Abstract
We previously identified Candida albicans Not5p as an immunogenic protein expressed during oropharyngeal candidiasis (OPC). In this study, we demonstrate that C. albicans NOT5 reverses the growth defects of a Saccharomyces cerevisiae not5 mutant strain at 37°C, suggesting that the genes share at least some functional equivalence. We implicate C. albicans NOT5 in the pathogenesis of disseminated candidiasis (DC) induced by intravenous infection among neutropenic and nonimmunosuppressed mice, as well as in that of OPC in mice immunosuppressed with corticosteroids. We find no role in virulence, however, among neutropenic and corticosteroid-suppressed mice with DC resulting from gastrointestinal translocation, nor do we implicate the gene in vulvovaginal candidiasis among mice in pseudoestrus. These findings suggest that the role of NOT5 in virulence depends on the specific in vivo environment and is influenced by diverse factors such as tissue site, portal of entry, and the status of host defenses. NOT5 is necessary for normal adherence to colonic and cervical epithelial cells in vitro, demonstrating that such assays cannot fully replicate disease processes in vivo. Lastly, antibody responses against Not5p do not differ in the sera of patients with OPC, patients with DC, and healthy controls, suggesting that the protein is associated with both commensalism and the pathogenesis of disease.
Candida albicans is a versatile opportunistic pathogen that causes a variety of infections, ranging from harmless colonization of mucosal surfaces to disease states associated with tissue invasion and destruction such as oropharyngeal, vulvovaginal, and disseminated candidiasis (OPC, VVC, and DC, respectively). The pathogenesis of candidal diseases is facilitated by a number of virulence factors involved in functions such as adherence to host cells, secretion of hydrolytic enzymes such as proteases and phospholipase, sequestration of iron, survival within phagocytes, yeast-to-hypha morphogenesis, and phenotypic switching (2). It is clear that no single virulence factor is dominant. Rather, pathogenesis depends on the coordinated expression of multiple genes in a manner most appropriate for the conditions of the local environment (19). Since these conditions differ greatly at different sites of infection, it is likely that selected virulence-associated genes are important during specific types of candidiasis but not others. This hypothesis, however, has not been extensively tested. Furthermore, the extent to which so-called virulence factors contribute to routine colonization as well as to disease processes is not clear.
We recently used pooled sera from human immunodeficiency virus-infected patients with active OPC to screen a C. albicans genomic DNA expression library, thereby identifying genes encoding in vivo-expressed proteins (13, 21). Among the genes identified was C. albicans IPF16198, which we named NOT5 based on sequence homology to a gene in Saccharomyces cerevisiae (3). S. cerevisiae NOT5 encodes a member of the CCR-NOT complex, which is a global regulator of transcription that influences diverse processes, including cell wall integrity, carbon catabolite repression, and filamentation (7, 8, 18, 22). Given the functions of S. cerevisiae NOT5, we hypothesized that its C. albicans homologue would be important in morphogenesis and virulence. To test this, we used the ura-blaster method to disrupt the C-terminal domains of both C. albicans NOT5 alleles and reintroduced the URA3 selection marker to its native locus (4). We demonstrated that disruption of NOT5 attenuated hyphal formation in vitro, adherence to human buccal epithelial cells (BECs), and mortality during murine DC resulting from intravenous (i.v.) inoculation (DC-IV) (4).
The goal of the present project was to more completely characterize NOT5 and its contributions to disease processes. In our previous studies, we could not exclude the possibility that the expression of an N-terminal Not5p-HisG fusion protein might have confounded our findings. In this study, we first created a new null mutant strain in which the complete NOT5 open reading frame is disrupted and URA3 reintroduced to its native locus. We used this strain to further characterize the contribution of NOT5 to the pathogenesis of diverse diseases, including DC-IV, DC resulting from gastrointestinal (GI) translocation (DC-GI), OPC, and VVC. We also evaluated the antibody responses against Not5p among individual patients with OPC and DC, as well as in healthy volunteers. This addresses a potential shortcoming of our original screening strategy, which did not distinguish between genes expressed exclusively during OPC and those expressed more generally during other disease states and routine colonization.
MATERIALS AND METHODS
Strains and growth condition.
C. albicans strains used or constructed in this study are described in Table 1. All strains were routinely grown in yeast extract-peptone-dextrose (YPD) medium (1% yeast extract, 1% Bacto peptone, 2% α-d-glucose) at 30°C unless otherwise noted. S. cerevesiae not5 deletion strains (purchased from the American Type Culture Collection [ATCC]) were grown in YPD containing G418 at a concentration of 200 μg/ml.
TABLE 1.
Candida albicans and Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| SC5314 | Candida albicans parental strain | 12 |
| CAI4 | Δura3::imm434/Δura3::imm434 | 11 |
| CAI-12 | Δura3::imm434/URA3 | 11 |
| Heterozygous mutant | Δnot5::hisG/NOT5 URA3/Δura3::imm434 | This study |
| not5-null mutant | Δnot5::hisG/Δnot5::hisG URA3/Δura3::imm434 | This study |
| NOT5 revertant | Δnot5::hisG/NOT5::URA3-hisG Δura3::imm434/Δ ura3::imm434 | This study |
| S288C | Wild-type S. cerevisiae | ATCC |
| 35491 | S. cerevisiae not5 homozygous diploid mutant | ATCC |
| 35491::C. albicans NOT5 | 35491 (yPB1-ADHp-C. albicans NOT5) | This study |
Complementation study.
Genomic DNA of C. albicans SC5314 was used as a template for PCR of NOT5 and flanking regions (corresponding to nucleotides −31 to +716 relative to the ATG and TGA, respectively, in the NOT5 sequence). The oligonucleotide primers used were NOT5Exp-F (5′-CGTACAGGATCCCAACATCGAAAAGAAGTGTCT-3′) and NOT5Exp-R (5′-CGTACAGGATCCCCATTCCCTCTTCAAATC-3′) (the introduced BamHI restriction sites are underlined). The PCR product was digested with BamHI and inserted into the BglII site of the C. albicans expression vector pYPB1-ADHpt; this vector contains the C. albicans ADH1 promoter and terminator regions (1), an autonomously replicating sequence, C. albicans URA3 as a selectable marker, and S. cerevisiae 2μm sequences (kindly provided by Malcolm Whiteway). The insert was confirmed by sequencing. The expression plasmid was then transformed into an S. cerevisiae not5-null mutant (strain 35491) by electroporation (Table 1). Ura+ transformants were selected from Sabouraud dextrose agar (SDA) plates lacking uridine. Both plasmid yPB1-ADHp alone and plasmid yPB1-ADHp containing C. albicans NOT5 in the reverse orientation were used as controls. Expression of C. albicans NOT5 was confirmed for the complemented strains and excluded for the control strains by reverse transcription-PCR. The complemented S. cerevisiae strain was then tested for growth in YPD at 37°C.
Construction of NOT5 mutants.
Disruptions of both alleles of C. albicans NOT5 were performed using the ura-blaster method (11). The proximal fragment at nucleotide −600 bp to −203 bp relative to ATG of NOT5 was amplified by PCR using primers NOT5-N-F1F (5′-GGAAGGCGACTGCAGGGAAAACAGAGTGTTTGAGT-3′) and NOT5-N-F1R (5′-GGCAAGAAGTCGACCACGACCCAAGAAGTCTATA-3′); the restriction sites PstI and SalI were introduced into NOT5-N-F1F and NOT5-N-F1R, respectively (underlined). The distal gene fragment at position +1681 bp to +1918 bp relative to the ATG of NOT5 was amplified using primers CA29F2-FOR (5′-AACCTCAGATGTTCGAAATACAATGCAGCC-3′) and CA29F2-REV (5′-TTCACCGAGCTCTCTGTCTGTTTTCAGTAG-3′), which contained the introduced BglII and SacI restriction sites (underlined). Following amplification, the fragments were digested with the appropriate restriction enzymes and ligated sequentially into plasmid pMB-7, flanking the hisG-URA3-hisG disruption cassette. The resulting plasmid was digested with PstI and SacI and transformed into C. albicans strain CAI4 by electroporation. Ura+ transformants were selected on SDA plates lacking uridine. After confirmation of disruption by Southern blot analysis, a Ura+ transformant was screened for segregants on 5-fluoroorotic acid plates. This ura− strain was transformed with the same disruption cassette to disrupt the second copy of NOT5. The ura− strains were obtained following 5-fluoroorotic acid selection and confirmed by Southern blotting. In the final step, URA3 was reintroduced in its original locus by transforming a 4.8-kb BglII-PstI fragment isolated from plasmid pUR3 (kindly provided by William Fonzi) into the ura− strains by electroporation (23). The success of transformation was confirmed using Southern blot analysis.
To reinsert one copy of Not5 at its own locus, NOT5 was amplified from nucleotide position 618 before the start codon to nucleotide position 693 after the stop codon by PCR using primers NOT5-Reinsert-For (5′-GGAAGGCGACTGCAGGACAATTAGTTTGGTGTTGG-3′ [PstI site underlined]) and NOT5-Reinsert-Rev (5′-GGCAAGAAGTCGACCCCTCTTCAAATCTAGAAGC-3′ [SalI site underlined]). This fragment was then subcloned into pMB7-1 following PstI and SalI digestion; pMB7-1 is a modified pMB7, in which one copy of hisG was eliminated by digestion with XbaI followed by religation (3). The reinsertion cassette was released by digestion with PstI and KpnI and used to transform a ura− not5-null mutant. The success of the reinsertion was confirmed by Southern blot analysis.
Phenotypic observations.
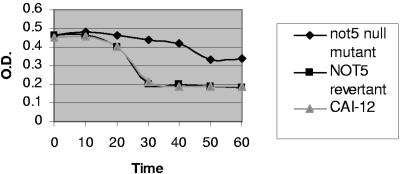
For sensitivity to cell wall agents, portions (10 μl) of serial 10-fold dilutions of an overnight growth culture were spotted onto YPD agar containing 20 μg/ml calcofluor white or 0.04% sodium dodecyl sulfate (SDS); plates were incubated at 37°C for 24 h. For the Zymolyase assay, either exponentially grown C. albicans cells or stationary-phase cells at an optical density at 599 nm (OD599) of ∼0.8 were incubated with 100 μg/ml of Zymolyase 100T in 10 ml of Tris-HCl, pH 7.5, in a shaking incubator at 35°C. An aliquot of cells was removed at timed intervals, and the OD599 was measured. The OD599 was plotted against the time of incubation.
Adherence assays. (i) Adherence to the colon adenocarcinoma cell line HT-29 and the cervical epithelial cell line HeLa.
HT-29 and HeLa cells (purchased from ATCC) were grown to confluence in a 12-well plate in ATCC complete growth medium (McCoy's 5a medium with 1.5 mM l-glutamine supplemented with 10% fetal bovine serum). After a wash in phosphate-buffered saline (PBS), cells were incubated with 1 × 104 CFU of C. albicans strains in the same medium at 37°C with 5% CO2 for 90 min. The wells were washed three times with PBS to remove nonadherent cells. SDA was then added to each well. The plates were incubated at 37°C for 24 h, at which time a colony count was performed. The adherence of the wild-type strain was defined as 100%. These assays were performed in duplicate, and experiments were repeated on at least three different occasions.
(ii) Adherence to endothelial cells.
Adherence to HUV-EC-C cells (purchased from ATCC) (25) was performed as for HT-29 and HeLa cells above, except that cells were grown to confluence in a six-well plate, and the inoculum used to infect cells in each well was 100 CFU. Hanks' balanced salt solution was used to wash off nonadherent cells.
Virulence determination. (i) DC-IV.
Seven-week-old male ICR mice (Harlan Sprague) were inoculated by i.v. injection of the lateral tail vein with C. albicans strains in 0.2 ml of normal saline solution. Mice were followed until they were moribund, at which point they were sacrificed, or for 30 days. For histopathological study, the kidneys from sacrificed mice were fixed with formalin and embedded in paraffin, after which thin sections were prepared and stained with Gomori methamine silver (GMS) stain. To determine tissue fungal burden, the kidneys, liver, and spleen were removed, homogenized, and quantitatively cultured on SDA containing 60 μg amikacin/ml. To induce neutropenia, the mice were given intraperitoneal injections of cyclophosphamide at 0.1 mg/g of body weight on days 4 and 1 before, and days 3 and 7 after, infection with C. albicans. Mice were given ciprofloxacin in their drinking water (250 mg/liter), starting a day before infection.
(ii) OPC.
For OPC (13), 7-week-old male ICR mice (Harlan Sprague) were immunosuppressed with 4 mg of cortisone acetate (Sigma Aldrich) in saline with 0.05% Tween 80 administered subcutaneously on the day before inoculation and 1 and 3 days after inoculation. Mice received tetracycline hydrochloride in their drinking water (0.5 mg/ml), starting the day before inoculation. For inoculation, mice were anesthetized by intraperitoneal injections with 3 mg of pentobarbital sodium solution (Abbott Laboratories, North Chicago, IL), and cotton wool balls (diameter, 3 mm) containing 108 CFU of C. albicans were placed sublingually in the oral cavity for 2 h. The mice were sacrificed at day 7 postinfection, and the mandibular soft tissue, including the tongue, was dissected free of the teeth and bone. The excised tissue was used for either tissue burden or histopathological evaluation.
(iii) VVC.
For VVC (26), CBA/J (H-2k) mice were given subcutaneous injections of 0.02 mg of estradiol valerate in 100 μl of sesame oil 3 days prior to and 4 days after intravaginal inoculation with 20 μl of PBS containing 5 × 104 CFU of the C. albicans strains. Vaginal lavages were performed on days 4 and 7 postinfection by applying 100 μl PBS intravaginally followed by constant aspiration for 30 s. The lavage fluid was serially diluted and plated for tissue burden enumeration.
(iv) DC-GI.
Outbred CFW (Swiss Webster) BR mice obtained from Charles River Farms (Wilmington, MA) were used to establish a breeding colony, and the offspring of these animals were used in this experiment. Six-day-old infant mice weighing 3 to 4 g were inoculated with 2 × 108 CFU by the oral-intragastric route (5, 6, 20). All mice were found to carry C. albicans in their fecal pellets at 9 days postinfection and were therefore selected for further study (5, 6, 20). After this point, mice were immunocompromised with intraperitoneal cyclophosphamide (0.2 mg/g body weight) and cortisone acetate (1.25 mg) on days 11 and 14 post-oral challenge. They were sacrificed at day 20 postinfection, and tissue burdens of C. albicans as well as histopathology of the stomachs, livers, kidneys, and spleen were assessed.
PMN phagocytosis.
Polymorphonuclear cells (PMNs) were isolated from heparinized blood of healthy volunteers by dextran sedimentation, followed by centrifugation through Ficoll-Hypaque. After removal of contaminating erythrocytes by hypotonic lysis, the PMNs were resuspended in RPMI 1640. Phagocytosis by PMNs was assessed by microscopy (24). After opsonization with human serum for 30 min, the C. albicans strains were incubated with 106 PMNs at a 1:1 ratio in 1 ml RPMI at 37°C for 15 min on a shaker. Three drops of the sample were then cytospun and Gram stained. Percent phagocytosis was calculated as the proportion of PMNs containing 1 or more blastoconidia after 100 PMNs were counted. A phagocytosis index was calculated as the average number of blastoconidia associated with each phagocytosing PMN. No attempt was made to differentiate the C. albicans cells that were attached to the surfaces of the PMNs from those that were phagocytosed. The fungicidal activity of PMNs was assessed by a CFU assay (24). Opsonized C. albicans strains were incubated with 106 PMNs at a 1:1 ratio in 1 ml of RPMI 1640 containing 5% human serum at 37°C for 2 h. After complete lysis of PMNs with sterile water, serial 10-fold dilutions were made, and a colony count was performed. The fungicidal activity was calculated as the percentage of C. albicans organisms killed after a 2-h incubation with PMNs. The phagocytosis and killing experiments were performed in triplicate and repeated at least twice.
Determination of antibody titer against Not5p.
Two peptides of 151 and 74 amino acids (aa) (amino acid positions 250 to 400 and 401 to 474 of Not5p) were expressed in Escherichia coli by using pET30 EK/LIC. These peptides were chosen because they have no homology to Not-related proteins in C. albicans or S. cerevisiae, or to any peptides identified in the NCBI gene bank.
The two respective NOT5 DNA fragments were amplified by PCR. Primers for the 151-aa fragment were Not5-Exp1-For (5′-GACGACGACAAGATGAATCCACCAAGGACG-3′) and Not5-Exp1-Rev (5′-GAGGAGAAGCCCGGTTTAATTATCGGTAGAAGC-3′); primers for the 74-aa fragment were Not5-Exp2-For (5′-GACGACGACAAGATGACTCATGCACCAGCAGCAGTGT-3′) and Not5-Exp2-Rev (5′-GAGGAGAAGCCCGGTTAAACAATT CGAGAAATGGTATCAC-3′). The fragments were cloned into pET30 using an EK/LIC cloning kit (EMD Biosciences, Inc.). The DNA sequences of the inserts were confirmed by DNA sequencing. Each plasmid was then transformed into E. coli BL21(DE3) (Novagen), and the recombinant protein was expressed after isopropyl β-d-thiogalactoside (IPTG) induction for 2 h at 37°C. The cell pellet was harvested, washed, resuspended in BugBuster solution (EMD Biosciences, Inc.), and incubated briefly at room temperature. Following clarification by centrifugation, the supernatant was filtered through a 0.45-μm-pore-size filter (Millipore). The supernatant was passed through the His-Bind column, followed by washing of the column with Binding Buffer and Wash Buffer (both from EMD Biosciences, Inc.). The peptide of interest was then eluted with 100 mM imidazole buffer. The eluted fractions were confirmed by 15% SDS-polyacrylamide gel electrophoresis, which showed a single band of the expected size, and by Western blot analysis with an anti-His monoclonal antibody (Invitrogen).
The purified protein fragments were reconstituted in 1 M sodium carbonate-sodium bicarbonate buffer (pH 9.6) and used to coat 96-well plates at a concentration of 0.5 μg per well. After incubation for 1 h at 37°C, the plates were washed with PBS with 0.1% Tween 20 (PBS-T). The wells were then blocked with 0.25% gelatin in PBS-T for 1 h at 37°C and washed with PBS-T. We used an enzyme-linked immunosorbent assay (ELISA) to determine serum antibody titers against each of the two protein fragments (10). The human serum samples were preadsorbed individually overnight at 4°C against whole cells and cell lysates of E. coli strain BL21(DE3) grown in LB at 37°C. The following were added in succession: diluted human serum (1/50 dilution; incubated for 1 h at 37°C), horseradish peroxidase-conjugated goat anti-human immunoglobulin M (IgM), IgG, or IgA (1/500 dilution in PBS-T; incubated for 1 h at 37°C), and, as a substrate, o-phenylenediamine (5 mg) in citrate buffer and 5% hydrogen peroxide (incubated at 37°C). The developing reaction was stopped with 1 M phosphoric acid. The OD was determined on an automated ELISA plate reader at a wavelength of 450 nm. Background was defined in wells coated with the protein to which the secondary antibody was added but the primary antibody was not. The reactive titer was defined as the inverse of the dilution at which the OD was twofold greater than background. All serum samples were tested in duplicate. In addition to wells lacking the primary antibody, wells that were not coated with protein were further included as negative controls.
Data and statistical analyses.
Survival curves were calculated according to the Kaplan-Meier method using the PRISM program (GraphPad Software) and compared using Newman-Keuls analysis. The statistical significances of the differences in adherence, PMN phagocytosis, and killing between the strains were determined by Student's t test. The tissue burdens were logarithmically transformed, and data were presented as log10 CFU/g tissue ± standard deviations; the differences in tissue burden between strains were calculated using Wilcoxon's test. The antibody data were expressed as log10 titers ± standard deviations; the differences in antibody titers against a specific protein fragment between the three groups of patients were determined by analysis of variance. For all experiments, P values of ≤0.05 were considered significant.
RESULTS
C. albicans NOT5 contributes to hyphal formation, cell wall integrity, and adherence to epithelial cells and shares functional equivalence with S. cerevisiae NOT5.
In our previous studies, we implicated C. albicans NOT5 in hyphal formation, resistance to the detergent SDS, and adherence to human BECs by using a null mutant strain in which the C-terminal domains of both alleles were disrupted using the ura-blaster method (3). To ensure that the expression of an N-terminal Not5p-HisG fusion protein did not confound our original findings, we began the present study by creating a new null mutant strain in which the complete NOT5 open reading frame was disrupted and URA3 reintroduced to its native locus. We also created a NOT5 revertant strain in the null mutant background (Fig. 1; Table 1). The disruption and revertant strains did not differ from the wild-type strain CAI-12 in growth rates at 37°C in YPD or in minimal media.
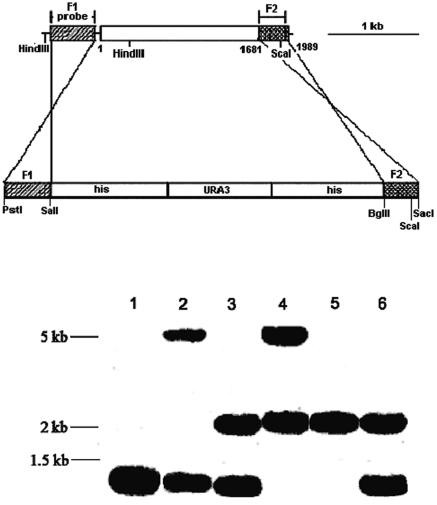
FIG. 1.
Targeted disruption of C. albicans NOT5. (Upper panel) Schematic diagram of the disruption protocol. Shown is the disruption cassette created in pMB7, in which hisG-URA3-hisG is flanked by C. albicans NOT5 fragments F1 (nucleotides −600 to −203 relative to ATG) and F2 (nucleotides +1681 to +1978). The cassette was released by digestion with PstI and SacI and transformed into strain CAI4 by electroporation. Recombination is indicated by the connecting lines. (Lower panel) Southern blot analysis. Genomic DNA was prepared for strain CAI4 (lane 1), the NOT5/Δnot5::hisG-URA3-hisG Δura3::imm434/Δura3::imm434 (lane 2), NOT5/Δnot5::hisG Δura3:: imm434/Δura3::imm434 (lane 3), Δnot5::hisG-URA3-hisG/Δnot5::hisG Δura3::imm434/Δura3::imm434 (lane 4), and Δnot5::hisG/Δnot5::hisG Δura3::imm434/Δura3::imm434 (lane 5) strains, and the Δcanot5::hisG/caNOT5::URA3-hisG Δura3::imm434/Δura3::imm434 revertant (lane 6). The DNA was digested with HindIII and ScaI, and the reactive bands were hybridized with radiolabeled fragment F1 as a probe.
The newly created null mutant was deficient in hyphal formation on solid agar (YPD plus 10% fetal calf serum, Spider agar, Lee's agar, and SLAD agar) and within liquid medium (YPD plus 10% fetal calf serum) (data not shown) and impaired in cell wall integrity, as evident by increased susceptibility to 0.04% SDS. Previously, we found that the C-terminal not5-null mutant strain was not more susceptible to low concentrations of the cell wall-perturbing agent calcofluor white (10 μg/ml) than strain CAI-12 (3). In the present study, however, both the C-terminal and complete-gene null mutant strains demonstrated increased susceptibility to higher concentrations of calcofluor white (20 μg/ml). We further corroborated the cell wall defect by testing for susceptibility to Zymolyase, a cell wall-degrading enzyme. When exponentially grown cells were tested, strain CAI-12 showed exquisite sensitivity to Zymolyase (100 μg/ml), with a marked reduction in OD evident within 30 min (Fig. 2). The newly created null mutant strain was more resistant, with the first reduction in OD not detected until 50 min and lesser overall reductions noted. Similar findings were observed with stationary-phase cells (data not shown). Of note, the aberrancies in hyphal formation and cell wall integrity were reversed in the revertant strain.
FIG. 2.
Sensitivity of C. albicans strains to Zymolyase. Cells in exponential phase were incubated with Zymolyase (100 μg/ml) at 37°C with shaking, and the OD was determined at the times indicated. Experiments were performed three times with similar results.
As in our previous study, we demonstrated that the newly created not5-null mutant was deficient in adherence to freshly harvested BECs compared to CAI-12 (data not shown). In addition, the null mutant exhibited diminished adherence to HT-29 colonic and HeLa cervical epithelial cells (41.5% ± 9.0% and 23.1% ± 14.8%, respectively, compared to 100%; P < 0.01 for both). There was no difference in adherence to HT-29 and HeLa cells between the heterozygous mutant (96.2% ± 19.5% and 92.3% ± 12.3%, respectively) and the revertant (111% ± 25% and 86.1% ± 10.9%, respectively). In contrast to our findings with the epithelial cells, the null mutant did not differ from CAI-12 in adherence to HUV-EC-C endothelial cells (92.3% ± 10.9% versus 100%; P = NS[not significant]).
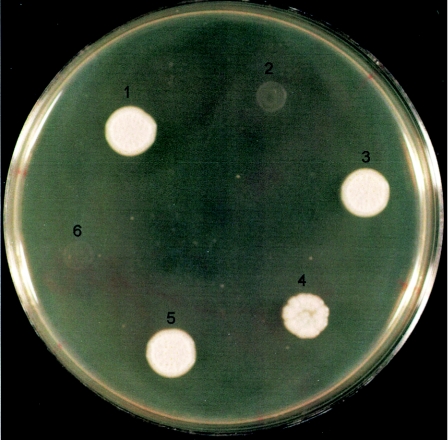
Finally, we transformed a plasmid carrying C. albicans NOT5 into a not5 mutant strain of S. cerevisiae. The complemented strains were able to grow as well as the S. cerevisiae wild-type strain S288C on YPD at 37°C (Fig. 3). The S. cerevisiae not5 mutant strain and the mutant strains complemented either with the vector alone or with NOT5 in reverse orientation were not able to grow at 37°C (Fig. 3).
FIG. 3.
Growth of S. cerevisiae strains on YPD medium at 37°C. Growth after 48 h is pictured for the following strains: 1, S288C (wild-type S. cerevisiae); 2, 35491 (S. cerevisiae not5 mutant); 3 to 5, independently created complemented strains (strain 35491::C. albicans NOT5); 6, 35491 transformed with plasmid yPB1-ADHp alone (vector control). Not pictured is strain 35491 transformed with a plasmid containing C. albicans NOT5 in the reverse orientation, which did not grow on YPD medium.
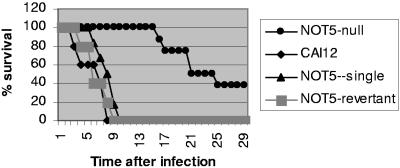
NOT5 is required for complete virulence during DC-IV in nonimmunosuppressed mice.
We inoculated 8 to 10 ICR mice via the lateral tail vein with 106 CFU of CAI-12, the newly created heterozygous mutant, the null mutant, or the revertant strain. Mice infected with the null mutant strain survived significantly longer than mice infected with CAI-12 (P < 0.001 by Newman-Keuls multiple-contrast analysis). There was no difference in mortality among mice infected with CAI-12, the heterozygous mutant, or the revertant strain (Fig. 4). Of further note, there were no differences in mortality rates and time to death among mice infected with the newly created complete-gene null mutant and the previously created C-terminal null mutant strain (3).
FIG. 4.
Survival plot of mice infected intravenously with C. albicans strains. The figure represents the cumulative results of two separate experiments.
We also determined whether NOT5 contributed to the burden of infection and extent of tissue damage within the kidneys. The kidneys of mice infected with the newly created not5-null mutant showed significantly reduced C. albicans burdens at 1 and 5 days postinfection (mean log10 CFU/g of tissue, 3.53 ± 0.30 at day 1 and 4.81 ± 0.36 at day 5) compared to the kidneys of nonmoribund mice infected with CAI-12 (mean log10 CFU/g of tissue, 4.65 ± 0.34 and 5.93 ± 0.32, respectively) (P < 0.001 for both days). The tissue burdens caused by the revertant strain at 5 days, on the other hand, were not significantly different from those observed with CAI-12 (mean log10 CFU/g, 5.2 ± 0.9 and 5.6 ± 0.5, respectively).
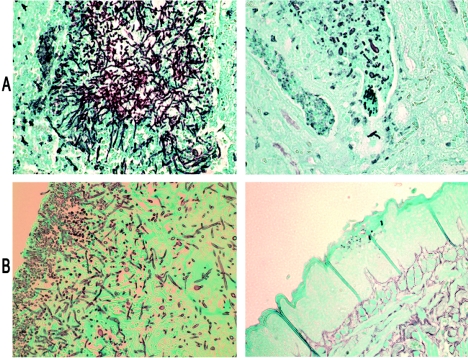
The histopathology of infected kidneys at days 1 and 5 paralleled the tissue burden data. We found that the kidneys of mice infected with CAI-12 or the revertant strain showed diffuse infection and damage of normal architecture, whereas the kidneys of mice infected with the null mutant showed fewer foci of infection and greater tissue preservation (Fig. 5A). The loci of infection due to the mutant were smaller and less well organized than those observed for CAI-12 and the revertant (Fig. 5A). Furthermore, while both yeast and hyphal morphologies were seen in the kidneys of mice infected with all strains, the hyphae associated with the mutant were notably shorter (Fig. 5A).
FIG. 5.
Histopathology of kidneys (A) and tongues (B) of mice infected with CAI-12 (left panels) and the not5-null mutant (right panels). Representative GMS stains of tissues are shown.
NOT5 also contributes to virulence during DC-IV among neutropenic mice.
We immunosuppressed ICR mice with cyclophosphamide before inoculating the respective strains into the lateral tail vein (10 mice/group). In preliminary experiments, we verified that our immunosuppressive regimen caused a reduction in absolute neutrophil counts from baseline levels of 2.5 × 103 to 4.0 × 103/mm3 to <500/mm3 at the time of infection and throughout the study period. All mice infected with 1 × 105 CFU of CAI-12 or the not5-null mutant were moribund and were sacrificed on day 3 postinfection. There were no significant differences in tissue burdens between the kidneys of mice infected with these strains (mean log10 CFU/g, 6.27 ± 0.47 versus 6.75 ± 0.22, respectively). The mice infected with the null mutant, however, had significantly lower tissue burdens in the liver and spleen (mean log10 CFU/g, 3.31 ± 0.02 and 3.00 ± 0.56, respectively) than those infected with CAI-12 (mean log10 CFU/g, 3.98 ± 0.23 and 4.08 ± 0.50, respectively) (P = 0.02 and 0.04, respectively).
When we repeated the experiments with lower inocula (1 × 103 CFU/mouse), none of the mice appeared ill by day 7 postinfection. Tissue burden assessment on this day showed that all 10 mice infected with CAI-12 harbored C. albicans in the kidneys (mean log10 CFU/g, 3.59 ± 1.0) whereas 9 of 10 mice infected with the null mutant had no organisms recovered; the 10th mouse had only 35 CFU recovered from both kidneys. Livers of mice infected with the mutant also had no organisms recovered (versus a mean log10 CFU/mouse of 0.31 ± 0.11 for mice infected with CAI-12). No organisms were recovered from the spleens of mice infected with either CAI-12 or the mutant. Upon repetition of the experiment using the same inocula, there was no difference in tissue burdens at day 7 between the kidneys of mice infected with CAI-12 and those of mice infected with the revertant strain (mean log10 CFU/g, 4.35 ± 0.49 and 4.98 ± 0.21, respectively). In findings similar to those of the initial experiment, five of seven mice infected with the null mutant had sterile kidneys; the kidneys from the other two mice harbored only 4 and 75 CFU.
NOT5 contributes to virulence during murine OPC.
Groups of 10 to 12 ICR mice immunocompromised with cortisone acetate were infected sublingually with 108 CFU of CAI-12, the not5-null mutant, or the revertant strain (13). There were no differences in the overall behavior, weight, or level of activity of mice infected with the three strains. At autopsy on day 7 following infection, however, thrush-like lesions were visible in the oropharynges and esophagi of mice infected with CAI-12 or the revertant strain but not in those of mice infected with the null mutant. We demonstrated that mice infected with CAI-12 or the revertant strain carried significantly higher burdens of C. albicans in their oral cavities and esophagi (log10 CFU/g of tissue, 6.17 ± 0.45 and 6.02 ± 0.31, respectively) than mice infected with the not5 mutant strain (log10 CFU/g, 5.18 ± 0.23) (P = 0.0003 and 0.0002, respectively). The experiment was repeated, and similar results were obtained.
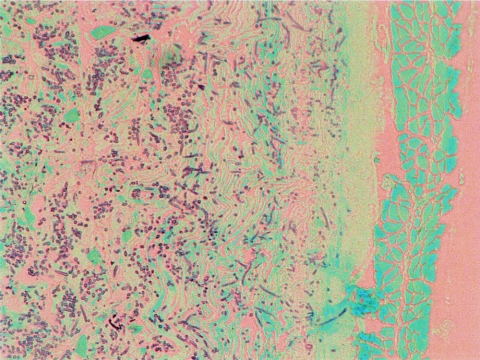
Histopathology of the oropharynges of mice infected with CAI-12 or the revertant strain showed destruction of the superficial keratotic epithelium and invasion of hyphae past the basal layer of the tongue (Fig. 5B). The tongues of the mice infected with the null mutant, on the other hand, showed intact epithelium with only a few surface loci of yeasts and, more rarely, hyphae (Fig. 5B); there were no organisms in the basal layer. In the esophagus, mice infected with CAI-12 or the revertant demonstrated diffuse hyphae throughout the lumen as well as the mucosal and, to a lesser extent, the submucosal layer (Fig. 6). The null mutant, on the other hand, caused no significant infection in the esophagus; despite numerous evaluations, we were not able to find a single focus of organisms.
FIG. 6.
Histopathology of the esophagus of a mouse infected orally with CAI-12. The GMS stain shown is representative of the esophagi of three mice.
NOT5 is not implicated in the pathogenesis of DC-GI or VVC.
Groups of 12 to 13 infant CFW (Swiss Webster) BR mice were inoculated with 2 × 108 CFU of CAI-12 or the not5-null mutant strain by the oral-intragastric route (5, 6) and immunosuppressed with cyclophosphamide and cortisone acetate. We demonstrated that 20 days after GI infection, (i) mice infected with CAI-12 or the null mutant strain had persistent infections in the stomachs that showed no significant differences in tissue burdens (Table 2); (ii) CAI-12 and the mutant strain disseminated to the same extents to the kidneys, livers, and spleens (Table 2); (iii) the stomachs, livers, spleens, and kidneys of mice infected with CAI-12 and of those infected with the null mutant could not be distinguished based on the depth of infection or tissue damage; and (iv) both hyphae and yeasts were evident in the histopathology of different organs (there were no differences in the morphologies of C. albicans strains in these tissues).
TABLE 2.
Tissue burdens of mice infected intragastrically with C. albicans strains
| C. albicans strain | Tissue burden (mean log10 CFU/g of tissue ± SEM) in:
|
|||
|---|---|---|---|---|
| Stomach | Liver | Kidney | Spleen | |
| CAI-12 | 6.58 ± 0.22 | 6.26 ± 0.41 | 3.86 ± 0.46 | 3.04 ± 0.37 |
| not5-null mutant | 6.55 ± 0.25 | 6.18 ± 0.35 | 3.69 ± 0.39 | 2.84 ± 0.34 |
For the model of VVC, female CBA/J (H-2k) mice in pseudoestrus were intravaginally inoculated with 5 × 104 CFU of the C. albicans strains (26). We detected no differences in morphology or tissue burdens between CAI-12 and the null mutant in vaginal lavages recovered on days 4 and 7 postinfection (log CFU/ml for CAI-12, 3.34 ± 0.35 at day 4 and 3.13 ± 0.34 at day 7; log CFU/ml for the null mutant, 3.96 ± 0.53 at day 4 and 2.54 ± 0.89 at day 7) (P = NS for both days). The experiment was repeated, and similar results were obtained.
Disruption of NOT5 does not result in increased phagocytosis of C. albicans by human PMNs.
We studied the effects of gene disruption on ingestion and killing by freshly harvested human PMNs (24). The percentage of PMNs that ingested at least one CAI-12 cell (i.e., percent phagocytosis) was significantly higher than the percentage ingesting the not5-null mutant strain (75.5% ± 3.4% versus 62.8% ± 8.9%, respectively) (P = 0.04). Similarly, the average number of C. albicans cells associated with each phagocytosing PMN (i.e., phagocytosis index) was significantly higher for CAI-12 than for the mutant (2.3 ± 0.3 versus 1.7 ± 0.1) (P = 0.006). The overall fungicidal activities of PMNs were similar against CAI-12 and the mutant strain (percentages of the original candidal inoculum killed were 60.5% ± 9.9% versus 50.0% ± 7.5%, respectively) (P = NS). These experiments were performed three times, and similar findings were obtained. We also repeated the experiments with CAI-12 and the revertant strain and found no differences in the percent phagocytosis (79.8% ± 0.9% versus 77.0% ± 2.1%, respectively), the phagocytosis index (2.66 ± 0.52 versus 2.68 ± 0.36, respectively), or fungicidal activity (60.7% ± 8.8% versus 61.6% ± 3.6%, respectively).
Antibody against recombinant Not5p is present in sera of patients with candidiasis as well as in sera of healthy volunteer controls.
We expressed and purified two peptide fragments of Not5p that do not share homology with any sequences in available databases (amino acid positions 250 to 400 and 401 to 474). We used these in an ELISA to assess antibody responses in the sera of 36 patients with candidiasis (18 with OPC and 18 with DC) and 24 healthy men and women without any evidence of candidiasis. We demonstrated that all individuals mounted significant antibody responses against both peptide fragments, regardless of whether they had candidiasis or not. For peptide fragment 250-400, there were no differences in serum IgG and IgM titers between patients with OPC, patients with DC, and healthy controls (Table 3). Serum IgA titers, however, were significantly higher in patients with candidiasis than in individuals without candidiasis (Table 3). For fragment 401-474, there were no significant differences in IgG, IgM, and IgA titers between patients with OPC, patients with DC, and controls (Table 3).
TABLE 3.
Not5p-specific IgG, IgM, and IgA titers against peptide fragments in sera of patients with OPC or DC and in sera of healthy controls
| Peptide fragment and antibody | Log10 antibody titer ± SD
|
P | ||
|---|---|---|---|---|
| OPC | DC | Controls | ||
| Fragment 250-400 | ||||
| IgG | 3.95 ± 0.16 | 4.00 ± 0.15 | 3.94 ± 0.10 | NS (0.5) |
| IgM | 3.57 ± 0.24 | 3.62 ± 0.27 | 3.54 ± 0.14 | NS (0.7) |
| IgA | 3.72 ± 0.12 | 3.62 ± 0.09 | 3.48 ± 0.06 | 0.0003 |
| Fragment 401-474 | ||||
| IgG | 2.87 ± 0.61 | 2.39 ± 0.47 | 2.48 ± 0.50 | NS (0.12) |
| IgM | 3.26 ± 0.51 | 3.32 ± 0.43 | 3.10 ± 0.50 | NS (0.6) |
| IgA | 3.60 ± 0.59 | 3.41 ± 0.59 | 3.57 ± 0.13 | NS (0.7) |
DISCUSSION
In this study, we demonstrate that C. albicans NOT5 makes complex contributions to candidal virulence. We conclusively implicate the gene in the pathogenesis of DC-IV and OPC, but not in that of DC-GI or VVC. Taken together, our findings suggest that the particular in vivo environment associated with a given type of candidiasis is an important determinant of whether NOT5 plays a role in virulence.
Numerous host factors differed between our murine models and might account for the differing contributions of NOT5 to virulence. First, the models assessed disease at a variety of tissue sites. It is conceivable, therefore, that Not5p is adapted to facilitate invasion and destruction of particular sites, such as the oral mucosa, but is not necessary for these processes to occur at other sites, such as the GI tract or vaginal mucosa. Second, the portals of entry of C. albicans to the sites of infection differed, a difference which, in turn, might have influenced virulence. During DC, for example, our data suggest that NOT5 is important for the disease process within the kidneys if the organism is inoculated directly into the bloodstream but not if it must first traverse the GI tract. Passage through the GI mucosa, therefore, might alter the subsequent mechanisms of virulence within the bloodstream and kidneys such that NOT5 is not necessary for disease to occur. Finally, the status of host defenses might influence the importance of NOT5 in different types of candidiasis. In the VVC model, mice have alterations of the vaginal mucosa caused by pseudoestrus but are not immunosuppressed. In this setting of intact immune function, the contribution of Not5p to virulence might not be sufficient to play a discernible role in establishing mucosal disease; alternatively, other factors may take greater precedence in vaginal tissue or in the presence of estrogen. In the OPC model, on the other hand, cortisone acetate causes pleiotropic disturbances in host cellular immunity (17); the virulence effects of Not5p might become “unmasked” and facilitate tissue destruction. During GI candidiasis, mice have defects in both PMN quantity and cellular immunity. In the face of such profound immunosuppression, it is possible that the host is not able to defend itself against even a relatively “hypovirulent” not5-null mutant strain, and invasion of the GI tract might be indistinguishable from that seen with a wild-type strain.
We must acknowledge that the differences in virulence we observed might also result from inadequacies of the murine models. The DC-IV model, for example, does not mimic the pathogenesis of the majority of cases of human candidemia, which most commonly arise from GI translocation. Furthermore, in those cases of DC in humans in which an i.v. catheter is implicated, the inoculum is generally small. The murine model we employed is popular not because of its physiologic relevance but primarily because of its interlaboratory reproducibility and relative simplicity. At the same time, our use of different strains of mice in the various models might also have influenced our findings. We chose these mouse strains because the respective models have been well established. Based on this study, therefore, we cannot definitively conclude that NOT5 never plays a role in the pathogenesis of diseases such as GI candidiasis or VVC. Nevertheless, the fact that we could not demonstrate a role in our models of these diseases supports our conclusion that the gene encodes a protein whose contribution to virulence is contingent on the appropriate host environment.
It is not clear if the differences in virulence in the models stem from the lack of NOT5 expression during GI candidiasis or VVC. It is noteworthy, however, that sera recovered from healthy young men and women with no history of candidal diseases contain anti-Not5p antibody titers that do not differ from those among patients with DC and OPC. As our ELISA experiments were performed against two unique protein fragments that do not share any homology with amino acid sequences identified in humans, S. cerevisiae, or other microbes, the antibodies are likely to be specifically directed against C. albicans Not5p. For this reason, our findings suggest that NOT5 is likely to be expressed during routine colonization of mucosal surfaces by C. albicans. As such, our study highlights the relationship between colonization and the pathogenesis of disease, and it suggests that a number of candidal genes might be involved in both processes. In its role as a commensal, for example, C. albicans must adhere to and persist at mucosal surfaces as well as resisting elimination by the host. In C. albicans' role as an opportunistic pathogen, the same properties might also contribute to disease states in a susceptible host environment. Genes such as NOT5 that are likely to be expressed during the colonization of tissues, therefore, can also encode virulence factors.
Interestingly, the attenuation in adherence of the not5 mutant to mucosal cell lines in vitro was not consistently associated with attenuated virulence at the corresponding tissue sites in the murine models. This observation highlights the complexity of in vivo environments and demonstrates that they are incompletely replicated by in vitro systems. At mucosal surfaces in vivo, for example, C. albicans must contend with numerous factors not included in adherence assays, such as antibodies, innate host defense factors, other aspects of immune function, and competing microorganisms. For these reasons, it is not surprising that specific genes implicated in adherence during in vitro studies might not be found to affect virulence during in vivo infections.
We believe that NOT5 is likely to mediate its effects on virulence indirectly. Our finding that C. albicans NOT5 complemented an S. cerevisiae not5-null mutant strain implies the presence of a candidal CCR-NOT complex. The functionally diverse S. cerevisiae CCR-NOT complex serves as a global transcriptional regulator involved in a wide range of cellular processes (16). The complex is also involved in several aspects of mRNA metabolism, including activation of initiation, control of elongation, and deadenylation and degradation (9). If C. albicans NOT5 encodes a component of a similar complex, it is likely that the gene influences the transcription of other virulence-associated genes. Indeed, coregulation of virulence genes has been demonstrated for several genes that regulate hyphal formation (15). It is further plausible that the genes and transcripts influenced by a CCR-NOT complex might encode factors that are adapted to particular host environments. Of note, we have previously shown that NOT3, encoding another member of the putative C. albicans CCR-NOT complex, does not contribute to the pathogenesis of DC-IV in nonimmunosuppressed mice (4). Other investigators, on the other hand, have demonstrated a role for NOT4 during DC-IV, which suggests that individual genes of the NOT family are likely to be responsible for a subset of functions within the entire complex (14). At the same time, specific aspects of the complex's function might remain operational in the absence of individual genes (14). It is also possible that NOT5 has functions outside of the larger complex, in which case the gene could influence virulence more directly.
Our findings lead to several conclusions. First, the role of C. albicans NOT5 in virulence is shaped by factors such as tissue location, host immune status, and portal of entry to the site of infection. At the same time, NOT5 might also play a role in the routine colonization of mucosal surfaces, highlighting the relationship between commensalism and the pathogenesis of disease. We are currently investigating the potential interactions between Not5p, putative members of the C. albicans CCR-NOT complex, and other virulence proteins. Finally, we will also be studying the expression patterns of NOT5 during different murine infections to determine if transcriptional regulation accounts for the gene's variable contributions to virulence.
Acknowledgments
We thank William Fonzi of Georgetown University and Malcolm Whiteway of Biotechnology Research Institute/National Research Council, who kindly provided plasmids and C. albicans strains used in this study. We thank John Wingard for insightful review of the manuscript.
Informed consent was obtained from patients before the sera were taken. Human experimentation guidelines of the U.S. Department of Health and Human Services and the University of Florida College of Medicine were followed in the conduct of this research. Animal experimentation guidelines were followed in animal studies. The authors do not have a commercial or other association that might pose a conflict of interest.
This project was supported in part by the National Institute of Dental and Craniofacial Research (NIH-RO1-DE13980-01 to M.H.N. and 5R21DE015069-02 to C.J.C.) and by the National Institute of Allergy and Infectious Diseases (5P01AI061537-02 to M.H.N.). In addition, M.H.N. is the recipient of a VA Advanced Research Career Development Award. The research was conducted in the laboratories of M. H. Nguyen at the North Florida/South Georgia VA Medical Center, Gainesville, Fla.
Editor: T. R. Kozel
REFERENCES
- 1.Bertram, G., R. K. Swoboda, N. A. Gow, G. W. Gooday, and A. J. P. Brown. 1996. Structure and regulation of the Candida albicans ADH1 gene encoding an immunogenic alcohol dehydrogenase. Yeast 12:115-127. [DOI] [PubMed] [Google Scholar]
- 2.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, S., C. J. Clancy, M. A. Checkley, M. Handfield, J. D. Hillman, A. Progulske-Fox, A. S. Lewin, P. L. Fidel, and M. H. Nguyen. 2003. Identification of Candida albicans genes induced during thrush offers insight into pathogenesis. Mol. Microbiol. 48:1275-1288. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, S., M. H. Nguyen, Z. Zhang, H. Jia, M. Handfield, and C. J. Clancy. 2003. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect. Immun. 71:6101-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, G. T., K. R. Seshan, L. M. Pope, and R. J. Yancey. 1988. Morphological aspects of gastrointestinal tract invasion by Candida albicans in the infant mouse. J. Med. Vet. Mycol. 26:173-185. [PubMed] [Google Scholar]
- 6.Cole, G. T., K. T. Lynn, K. R. Seshan, and L. M. Pope. 1989. Gastrointestinal and systemic candidosis in immunocompromised mice. J. Med. Vet. Mycol. 27:363-380. [DOI] [PubMed] [Google Scholar]
- 7.Collart, M. A., and K. Struhl. 1994. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8:525-537. [DOI] [PubMed] [Google Scholar]
- 8.Collart, M. A. 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313:1-16. [DOI] [PubMed] [Google Scholar]
- 9.Denis, C. L., and J. Chen. 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73:221-250. [DOI] [PubMed] [Google Scholar]
- 10.Ebersole, J. L., M. A. Taubman, D. J. Smith, D. E. Frey, A. D. Haffajee, and S. S. Socransky. 1987. Human serum antibody responses to oral microorganisms. IV. Correlation with homologous infection. Oral Microbiol. Immunol. 2:53-59. [DOI] [PubMed] [Google Scholar]
- 11.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198: 179-182. [DOI] [PubMed] [Google Scholar]
- 13.Kamai, Y., M. Kubota, Y. Kamai, T. Hosokawa, T. Fukuoka, and S. G. Filler. 2002. Contribution of Candida albicans ALS1 to the pathogenesis of experimental oropharyngeal candidiasis. Infect. Immun. 70:5256-5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krueger, K. E., A. K. Ghosh, B. P. Krom, and R. L. Cihlar. 2004. Deletion of the NOT4 gene impairs hyphal development and pathogenicity in Candida albicans. Microbiology 150:229-240. [DOI] [PubMed] [Google Scholar]
- 15.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 16.Lenssen, E., U. Oberholzer, J. Labarre, C. De Virgilio, and M. A. Collart. 2002. Saccharomyces cerevisiae Ccr4-not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol. 43:1023-1037. [DOI] [PubMed] [Google Scholar]
- 17.Lionakis, M. S., and D. P. Kontoyiannis. 2003. Glucocorticoids and invasive fungal infections. Lancet 362:1828-1838. [DOI] [PubMed] [Google Scholar]
- 18.Liu, H. Y., V. Badarinarayana, D. C. Audino, J. Rappsilber, M. Mann, and C. L. Denis. 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17:1096-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259: 686-688. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee, P. K., K. R. Seshan, S. D. Leidich, J. Chandra, G. T. Cole, and M. A. Ghannoum. 2001. Reintroduction of the PLB1 gene into Candida albicans restores virulence in vivo. Microbiology 147:2585-2597. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen, M. H., S. Cheng, and C. J. Clancy. 2004. Assessment of Candida albicans genes induced during infections as a tool to understand pathogenesis. Med. Mycol. 42:293-304. [DOI] [PubMed] [Google Scholar]
- 22.Oberholzer, U., and M. A. Collart. 1998. Characterization of NOT5 that encodes a new component of the Not protein complex. Gene 207:61-69. [DOI] [PubMed] [Google Scholar]
- 23.Porta, A., A. M. Ramon, and W. A. Fonzi. 1999. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J. Bacteriol. 181:7516-7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roilides, E., P. Paschalides, A. Freifeld, and P. A. Pizzo. 1993. Suppression of polymorphonuclear leukocyte bactericidal activity by suramin. Antimicrob. Agents Chemother. 37:495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchimori, N., L. L. Sharkey, W. A. Fonzi, S. W. French, J. E. Edwards, Jr., and S. G. Filler. 2000. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect. Immun. 68:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wozniak, K. L., F. L. Wormley, Jr., and P. L. Fidel, Jr. 2002. Candida-specific antibodies during experimental vaginal candidiasis in mice. Infect. Immun 70:5790-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]