattered substitutions and rare short deletions to large segments of sequence diversity that are shared in other MSA-2a/b proteins suggestive of genetic exchange. The most variable part of the molecule, though, is the HVR; it has no regions of complete conservation and contains large indels. Much of this length variation can be attributed to the presence of a series of variable numbers of degenerate repeats in the 5′ end of the HVR. This is reminiscent of the central region II of the Plasmodium CSP, which is composed of 25 to 40 short proline-containing, tandemly arranged, repeating motifs (3). Slipped-strand mispairing, occurring during clonal expansion, has been implicated in generating the differences in the number of repeats between different clones of Plasmodium falciparum and Plasmodium reichenowi (21). The types of changes generated by this method have yet to be observed in the B. bovis VMSAs. As these HVR polymorphisms likely lead to changes in surface epitopes, a clearly useful application of this method of diversification, as well as gene conversion, would be during clonal expansion within the mammalian host. Finely targeted unequal crossover events within the HVR can also lead to a series of repeats in this region. As the sexual stages of B. bovis occur in Boophilus microplus, variation in the HVR produced by this method would occur during passage of the parasite through the tick vector. Studies designed to determine in what part of the life cycle and how these changes arise, as well as how these changes may affect escape from immune recognition, are currently under way.
Initial characterization of the msa-2 locus in the biological clone Mo7 demonstrated that the locus contained three msa-2a/b genes (-2a1, -2a2, and -2b) and one msa-2c gene (10). While another American strain, T2Bo, similarly has four msa-2 genes in the locus, the msa-2 locus from 12 Australian strains and isolates has only one msa-2a/b and one msa-2c gene. How the presence of only a solitary msa-2a/b gene in the Australian loci affects the generation of sequence diversity, compared to multigene loci such as Mo7 and T2Bo, is unknown. Several mechanisms of gene diversification, such as gene conversion and homologous recombination, depend a priori on pools of similar genetic material. Because the Australian msa-2 loci have only one msa-2a/b gene, the pool of genetic material available for these mechanisms is limited in comparison to multigene loci. Nevertheless, the ability of these isolates to persist in the mammalian host and in the population, coupled with their postulated role in host immune system evasion, indicates that mechanisms of generating diversity in these loci provide sufficient variability. While this could be accomplished by homologous recombination or gene conversion among msa-2a/b genes from different isolates, occurring in the tick vector and/or mammalian host, sequence similarities among MSA-2a/b, MSA-2c, and MSA-1 suggest that additional sources of genetic material available for genetic exchange may include msa-1 and msa-2c.
In summary, the msa-2 locus of all Australian B. bovis strains and isolates examined, unlike the American strains, contains only two msa-2 genes: one msa-2c and one msa-2a/b gene. Analysis of 22 msa-2a/b genes from 16 different American and Australian B. bovis strains and isolates indicates that these genes encode a diverse set of proteins. As hypothesized, the encoded MSA-2 proteins of Australian vaccine breakthrough strains are different from the MSA-2 proteins of their vaccine counterpart, suggesting that these proteins may be involved in strain-specific protective immunity. The greatest diversity among MSA-2a/b proteins is in a hypervariable region, upstream of the GPI anchor signal sequence, which arises from variable numbers of proline-rich, degenerate repeats, similar to the repeat regions of Plasmodium CSP and T. brucei procyclins. Whether changes in the HVR arise only during the sexual stages, occurring in the tick vector, or also during passage through the mammalian host is under investigation.
Acknowledgments
The skillful technical assistance of Bev Hunter and Deb Alperin is acknowledged. David Herndon and Jeanne Howell are gratefully acknowledged for provision of the 1K11 BAC clone and sequencing of the T2Bo msa-2 locus in this clone. We thank Carlos Suarez for valuable technical input and Guy Palmer and Wendy Brown for critical discussions and review of the manuscript.
This work was supported by NIH K08 Award 1 K08 AI060630-01 and USDA SCA58-5348-2-683.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Acosta-Serrano, A., E. Vassella, M. Liniger, C. Kunz Renggli, R. Brun, I. Roditi, and P. T. Englund. 2001. The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc. Natl. Acad. Sci. USA 98:1513-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anziani, O. S., A. A. Guglielmone, A. A. Abdala, D. H. Aguirre, and A. J. Mangold. 1993. Acquired protection in Frisian steers with Babesia bovis attenuated vaccine. Rev. Med. Vet. 74:47-49. [Google Scholar]
- 3.Ayala, F. J., A. A. Escalante, and S. M. Rich. 1999. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parasitologia 41:55-68. [PubMed] [Google Scholar]
- 4.Bock, R. E., A. J. de Vos, T. G. Kingston, I. A. Shiels, and R. J. Dalgliesh. 1992. Investigations of breakdowns in protection provided by living Babesia bovis vaccine. Vet. Parasitol. 43:45-56. [DOI] [PubMed] [Google Scholar]
- 5.Bock, R. E., A. J. de Vos, A. Lew, T. G. Kingston, and I. R. Fraser. 1995. Studies on failure of T strain live Babesia bovis vaccine. Aust. Vet. J. 72:296-300. [DOI] [PubMed] [Google Scholar]
- 6.Cowman, A. F., O. Bernard, N. Stewart, and D. J. Kemp. 1984. Genes of the protozoan parasite Babesia bovis that rearrange to produce RNA species with different sequences. Cell 37:653-660. [DOI] [PubMed] [Google Scholar]
- 7.Eichinger, D. J., D. E. Arnot, J. P. Tam, V. Nussenzweig, and V. Enea. 1986. Circumsporozoite protein of Plasmodium berghei: gene cloning and identification of the immunodominant epitopes. Mol. Cell. Biol. 6:3965-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erp, E. E., S. M. Gravely, R. D. Smith, M. Ristic, B. M. Osorno, and C. A. Carson. 1978. Growth of Babesia bovis in bovine erythrocyte cultures. Am. J. Trop. Med. Hyg. 27:1061-1064. [DOI] [PubMed] [Google Scholar]
- 9.Fang, X. D., D. C. Kaslow, J. H. Adams, and L. H. Miller. 1991. Cloning of the Plasmodium vivax Duffy receptor. Mol. Biochem. Parasitol. 44:125-132. [DOI] [PubMed] [Google Scholar]
- 10.Florin-Christensen, M., C. E. Suarez, S. A. Hines, G. H. Palmer, W. C. Brown, and T. F. McElwain. 2002. The Babesia bovis merozoite surface antigen 2 locus contains four tandemly arranged and expressed genes encoding immunologically distinct proteins. Infect. Immun. 70:3566-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jasmer, D. P., D. W. Reduker, S. A. Hines, L. E. Perryman, and T. C. McGuire. 1992. Surface epitope localization and gene structure of a Babesia bovis 44-kilodalton variable merozoite surface antigen. Mol. Biochem. Parasitol. 55:75-83. [DOI] [PubMed] [Google Scholar]
- 12.LeRoith, T., K. A. Brayton, J. B. Molloy, R. E. Bock, S. A. Hines, A. E. Lew, and T. F. McElwain. 2005. Sequence variation and immunologic cross-reactivity among Babesia bovis merozoite surface antigen 1 proteins from vaccine strains and vaccine breakthrough isolates. Infect. Immun. 73:5388-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lew, A. E., R. E. Bock, J. M. Croft, C. M. Minchin, T. G. Kingston, and R. J. Dalgliesh. 1997. Genotypic diversity in field isolates of Babesia bovis from cattle with babesiosis after vaccination. Aust. Vet. J. 75:575-578. [DOI] [PubMed] [Google Scholar]
- 14.Macias, M. J., S. Wiesner, and M. Sudol. 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 513:30-37. [DOI] [PubMed] [Google Scholar]
- 15.Mahoney, D. F., and I. G. Wright. 1976. Babesia argentina: immunization of cattle with a killed antigen against infection with a heterologous strain. Vet. Parasitol. 2:273-282. [Google Scholar]
- 16.Mosqueda, J., T. F. McElwain, and G. H. Palmer. 2002. Babesia bovis merozoite surface antigen 2 proteins are expressed on the merozoite and sporozoite surface, and specific antibodies inhibit attachment and invasion of erythrocytes. Infect. Immun. 70:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosqueda, J., T. F. McElwain, D. Stiller, and G. H. Palmer. 2002. Babesia bovis merozoite surface antigen 1 and rhoptry-associated protein 1 are expressed in sporozoites, and specific antibodies inhibit sporozoite attachment to erythrocytes. Infect. Immun. 70:1599-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamoto, W., M. A. Marteline, P. E. Machado, E. M. Rubio-Colauto, F. A. Schimidt, S. M. Tsai, and D. H. Moon. 1998. Duffy blood group antigens (Fy(a) and Fy(b)) on cattle erythrocytes. Ann. N. Y. Acad. Sci. 849:490-493. [DOI] [PubMed] [Google Scholar]
- 19.Palmer, G. H., T. F. McElwain, L. E. Perryman, W. C. Davis, D. R. Reduker, D. P. Jasmer, V. Shkap, E. Pipano, W. L. Goff, and T. C. McGuire. 1991. Strain variation of Babesia bovis merozoite surface-exposed epitopes. Infect. Immun. 59:3340-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reduker, D. W., D. P. Jasmer, W. L. Goff, L. E. Perryman, W. C. Davis, and T. C. McGuire. 1989. A recombinant surface protein of Babesia bovis elicits bovine antibodies that react with live merozoites. Mol. Biochem. Parasitol. 35:239-247. [DOI] [PubMed] [Google Scholar]
- 21.Rich, S. M., and F. J. Ayala. 2000. Population structure and recent evolution of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 97:6994-7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rich, S. M., M. U. Ferreira, and F. J. Ayala. 2000. The origin of antigenic diversity in Plasmodium falciparum. Parasitol. Today 16:390-396. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez, S. D., G. M. Buening, T. J. Green, and C. A. Carson. 1983. Cloning of Babesia bovis by in vitro cultivation. Infect. Immun. 42:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Singh, S. K., A. P. Singh, S. Pandey, S. S. Yazdani, C. E. Chitnis, and A. Sharma. 2003. Definition of structural elements in Plasmodium vivax and P. knowlesi Duffy-binding domains necessary for erythrocyte invasion. Biochem. J. 374:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suarez, C. E., M. Florin-Christensen, S. A. Hines, G. H. Palmer, W. C. Brown, and T. F. McElwain. 2000. Characterization of allelic variation in the Babesia bovis merozoite surface antigen 1 (MSA-1) locus and identification of a cross-reactive inhibition-sensitive MSA-1 epitope. Infect. Immun. 68:6865-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanabe, K., N. Sakihama, A. Farnert, I. Rooth, A. Bjorkman, D. Walliker, and L. Ranford-Cartwright. 2002. In vitro recombination during PCR of Plasmodium falciparum DNA: a potential pitfall in molecular population genetic analysis. Mol. Biochem. Parasitol. 122:211-216. [DOI] [PubMed] [Google Scholar]
- 28.Wilkowsky, S. E., M. Farber, I. Echaide, S. Torioni de Echaide, P. I. Zamorano, M. Dominguez, C. E. Suarez, and M. Florin-Christensen. 2003. Babesia bovis merozoite surface protein-2c (MSA-2c) contains highly immunogenic, conserved B-cell epitopes that elicit neutralization-sensitive antibodies in cattle. Mol. Biochem. Parasitol. 127:133-141. [DOI] [PubMed] [Google Scholar]
- 29.Ziegelbauer, K., B. Stahl, M. Karas, Y. D. Stierhof, and P. Overath. 1993. Proteolytic release of cell surface proteins during differentiation of Trypanosoma brucei. Biochemistry 32:3737-3742. [DOI] [PubMed] [Google Scholar]