Abstract
Pseudomonas aeruginosa, an opportunistic respiratory pathogen that infects the majority of patients with cystic fibrosis, initiates host inflammatory responses through interaction with airway epithelial cells. The Toll-like receptors (TLRs) are a family of pathogen pattern recognition receptors that play key roles in host innate immunity. In this study we aimed to determine whether TLRs mediate the interaction between P. aeruginosa and airway epithelial cells. Individual murine TLRs (TLR1 to TLR9) and dual combinations of these TLRs that activate an NF-κB-driven luciferase reporter in response to PAO1 were screened in HEK 293 cells. TLR5, TLR2, a combination of TLR1 and TLR2, or a combination of TLR2 and TLR6 responded to PAO1. Another P. aeruginosa strain, strain PAK, activated TLR5 similarly, while the isogenic flagellin-deficient strain PAK/fliC and the flagellum-free bacterium Haemophilus influenzae failed to activate TLR5. Reverse transcription-PCR was used to probe the presence of multiple TLRs (including TLR5) in primary human airway epithelial cells (HAECs). Immunostaining with TLR5 antibodies showed that TLR5 was expressed in HAECs and on the apical surface of the human trachea epithelium. In HAECs, PAO1, PAK, and Burkholderia cepacia, but not flagellin-deficient strain PAK/fliC or a B. cepacia fliC mutant, activated the NF-κB reporter. Dominant negative TLR5 specifically blocked the response to P. aeruginosa but not to the response to lipoteichoic acid, a specific ligand of TLR2. We also determined that MyD88, IRAK, TRAF6, and Toll-interacting protein (Tollip), but not TIRAP, were involved in the TLR-mediated response to P. aeruginosa in HAECs. These findings demonstrate that the airway epithelial receptor TLR5 senses P. aeruginosa through its flagellin protein, which may have an important role in the initiation of the host inflammatory reaction to clear the invading pathogen.
Pseudomonas aeruginosa is a virulent gram-negative pathogen that infects patients through the respiratory tract and colonizes the surface of the airway epithelium. In cystic fibrosis patients, P. aeruginosa infection occurs in 70% of the patients at an early age and contributes to the chronic lung destruction responsible for mortality; P. aeruginosa infection rarely occurs in healthy hosts due to efficient clearance of the pathogen by the innate immune response.
Human airway epithelia form the first line in the pulmonary defense against invading pathogens. Both the mechanical mucociliary clearance and the surface fluid, which contains a mixture of molecules that have antimicrobial activity secreted by the epithelial cells, contribute to the elimination of pathogens in nonspecific ways (4). Once P. aeruginosa penetrates the initial barriers, a direct interaction between the pathogen and the epithelial cells occurs, which initiates a series of inducible cellular host immune responses for more specific bacterial killing. These responses include secretion of antimicrobial peptides like beta-defensins into the airway surface fluid and release of cytokines and chemokines to attract immune active cells to the site of infection (16, 38). A recent study also showed that human epithelial cells provide a link to T helper 2-type allergic inflammation by producing cytokines to activate dendritic cells, the cellular gate to adaptive immunity (37). Thus, efficient clearance of P. aeruginosa relies critically on recognition of the pathogen, which can further mount intracellular signaling pathways responsible for sensing and responding to the danger signal in the epithelial cells.
Mammalian Toll-like receptors (TLRs) are homologues of the Drosophila Toll protein, a single transmembrane receptor important for recognition of pathogens and mediation of innate immunity in insects. At present, 9 murine TLRs and 11 human homologues have been cloned. These molecules are expressed differentially in a wide range of immunoactive cells, such as macrophages, dendritic cells, and epithelial cells (24). TLRs mediate responses to bacteria and viruses and their components through specifically binding to the structural motifs unique to the pathogens called pathogen-associated molecular patterns (PAMPs). Upon recognition of PAMPs, TLRs provoke rapid activation of innate immunity by inducing production of proinflammatory cytokines or antimicrobial peptides or upregulation of costimulatory molecules that share the signaling pathways of the interleukin receptor (IL-1R) featured in activation of the central transcriptional factor NF-κB (8, 19, 42). At present, the ligand profile of TLRs has been partially determined. TLR2 responds to a variety of microbial products, including the gram-positive bacterial components lipoteichoic acid (LTA) and peptidoglycans; TLR4 plays a major role in the host response to lipopolysaccharide (LPS); TLR3 recognizes double-stranded RNA released during viral proliferation; TLR5 has a ligand identified as flagellin, the protein component of bacterial flagella; and TLR9 binds bacterial DNA that has unmethylated CpG repeats, as reviewed by Kaisho and Akira (24). Recent studies have shown that TLRs also form heterodimers to recognize different pathogens (15, 39). By this mechanism, the spectrum of PAMPs to which TLRs can respond has been greatly expanded.
Recently, using transformed intestinal or airway epithelial cell lines, workers have shown that TLRs are indeed involved in cellular responses, such as activation of the NF-κB signaling pathway and proinflammatory genes in response to P. aeruginosa (1, 13, 22, 40). Our previous in vivo study showed that pulmonary clearance of Haemophilus influenzae was significantly delayed in TLR4-deficient mice and that inflammatory cytokine secretion in the airway epithelial cells was correspondingly diminished (44), while the ability to clear P. aeruginosa in these mice remained intact. These findings led us to the hypothesis that there are receptors in addition to TLR4, probably other TLRs, in the human airway epithelial cells (HAECs) that sense the pathogen and mediate its killing. To test this hypothesis, we performed a direct screening analysis of all the TLRs and dual combinations of the TLRs reconstituted with an NF-κB promoter-driven luciferase reporter in HEK 293 cells to search for the molecules that sense P. aeruginosa. TLRs that gave positive signals in response to P. aeruginosa stimulation in this in vitro system were further verified with respect to their functional role in primary airway epithelial cells.
MATERIALS AND METHODS
Reagents.
Lipoteichoic acid (LTA) from Bacillus subtilis (L3265) and Staphylococcus aureus (L2515), chromatographically purified LPSs from P. aeruginosa (L8643) and Escherichia coli O111:B4 (L3012), dithiothreitol (DTT) (D8255), protease from Streptomyces griseus (P5147), DNase I (DN25), amphotericin B (A2942), and fluorescein isothiocyanate (FITC)- and tetramethyl rhodamine isothiocyanate (TRITC)-labeled anti-goat and anti-rabbit secondary antibodies were all purchased from Sigma (St. Louis, MO). Ceftazidime was purchased from GlaxoWellcome. Tobramycin was purchased from Eli Lilly and Company. Goat antibodies against the human TLR5 C and N termini (C14 and N15), rabbit antibody against Mucin1, and blocking peptides against TLR5 antibody C14 and N15 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Bacterial strains.
P. aeruginosa strain PAO1 is a well-characterized laboratory strain. PAK is a wild-type, nonmucoid, piliated, motile strain. Strain PAK/fliC is a nonmotile derivative of PAK in which the fliC gene encoding flagellin was replaced by homologous recombination with a mutant gene interrupted by a gentamicin resistance cassette (36). Burkholderia cepacia and the isogenic strain Burkholderia cepacia BC/fliC were provided by Joanna B. Goldberg (University of Virginia Health System, Charlottesville). B. cepacia is a gram-negative bacterium that has flagella. BC/fliC is a derivative of B. cepacia in which the fliC gene encoding flagellin was replaced by homologous recombination with a mutant gene interrupted by a gentamicin resistance cassette (unpublished data). All the strains described above were grown in tryptic soy broth (Difco Laboratories, Detroit, MI) supplemented with 10 μg/ml kanamycin for 6 h until the optical density at 600 nm was 0.5 or the concentration was about 1 × 108 CFU/ml. A kanamycin-resistant encapsulated type b strain, H. influenzae H338, was cultured as previously described (44). The bacteria were collected by centrifugation at 3,000 rpm for 10 min, washed twice by resuspension in sterile phosphate-buffered saline (PBS) (GIBCO, Grand Island, NY), and finally suspended at the desired dilution in PBS. The suspension was immediately incubated in a 60°C water bath for 30 min to heat kill the bacteria. Samples of the preparation were cultured on tryptic soy broth plates overnight to monitor complete killing of the bacteria.
Purification of flagellin.
P. aeruginosa PAK flagellin was purified by the procedure of Allison et al. (3). Briefly, overnight liquid cultures in mineral salts medium [7.0 g K2HPO4 per liter, 3.0 g KH2PO4 per liter, 1.0 g (NH4)2SO4 per liter, 50 mg MgSO4 · 7H2O per liter, 2.5 mg FeCl3 · H2O per liter, 7.4 mg l-methionine per liter, and 4.0 g sodium succinate per liter, pH 7.0] were centrifuged at 5,000 × g for 15 min at 4°C, and the bacterial pellet was resuspended in 10 mM potassium phosphate (pH 7.0) and blended at a low speed in a commercial blender (Black and Decker, Towson, MD) for 30 s at room temperature to shear the flagella. The suspension was centrifuged at 16,000 × g for 15 min at 4°C to remove the bacteria, and the resulting supernatant was centrifuged at 100,000 × g in a centrifuge (model L8-70; Beckman, Duarte, CA) with a 42.1 rotor for 3 h at 4°C. Protein concentrations were determined by the Coomassie blue technique of Bradford (6) with crystalline bovine serum albumin as the standard (Bio-Rad, Richmond, CA). A mock flagellin preparation was prepared in an identical manner using an extract of PAK/fliC bacteria.
Plasmid construction.
cDNAs of murine TLR1 to TLR7 were amplified by PCR from a mouse lung cDNA library. cDNAs of murine TLR8 and TLR9 were amplified from a mouse spleen cDNA library. A dominant negative mutant of human TLR5 (amino acids 1-664) was constructed by PCR amplification of the fragments lacking the intracellular signaling TIR domain from the full-length human TLR5 cDNA. Dominant negative mutants of human MyD88 (amino acids 155-296), IRAK (amino acids 1-208), and TRAF6 (amino acids 289-530) were constructed by PCR amplification of the corresponding DNA fragments from the wild-type template cDNAs (29). A dominant negative mutant of human TIRAP (P125H) was created by PCR mutagenesis of wild-type TIRAP (21). All the human full-length templates for construction of the dominant negative mutants and human Toll-interacting protein (Tollip) cDNA were obtained from a human lung cDNA library by PCR cloning. All the PCR-amplified cDNAs were subcloned into the pIRESpuro vector (catalog no. 6031-1; Clonetech), and sequences were confirmed by sequencing. The cDNA libraries used were purchased from Clonetech (Palo Alto, CA). The NF-κB promoter-driven luciferase reporter was generated by insertion of a 1.0-kb promoter region of the human beta-defensin 2 (hBD2) gene into the pGL3 vector (Promega, Madison, WI) as described previously (45).
Cell culture.
Human embryonic kidney (HEK) 293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Mediatech Cellgro, Virginia), and CHO-K1 cells were cultured in F-12 (Invitrogen). Both media were supplemented with 10% fetal bovine serum (HyClone, Logan, Utah), 100 IU/ml penicillin, and 100 μg/ml streptomycin.
HAECs were prepared by protease digestion from the large airways of a human lung that could not be utilized for transplantation (25). Freshly isolated trachea and central bronchi were rinsed with ice-cold minimal essential medium (GIBCO) and washed three times (4 h each) in 250 ml minimal essential medium (supplemented with 0.5 mg/ml DTT, 10 μg/ml DNase I, 50 μg/ml ceftazidime, 2.5 μg/ml amphotericin B, and 40 μg/ml tobramycin) with shaking at 200 rpm at 4°C. The tissue was digested in 250 ml of 0.1% protease-supplemented minimal essential medium (including DNase and antibiotics but lacking DTT) for an additional 30 to 36 h with rocking at 4°C. The media were collected and centrifuged at 1,500 rpm for 5 min to pellet the cells. The cells were resuspended in bronchial epithelial cell growth medium (BEGM) (Clonetics Biowhittaker) and spun briefly at <600 rpm to collect mucus, and supernatant was plated in 100-mm plates. The epithelial cells grew to 70% to 80% confluence after 5 to 7 days and then were used in experiments. Early passages of HAECs (passage 3 to 4) were used.
Transfection.
Transfection with HEK 293 cells and CHO-K1 cells was performed in 96-well plates using LipofectAmine2000 reagent (Invitrogen) according to the manufacturer's instructions. Briefly, 1 × 10 5 cells were seeded in 100 μl medium in each well of 96-well ViewPlates (Perkin-Elmer Life Sciences Inc., Boston, MA). The cells were approximately 80% confluent at the time of transfection. Twenty-five nanograms of each TLR plasmid (or a dual combination) was mixed with 25 ng firefly luciferase driven by the NF-κB promoter of hBD2 and 1 ng Renilla luciferase in 25 μl serum-free DMEM (43, 45). In addition, 0.5 μl LipofectAmine2000 reagent was mixed in another 25 μl serum-free DMEM. The two mixtures were combined and incubated at room temperature for 20 min. The resulting mixture was added to the cell culture and incubated for 24 h before bacterial challenge. Effectene reagent (QIAGEN Inc., Valencia, CA) was used to transfect HAECs by following the manufacturer's instructions. HAECs were seeded into 24-well plates and cultured in BEGM for more than 48 h to obtain 90% confluence at the time of transfection. Plasmids that included 0.3 μg of the hBD2 promoter-driven firefly luciferase gene and 10 ng of the Renilla luciferase reporter gene were diluted in EC buffer, and an appropriate amount of enhancer was added. Effectene was added to the mixture 5 min after incubation was started. Samples were incubated at room temperature for 10 min. The DNA transfection complex was diluted with BEGM and added to the cells. The precipitates were removed 10 h later by replacement with fresh medium. Then 14 h later, cells were challenged with bacteria or agonists. Empty pIRES vectors were used to normalize the total DNA content.
Luciferase assay.
A dual-luciferase reporter assay system was used as described by the manufacturer (Promega, Madison, WI). The transfected cell samples in 96-well ViewPlates were directly lysed in situ. Samples were measured using a Wallac 1420 multilabel counter (Perkin-Elmer). Data are reported below as relative luciferase activity, as determined by the firefly luciferase activity normalized by the Renilla luciferase activity; the latter served as an endogenous systematic control.
RT-PCR.
Total mRNA of HAECs was isolated by a magnetic separation mechanism using an mRNA isolation kit (Roche, Germany). In brief, HAECs were lysed and incubated with biotin-labeled oligo(dT)20 and streptavidin-conjugated magnetic particles. mRNA with a poly(A) tail bound to oligo(dT) was separated with the particles from the fluid. The beads were washed, and total mRNA was eluted with distilled water. A one-tube reverse transcription (RT)-PCR kit (Titan, Roche, Germany) was used to amplify TLR mRNA in total mRNA of HAECs according to the manufacturer's protocol. The primers used to probe human TLRs were TLR1-5′ (ACGGTCTCATCCACGTTCCTAAAGA), TLR1-3′ (CGCCAGAATACTTAGGAAGTAAGAAC) (covering the 5′-terminal 615 bp of TLR1), TLR2-5′ (GTCTTGTGACCGCAATGGTATCTGC), TLR2-3′ (CTCGCAGTTCCAAACATTCCACG) (612 bp), TLR5-5′ (GTGCTCATGGCCGGTCCTGTGTTTG), TLR5-3′ (CTCTGCTATACAAGCTATTAGCTGCG) (600 bp), TLR6-5′ (GCTTCCATTTTGTTTGCCTTATGATC), and TLR6-3′ (GTATTAACTGATATGTTCACTTGGA) (625 bp). The specificity of the primers was tested by a GenBank BLAST search. A T3 thermocycler PCR system (Biometra Whatman, Germany) was used for the reverse transcription reaction, and this was followed by a 30-cycle PCR program. PCR products were separated on a 1% agarose gel containing ethidium bromide. A MassRuler DNA ladder (Fermentas, Hanover, MD) was used as a size marker.
Immunostaining.
HAECs were fixed in acetone-methanol (1:1) for 5 min. Ten-microliter serial cryosections of human trachea were not fixed or were fixed in acetone for 3 min. The fixed cells or tissue was blocked with 10% goat serum for 1 h and then incubated with human TLR5 antibodies (C14 and N15) and/or with rabbit antibody against Mucin1 for 1 h. For neutralization experiments, TLR5 antibodies mixed with blocking peptides were incubated with the samples for 2 h according to the manufacturer's protocol. After thorough washing with PBS, samples were incubated with FITC- or TRITC-labeled secondary antibodies for 1 h, followed by thorough washing with PBS. Specimens were imaged using a Nikon Microphot-FXA fluorescence microscope (Nikon Inc., Garden City, NY). The negative controls consisted of preincubation with PBS, omission of the primary antibody, and replacement of the primary antibody by a nonimmune isotype-matched immunoglobulin G (IgG). Each control was performed for each experiment on the same day.
Statistical analysis.
The data were analyzed using analysis of variance, and differences were considered significant when the P value was <0.05. The results were expressed as means ± standard errors.
RESULTS
A screening analysis of TLR1 to TLR9 and all dual combinations with transfected 293 cells was performed. Cells were exposed to the PAO1 strain of P. aeruginosa and analyzed for activation of an NF-κB-dependent luciferase reporter gene. The individual TLRs that showed a positive response, as well as any heterologous pair that produced synergistic activation, were studied further, as shown in Fig. 1. Based on NF-κB activation, PAO1 stimulated TLR5 and TLR2 (Fig. 1A). The stimulation occurred in a time-dependent manner and peaked at 18 h after bacterial challenge (data not shown). The intensity of the TLR5- and TLR2-mediated responses to P. aeruginosa was dependent on the bacterial dose used for challenge (Fig. 1A). Similar results for TLR5 were obtained with the CHO-K1 cell system (Fig. 1B). All dual combinations that involved TLR5 produced the activation seen with TLR5 alone. In contrast, when TLR1 was coexpressed with TLR2, there was a synergistic response to PAO1 (Fig. 1C). When TLR6 was coexpressed with TLR2, there was constitutive activity for NF-κB activation independent of bacterial challenge, which has been reported by other workers (31). Even based on this basal activity, PAO1 induced significant activation when TLR2 and TLR6 were coexpressed (Fig. 1D). These findings are consistent with recent findings that TLR2 can interact with TLR1 or TLR6 to sense microbial components such as lipoproteins or the bacterium-secreted molecule modulin through heterodimerization (15, 39).
FIG. 1.
P. aeruginosa strongly stimulates NF-κB activation through TLR receptors in 293 cells. A screening analysis of TLRs and dual combinations of TLRs that respond to P. aeruginosa was performed with 293 cells. 293 cells were grown in 96-well plates and transiently transfected with 25 ng murine TLR1 to TLR9 DNA or dual combinations of TLRs together with 25 ng plasmid encoding firefly luciferase driven by the NF-κB promoter of human β-defensin 2 and 1 ng Renilla luciferase. Empty pIRES vector was used to ensure that equal quantities of DNA were used. Cells were challenged 24 h after transfection with PAO1 or were not challenged (medium) for 18 h, and then dual-luciferase assays were performed to measure the induction of the luciferase activity. (A) PAO1 activated NF-κB activation via TLR5 in a dose-dependent manner in 293 cells. (B) TLR5 mediated PAO1 (10 CFU/cell)-stimulated luciferase activity in CHO cells. (C) Synergetic activation of NF-κB through the TLR1/TLR2 combination in response to PAO1 in 293 cells. (D) TLR2/TLR6 combination mediated NF-κB activation by PAO1 in 293 cells. The data are means ± standard errors for at least two independent experiments. Samples were compared to a control. One asterisk indicates that the P value is <0.05; two asterisks indicate that the P value is <0.001.
P. aeruginosa PAK and PAO1 are genetically characterized, structurally similar laboratory stains. Studies have shown that PAK might be less virulent than PAO1, which may due to the fact that PAK is less piliated than PAO1 (7, 18, 20). However, in vivo studies using BALB/cByJ mice showed that virulence factors other than pili are more important in the development of acute pneumonia (41). To further explore the results obtained in the screening analysis, we employed two cognate strains of P. aeruginosa, PAK and PAK/fliC. PAK/fliC is a nonmotile derivative of PAK in which the fliC gene encoding flagellin was replaced by homologous recombination with a mutant gene interrupted by a gentamicin resistance cassette (36). Both PAK and PAK/fliC stimulated TLR2 in a dose-dependent manner with a maximum response similar to that seen with PAO1, while TLR5 had a very different response to the two strains (Fig. 2A). TLR5 responded to PAK as well as it responded to PAO1, but it could not be stimulated by the flagellin-deficient strain PAK/fliC, which indicated that TLR5 responded to P. aeruginosa through recognition of the flagellum protein flagellin (Fig. 2B). Consistent with this result, H. influenzae, a flagellum-free bacterium, also failed to activate TLR5, although it significantly stimulated TLR2 (Fig. 2C). Consistently, flagellin protein purified from PAK, but not the mock preparation from PAK/fliC, stimulated TLR5 in 293 cells (Fig. 2D). Similarly, for TLR2, combinations of TLR1 and TLR2 and of TLR6 and TLR2 responded to all bacteria equally well (data not shown), indicating that there is a flagellin-independent mechanism for TLR2-dependent responses.
FIG. 2.
TLR5 senses P. aeruginosa through flagellin in 293 cells. 293 cells were mock transfected or transfected with TLR2 or TLR5 DNA together with firefly and Renilla reporter genes as described in the legend to Fig. 1. Twenty-four hours after transfection, cells were incubated for 18 h with medium or 1, 10, or 100 CFU/cell of heat-killed PAK (A), PAK/fliC (B), or H. influenzae (C), and dual-luciferase assays were performed to measure induction of the luciferase activity in the cells. (D) Effect of PAK flagellin (1 μg/ml) or PAK/fliC mock flagellin preparation on TLR5 in 293 cells. The data are means ± standard errors for at least two independent experiments. Samples were compared to a control. One asterisk indicates that the P value is <0.05; two asterisks indicate that the P value is <0.001.
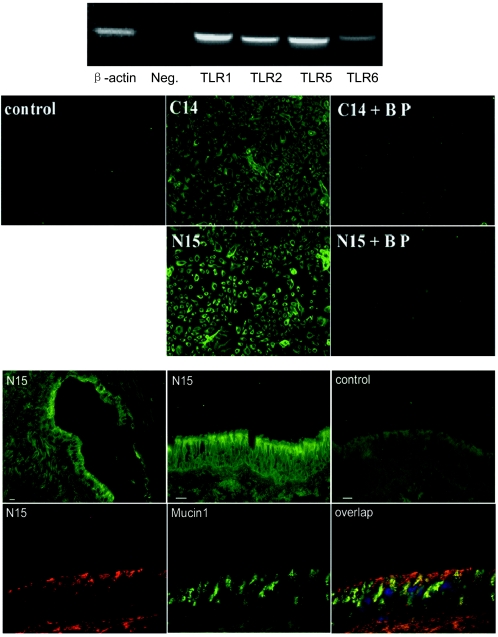
We next used primary HAECs that were freshly isolated from a human lung to verify the screening results obtained with the 293 cell system, focusing on the role of human TLR5. Using specific primers, human TLR1, TLR2, TLR5, and TLR6 could be detected by RT-PCR in mRNA from HAECs (Fig. 3A). The results for human β-actin (587 bp) and a negative control (no primers) were the results that were predicted. Immunocytochemistry for HAECs with human TLR5 antibodies generated against either the TLR5 C terminus (C14) or the TLR5 N terminus (N15) showed positive staining, and the specific blocking peptide of C14 or N15, which resembled the corresponding TLR5 epitopes, completely blocked the staining (Fig. 3B). If the peptides were interchanged, the staining signal was not affected (data not shown). This experiment specifically identified the expression of TLR5 in HAECs. Immunohistology for cryosections of human trachea with TLR5 antibody further localized the expression of TLR5 to the apical surface of the airway epithelium (Fig. 3C). Furthermore, TLR5 did not colocalize with Mucin1, which is expressed in goblet cells of airway epithelium (Fig. 3C). This suggests that TLR5 localizes on the apical surface of pseudostratified ciliated epithelial cells lining the airway. Recently, TLR5 has been shown to be expressed on the basolateral surface of an in vitro culture of polarized epithelial cells (22, 30), and TLR2 has been shown to be expressed on the apical surface of both a polarized epithelial cell culture and human trachea epithelium (30, 45). The difference in TLR polarization could be due to differences in the human tissue from the cell line culture and/or different TLRs studied. An absence of immunostaining was observed when TLR5 antibody was replaced with a control nonimmune isotype-matched IgG and when TLR5 antibody was preabsorbed with excessive corresponding blocking peptide (data not shown).
FIG.3.
TLR5 is expressed in HAECs and on the apical surface of human airway epithelium. (Upper panel) RT-PCR using TLR-specific primers was performed to test the expression of TLRs in total mRNA of HAECs. The samples were a human β-actin positive control (587 bp), a negative control (Neg.) in which primers were omitted from the reaction mixture, human TLR1 (615 bp), human TLR2 (612 bp), human TLR5 (600 bp), and human TLR6 (625 bp). (Middle panel) Expression of TLR5 in HAECs. HAECs cultured on glass slides were stained with human TLR5 antibodies generated against either the TLR5 C terminus (C14) or the TLR5 N terminus (N15). Specific blocking peptide (BP) of C14 or N15 was used to neutralize the staining, as described in Materials and Methods. The control shows staining after replacement of the primary antibody by an isotype-matched IgG. (Lower panel) Expression of TLR5 in airway epithelium. Cryosections of human trachea were blocked with 10% goat serum and stained with primary human TLR5 antibody (N15) and then FITC-labeled secondary antibodies. Images stained with N15 were obtained at low amplification (top left image) or high amplification (top middle image). The control (top right image) shows staining after antibodies were replaced by an isotype-matched IgG. Cryosections of human trachea were also stained with TLR5 N15 antibody (recognized by TRITC-labeled secondary antibody) (bottom left image), human Mucin1 antibody (recognized by FITC-labeled secondary antibody) (bottom middle image), and 4′,6′-diamidino-2-phenylindole (DAPI) at the same time. Sections were observed using a triple filter (bottom left image). Bars = 15 μm.
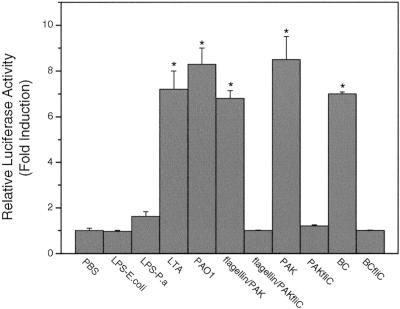
Efficient transfection of the primary HAECs was obtained by using the Effectene reagent. Firefly and Rinilla luciferase reporter genes were introduced into the cells as reporters for the dual-luciferase assay. When stimulated with bacteria or components, the endogenous TLR receptor response may be activated, leading to activation of NF-κB. Figure 4 shows that both wild-type P. aeruginosa PAO1 and PAK strongly activated NF-κB, while the flagellin knockout strain PAK/fliC did not have any stimulatory effect, even at a concentration of 100 CFU/cell. Similar results were obtained with the congenic organisms B. cepacia and BC/fliC. Consistently, flagellin protein purified from PAK, but not the mock preparation from PAK/fliC, showed a stimulation effect in HAECs (Fig. 2D). This indicates that P. aeruginosa may be sensed in a flagellin-dependent manner. LPS from either E. coli or P. aeruginosa at different doses (0.2 to 20 μg/ml) did not have a significant effect on HAECs. Figure 4 shows the results obtained at a concentration of 20 μg/ml, the highest concentration tested. LPS from P. aeruginosa seemed not to behave significantly differently from LPS from E. coli, although at a concentration of 20 μg/ml the stimulation by LPS from P. aeruginosa seemed to be slightly greater than that by LPS from E. coli. The gram-positive bacterial component LTA, a ligand of TLR2 (34), indeed activated the HAECs. These findings demonstrated that flagellin, not LPS, was the component recognized by HAECs.
FIG. 4.
Activation of NF-κB by P. aeruginosa is dependent on flagellin protein. HAECs were prepared and transfected in 24-well plates as described in Materials and Methods. Using the Effectene reagent, plasmids containing 0.3 μg of the hBD2 promoter-driven firefly luciferase gene and 10 ng of the Renilla luciferase reporter gene were used for each well. Cells were challenged 24 h after transfection with 20 μg/ml LPS from E. coli, 20 μg/ml LPS from P. aeruginosa, 10 μg/ml LTA, 1 μg/ml PAK flagellin or a mock flagellin preparation of PAK/fliC or 100 CFU/cell of heat-killed PAO1, PAK, PAK/fliC, B. cepacia (BC), or BC/fliC for 18 h, and then dual-luciferase assays were performed to measure induction of the luciferase activity The data are means ± standard errors for at least two independent experiments. Samples were compared to a control (medium). An asterisk indicates that the P value is <0.001. P.a LPS, P. aeruginosa LPS.
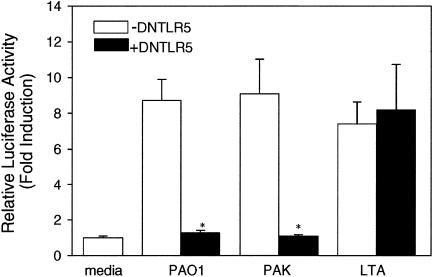
Dominant negative TLRs are truncated versions of TLRs that lack the intracellular signaling TIR domain. To prove that TLR5 plays a role in sensing P. aeruginosa in HAECs, a dominant negative TLR5 was cotransfected with the reporter genes into the cells. In this experiment, both PAO1- and PAK-induced NF-κB activation were blocked, while LTA-induced stimulation through TLR2 remained intact (Fig. 5). The effect of dominant negative TLR2 on the response to P. aeruginosa in HAECs was also studied. Although HAECs transfected with dominant negative TLR2 showed some reduction in the response to P. aeruginosa, the response was not significantly different from that observed with the non-dominant negative TLR2-transfected group (data not shown). The results clearly demonstrated that the sensing of P. aeruginosa by HAECs was specifically mediated by TLR5.
FIG. 5.
HAECs sense P. aeruginosa via TLR5. HAECs were seeded in 24-well plates. As described in Materials and Methods, 0.5 μg dominant negative human TLR5 (DNTLR5) (solid bars) or empty vector DNA (open bars) together with 0.3 μg of the hBD2 promoter-driven firefly luciferase gene and 10 ng of the Renilla luciferase reporter gene was used for each well. Cells were challenged 24 h after transfection with 10 μg/ml LTA or 100 CFU/cell of heat-killed PAO1 or PAK for 18 h, and then dual-luciferase assays were performed to measure induction of the luciferase activity. The data are means ± standard errors for at least two independent experiments. Samples were compared to a control. The group containing dominant negative human TLR5 (+DNTLR5) was compared to the group not containing dominant negative human TLR5 (−DNTLR5). An asterisk indicates that the P value is <0.001.
Our attention next focused on the intracellular signaling pathway of TLR5 in response to P. aeruginosa in HAECs. Activated IL-1R associates with a cytoplasmic adaptor protein, MyD88, through the homophilic interaction between their TIR domains. MyD88 also possesses the death domain, which mediates the association with a serine-threonine kinase IRAK. Subsequently, another adaptor molecule, TRAF6, is activated and in turn evokes diverse signaling, leading to activation of transcriptional factor NF-κB. TLRs, including TLR2, TLR4, and TLR9, employ signaling mechanisms similar to those of their homologous IL-1R (2). Here we used the dominant negative forms of human MyD88, IRAK, and TRAF6 to test the signaling of TLR5 in HAECs. When the results were expressed as percentages of the original signal (Fig. 6), all three dominant negative molecules reduced the response to PAO1 or PAK in a dose-dependent manner. Tollip is a newly identified component of the IL-1R pathway that binds IRAK and prevents it from associating with MyD88 (9). We cloned Tollip from a human lung cDNA library and overexpressed it in HAECs. Significant blocking of TLR5 signaling also occurred (Fig. 6). As shown in Fig. 6, dominant negative TIRAP had no effect on P. aeruginosa-induced NF-κB activation, which was by no means unexpected because TIRAP has been identified as a specific adaptor protein for TLR4 signaling. As the burden of DNA transfection in HAECs tends to reduce the general response to stimuli, TIRAP served as a negative control in this experimental setting. These results revealed a common initial part of the IL-1R signaling pathway involving MyD88, IRAK and its regulating protein Tollip, and TRAF6, which were utilized by TLR5 in response to P. aeruginosa.
FIG. 6.
Intracellular signaling pathway of P. aeruginosa-stimulated NF-κB activation in HAECs. Dominant negative (DN) human TIRAP (0.25 μg) or Tollip and dominant negative human MyD88, IRAK, or TRAF6 at a concentration of 0.25 or 0.5 μg, together with 0.3 μg of the hBD2 promoter-driven firefly luciferase gene and 10 ng of the Renilla luciferase reporter gene, was used for each well. Empty vector plasmids were used to make total amounts of DNA in the transfectants equal. Cells were challenged with 100 CFU/cell of heat-killed PAO1 or PAK for 18 h, and dual-luciferase assays were performed to measure induction of the luciferase activity. The values are percentages of the bacterium-stimulated induction in mock transfection experiments. The data are means ± standard errors for at least two independent experiments. Samples were compared to stimulation with mock transfection. An asterisk indicates that the P value is <0.001.
In summary, our findings showed that HAECs utilize a TLR5-mediated signaling pathway to specifically sense P. aeruginosa via its flagellin.
DISCUSSION
In this study, we sought to find the TLR receptor expressed in HAECs which functions as the P. aeruginosa sensor in the airway's innate immune defense. After a screening analysis with 293 cells, we evaluated the results obtained with primary HAECs. Finally, we determined that TLR5 is the epithelial TLR that mediates the response to P. aeruginosa.
P. aeruginosa is a gram-negative, piliated, mobile bacterium that is commonly found in immunocompromised hosts and the lungs of cystic fibrosis patients. Normally, the airway epithelium forms a mechanical and biological barrier to clear P. aeruginosa efficiently, before the bacterium colonizes the epithelium and leads to chronic infection and severe destruction of the lung. P. aeruginosa attaches to the apical surface of the epithelial cells through interaction with the cell surface adhesion molecules. There are multiple adhesion sites on the epithelial cell surface that are responsible for the differential binding sites for P. aeruginosa. Asialylated glycolipid receptors, such as asialo-GM1, have been shown to be responsible for binding pilin of pili and also the outer membrane LPS (14), and recently, cell-associated Muc1 mucin has been identified as an adhesion molecule for flagellin protein of flagella (28). Adhesion molecules are important for bacterial attachment, but they lack many intracellular signaling components. Therefore, they are not “professional” receptors for sensing and responding to environmental signals and by themselves cannot account for the immune response induced by the pathogen (27).
The receptors in the TLR family have been very well characterized as pathogen-recognizing receptors that signal specifically in response to a potentially very wide range of pathogen structures from bacteria, fungi, and viruses (24). Different profiles of TLRs are present in different subsets of immune cells, including macrophages and dendritic cells, and are directly enrolled in innate immune responses to pathogen killing and modulation of adaptive immunity (33, 37, 42). Although the human airway epithelium is among the first sites to see inhaled pathogens and our results have shown that multiple TLRs are expressed in the cells at this site (Fig. 3A), studies on the roles of TLRs in these cells are limited. Our previous in vivo study with mice showed that TLR4 senses H. influenzae and mediates its clearance, and the mouse airway epithelial cells were one of the recourses for inflammatory cytokine and chemokine secretion (44). Interestingly, clearance of P. aeruginosa in TLR4-deficient mice was barely affected, suggesting that TLRs other than TLR4 respond to this pathogen (unpublished data). Our findings in this study demonstrated that there was expression of TLR5 in HAECs and on the apical surface of human airway epithelium. TLR5 sensed P. aeruginosa and led to activation of transcriptional factor NF-κB in human airway epithelial cells. Consistently, TLR5 did not respond to stimulation with H. influenzae. These results not only may provide reasonable explanations for the intact P. aeruginosa clearance capacity in TLR4-deficient mice but also may indicate that the human airway epithelium provides a defense beyond being a barrier; it also functions as an integral part of the cell-mediated innate immunity in being a pathogen-sensor and the clearance initiator. This concept was supported by observations that human epithelial cells triggered a dendritic cell-mediated immune response by chemokine expression (37).
TLR4 signaling in 293 cells has been studied by other workers previously (10). Consistent with our observations, transfection of TLR4 in 293 cells resulted in constitutive activation of the NF-κB pathway and stimulation of TLR4 with LPS, leading to significant elevated activation of the pathway. However, stimulation with heat-killed P. aeruginosa on TLR4 did not show any activation (data not shown). Epithelial cell systems have shown differential TLR expression and signaling in response to pathogens or components (1, 5, 23, 35). In transformed human airway epithelial cell line 1HAEo- cells, which do not form cilia in transwell cultures, LPS stimulation induced no response (1), while in airway epithelial cell line BEAS-2B it induced inflammatory cytokine secretion (35). In primary human tracheobronchial epithelial cells, LPS induced beta-defensin 2 expression in a CD14-dependent manner (5), while in the primary human airway epithelial cells the response to LPS seemed to be limited by low expression of MD-2 (22). These observations suggest that the function of TLR4 and probably also the functions of other TLR signaling pathways heavily depend on the cellular environment, where cofactors such as adaptor proteins, coreceptors, or downstream signaling components might be differentially expressed.
A few studies using transformed epithelial cell lines have revealed TLR2 responses to P. aeruginosa through recognition of flagellin (1), while data obtained from primary airway epithelial cells suggest that TLR2 played no or little role in the response to P. aeruginosa. We speculate that this might due to the fact that different cell systems were used in these studies. Interestingly, the results of Adamo et al. agree with our finding (unpublished data) that neither pulmonary clearance of P. aeruginosa nor neutrophil infiltration induced by the bacteria was affected in the TLR2 knockout mice, suggesting that TLR2 signaling plays little role in mice (1). Considering that current information on TLR signaling in response to P. aeruginosa is based extensively on transformed epithelial cell lines, our study using primary human airway epithelium cells should add information that is more relevant to the function of airway epithelial cells of the human lung.
Besides recognizing pattern molecules from gram-positive bacteria such as LTA and peptidoglycans, TLR2 can also recognize lipoproteins and certain LPSs from gram-negative bacteria (8, 12). Erridge et al. recently found that LPS from P. aeruginosa PAC-611 signals via TLR2 in HeLa cells, while, interestingly, it shows little stimulation of monocytes isolated from human peripheral blood. The structure of LPS seems to have an impact on differential activation of TLR2 versus TLR4 (12). Ernst et al. also found that the structure of LPS in P. aeruginosa was affected by the conditions in which the bacteria were cultured and that the structural heterogeneity affected the properties of the immunostimulatory capabilities (11). Our data showed that LPSs from different sources indeed had different effects not only in 293 cells but also when 293 cells and HAECs were compared. In our study, TLR2-transfected 293 cells responded to P. aeruginosa efficiently, which is consistent with TLR2 responses to P. aeruginosa and LPS in CHO cells (17). However, in HAECs, TLR2 appeared to play little role in the cellular response to P. aeruginosa.
Flagella are highly conserved among diverse bacterial species. Like many other mucosal pathogens, P. aeruginosa cells express flagella, which provide motility and chemotaxis toward preferred substrates. Flagella are also major virulence components of the pathogen. The main structural and virulence protein in flagella is flagellin encoded by the fliC gene. A recent study showed that TLR5 recognizes flagellin purified from both gram-positive bacteria (Listeria monocytogenes) and gram-negative bacteria (P. aeruginosa) in a TLR5-luciferase reconstituted cell system (17). The results are consistent with our finding that flagellin-deficient P. aeruginosa failed to activate NF-κB in both a TLR5-transfected system and HAECs. B. cepacia also activated NF-κB in HAECs in a flagellin-dependent way. Interestingly, although there is LPS in the outer membrane of P. aeruginosa, the activation of NF-κB seems to be mostly dependent on TLR5, since even a high dose of purified LPS from P. aeruginosa did not induce significant stimulation of NF-κB. This observation was consistent with previous observations that LPS had no effect on either stimulation of Ca2+ release or cytokine production in primary airway epithelial cells or some airway epithelial cell lines (27, 32).
Polarized expression and function of TLRs have been reported in a few studies. The results depend extensively on the type of cell model used (1, 22). It has been suggested that TLR5 is expressed at the basal surface of 1HAEo- cells, while it has also been shown that addition of P. aeruginosa to the apical surface of human nasal cystic fibrosis epithelial CF15 cells elicited a considerable amount of gene upregulation (1, 22). Studies also have suggested that there is translocation of TLR5 from the basal to the apical surface after P. aeruginosa challenge; however, the underlying mechanism remains to be established (1, 22). Interestingly, immunostaining of human trachea tissue sections illustrated apical expression of TLR5.
Molecules involved in TLR-dependent signaling have been identified, although the networks of the pathway that results in an integrated host response to bacteria have not been delineated. In our study we utilized dominant negative molecules for several molecules involved in the TLR signaling pathways to address this question in HAECs. Signaling pathways of the TLR family are homologous to IL-1R because they share an endogenous TIR domain, which signals mainly through a similar mechanism using adaptor protein MyD88 (29). MyD88-dependent signaling is conveyed through sequential association of IRAK and TRAF6, which further leads to activation of NF-κB (2). In HAECs, we found that TLR5 sensed P. aeruginosa and transduced the signal into the nucleus using the classical components of this pathway involving MyD88, IRAK, and TRAF6. However, recent studies showed that more complicated downstream signaling mediated by TLRs is involved. TIRAP has been shown to be another adaptor protein for TLR4 signaling in addition to MyD88 (21). The data showed that dominant negative TIRAP did not affect P. aeruginosa-stimulated NF-κB activation in HAECs. However, it has been shown that there are other downstream signaling pathways in addition to NF-κB pathway for TLR4 to mediate gene regulation, such as the IRF3 pathway (2). It has also been reported that hBD2 secretion in gingival epithelial cells is regulated through a pathway involving the mitogen-activated protein kinase pathway but is independent of the NF-κB transcriptional factor family (26). The observations suggested that NF-κB activation is by no means a complete indicator of activation of all TLRs, so it is necessary to take the cellular context in which TLRs are expressed into consideration, and other signaling pathways also deserve further exploration.
Acknowledgments
This work was supported by NIH grant 5R01HL049040-13 and by the Cystic Fibrosis Foundation.
Editor: F. C. Fang
REFERENCES
- 1.Adamo, R., S. Sokol, G. Soong, M. I. Gomez, and A. Prince. 2004. Pseudomonas aeruginosa flagella activate airway epithelial cells through asialoGM1 and Toll-like receptor 2 as well as Toll-like receptor 5. Am. J. Respir. Cell Mol. Biol. 30:627-634. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 3.Allison, J. S., M. Dawson, D. Drake, and T. C. Montie. 1985. Electrophoretic separation and molecular weight characterization of Pseudomonas aeruginosa H-antigen flagellins. Infect. Immun. 49:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bals, R., D. J. Weiner, and J. M. Wilson. 1999. The innate immune system in cystic fibrosis lung disease. J. Clin. Investig. 103:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, M. N., G. Diamond, M. W. Verghese, and S. H. Randell. 2000. CD14-dependent lipopolysaccharide-induced beta-defensin-2 expression in human tracheobronchial epithelium. J. Biol. Chem. 275:29731-29736. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Bradley, D. E. 1974. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with non-retractile pili. Virology 58:149-163. [DOI] [PubMed] [Google Scholar]
- 8.Brightbill, H. D., D. H. Libraty, S. R. Krutzik, R. B. Yang, J. T. Belisle, J. R. Bleharski, M. Maitland, M. V. Norgard, S. E. Plevy, S. T. Smale, P. J. Brennan, B. R. Bloom, P. J. Godowski, and R. L. Modlin. 1999. Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science 285:732-736. [DOI] [PubMed] [Google Scholar]
- 9.Burns, K., J. Clatworthy, L. Martin, F. Martinon, C. Plumpton, B. Maschera, A. Lewis, K. Ray, J. Tschopp, and F. Volpe. 2000. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat. Cell Biol. 2:346-351. [DOI] [PubMed] [Google Scholar]
- 10.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 11.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 12.Erridge, C., A. Pridmore, A. Eley, J. Stewart, and I. R. Poxton. 2004. Lipopolysaccharides of Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa signal via Toll-like receptor 2. J. Med. Microbiol. 53:735-740. [DOI] [PubMed] [Google Scholar]
- 13.Gewirtz, A. T., T. A. Navas, S. Lyons, P. J. Godowski, and J. L. Madara. 2001. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167:1882-1885. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, S. K., R. S. Berk, S. Masinick, and L. D. Hazlett. 1994. Pili and lipopolysaccharide of Pseudomonas aeruginosa bind to the glycolipid asialo-GM1. Infect. Immun. 62:4572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajjar, A. M., D. S. O'Mahony, A. Ozinsky, D. M. Underhill, A. Aderem, S. J. Klebanoff, and C. B. Wilson. 2001. Cutting edge: functional interactions between Toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J. Immunol. 166:15-19. [DOI] [PubMed] [Google Scholar]
- 16.Harder, J., U. Meyer-Hoffert, L. M. Teran, L. Schwichtenberg, J. Bartels, S. Maune, and J. M. Schroder. 2000. Mucoid Pseudomonas aeruginosa, TNF-alpha, and IL-1beta, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am. J. Respir. Cell Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 18.Hazlett, L. D., M. M. Moon, A. Singh, R. S. Berk, and X. L. Rudner. 1991. Analysis of adhesion, piliation, protease production and ocular infectivity of several P. aeruginosa strains. Curr. Eye Res. 10:351-362. [DOI] [PubMed] [Google Scholar]
- 19.Hertz, C. J., S. M. Kiertscher, P. J. Godowski, D. A. Bouis, M. V. Norgard, M. D. Roth, and R. L. Modlin. 2001. Microbial lipopeptides stimulate dendritic cell maturation via Toll-like receptor 2. J. Immunol. 166:2444-2450. [DOI] [PubMed] [Google Scholar]
- 20.Holloway, B. W. 1969. Genetics of pseudomonads. Bacteriol. Rev. 33:419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horng, T., G. M. Barton, and R. Medzhitov. 2001. TIRAP: and adapter molecule in the Toll signaling pathway. Nat. Immunol. 2:835-841. [DOI] [PubMed] [Google Scholar]
- 22.Hybiske, K., J. K. Ichikawa, V. Huang, S. J. Lory, and T. E. Machen. 2004. Cystic fibrosis airway epithelial cell polarity and bacterial flagellin determine host response to Pseudomonas aeruginosa. Cell. Microbiol. 6:49-63. [DOI] [PubMed] [Google Scholar]
- 23.Jia, H. P., J. N. Kline, A. Penisten, M. A. Apicella, T. L. Gioannini, J. Weiss, and P. B. McCray, Jr. 2004. Endotoxin responsiveness of human airway epithelia is limited by low expression of MD-2. Am. J. Physiol. Lung Cell. Mol. Physiol. 287:L428—L437. [DOI] [PubMed] [Google Scholar]
- 24.Kaisho, T., and S. Akira. 2002. Toll-like receptors as adjuvant receptors. Biochim. Biophys. Acta 1589:1-13. [DOI] [PubMed] [Google Scholar]
- 25.Klockmann, M. T., H. U. Jahn, S. Hippenstiel, H. J. Kramer, and N. Suttorp. 1998. Interaction of human neutrophils with airway epithelial cells: reduction of leukotriene B4 generation by epithelial cell derived prostaglandin E2. J. Cell Physiol. 175:268-275. [DOI] [PubMed] [Google Scholar]
- 26.Krisanaprakornkit, S., J. R. Kimball, and B. A. Dale. 2002. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-kappaB transcription factor family. J. Immunol. 168:316-324. [DOI] [PubMed] [Google Scholar]
- 27.Kube, D., U. Sontich, D. Fletcher, and P. B. Davis. 2001. Proinflammatory cytokine responses to P. aeruginosa infection in human airway epithelial cell lines. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L493-L502. [DOI] [PubMed] [Google Scholar]
- 28.Lillehoj, E. P., B. T. Kim, and K. C. Kim. 2002. Identification of Pseudomonas aeruginosa flagellin as an adhesin for Muc1 mucin. Am. J. Physiol. Lung Cell. Mol. Physiol. 282:L751-L756. [DOI] [PubMed] [Google Scholar]
- 29.Medzhitov, R., P. Preston-Hurlburt, E. Kopp, A. Stadlen, C. Chen, S. Ghosh, and C. A. Janeway, Jr. 1998. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2:253-258. [DOI] [PubMed] [Google Scholar]
- 30.Muir, A., G. Soong, S. Sokol, B. Reddy, M. Gomez, A. Van Heeckeren, and A. Prince. 2004. Toll like receptors in normal and cystic fibrosis airway epithelial cells. Am. J. Respir. Cell. Mol. Biol. 30:777-783. [DOI] [PubMed] [Google Scholar]
- 31.Ozinsky, A., D. M. Underhill, J. D. Fontenot, A. M. Hajjar, K. D. Smith, C. B. Wilson, L. Schroeder, and A. Aderem. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. USA 97:13766-13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratner, A. J., R. Bryan, A. Weber, S. Nguyen, D. Barnes, A. Pitt, S. Gelber, A. Cheung, and A. Prince. 2001. Cystic fibrosis pathogens activate Ca2+-dependent mitogen-activated protein kinase signaling pathways in airway epithelial cells. J. Biol. Chem. 276:19267-19275. [DOI] [PubMed] [Google Scholar]
- 33.Schnare, M., G. M. Barton, A. C. Holt, K. Takeda, S. Akira, and R. Medzhitov. 2001. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2:947-950. [DOI] [PubMed] [Google Scholar]
- 34.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 35.Sha, Q., A. Q. Truong-Tran, J. R. Plitt, L. A. Beck, and R. P. Schleimer. 2004. Activation of airway epithelial cells by Toll-like receptor agonists. Am. J. Respir. Cell Mol. Biol. 31:358-364. [DOI] [PubMed] [Google Scholar]
- 36.Simpson, D. A., R. Ramphal, and S. Lory. 1992. Genetic analysis of Pseudomonas aeruginosa adherence: distinct genetic loci control attachment to epithelial cells and mucins. Infect. Immun. 60:3771-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soumelis, V., P. A. Reche, H. Kanzler, W. Yuan, G. Edward, B. Homey, M. Gilliet, S. Ho, S. Antonenko, A. Lauerma, K. Smith, D. Gorman, S. Zurawski, J. Abrams, S. Menon, T. McClanahan, R. de Waal-Malefyt, F. Bazan, R. A. Kastelein, and Y. J. Liu. 2002. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat. Immunol. 3:673-680. [DOI] [PubMed] [Google Scholar]
- 38.Suntres, Z. E., A. Omri, and P. N. Shek. 2002. Pseudomonas aeruginosa-induced lung injury: role of oxidative stress. Microb. Pathog. 32:27-34. [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi, O., S. Sato, T. Horiuchi, K. Hoshino, K. Takeda, Z. Dong, R. L. Modlin, and S. Akira. 2002. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169:10-14. [DOI] [PubMed] [Google Scholar]
- 40.Tallant, T., A. Deb, N. Kar, J. Lupica, M. J. de Veer, J. A. DiDonato. 2004. Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-kappa B and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol. 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang, H., M. Kays, and A. Prince. 1995. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect. Immun. 63:1278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoma-Uszynski, S., S. Stenger, O. Takeuchi, M. T. Ochoa, M. Engele, P. A. Sieling, P. F. Barnes, M. Rollinghoff, P. L. Bolcskei, M. Wagner, S. Akira, M. V. Norgard, J. T. Belisle, P. J. Godowski, B. R. Bloom, and R. L. Modlin. 2001. Induction of direct antimicrobial activity through mammalian Toll-like receptors. Science 291:1544-1547. [DOI] [PubMed] [Google Scholar]
- 43.Tsutsumi-Ishii, Y., and I. Nagaoka. 2002. NF-kappa B-mediated transcriptional regulation of human beta-defensin-2 gene following lipopolysaccharide stimulation. J. Leukoc. Biol. 71:154-162. [PubMed] [Google Scholar]
- 44.Wang, X., C. Moser, J. P. Louboutin, E. S. Lysenko, D. J. Weiner, J. N. Weiser, and J. M. Wilson. 2002. Toll-like receptor 4 mediates innate immune responses to Haemophilus influenzae infection in mouse lung. J. Immunol. 168:810-815. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X., Z. Zhang, J. P. Louboutin, C. Moser, D. J. Weiner, and J. M. Wilson. 2003. Airway epithelia regulate expression of human beta-defensin 2 through Toll-like receptor 2. FASEB J. 17:1727-1729. [DOI] [PubMed] [Google Scholar]