Abstract
We report that point mutations causing alteration of the fourth alpha-helix (α4-helix) of the CAP homology domain of eukaryotic (Saccharomyces cerevisiae) type II topoisomerases (Ser740Trp, Gln743Pro, and Thr744Pro) change the selection of type II topoisomerase-mediated DNA cleavage sites promoted by Ca2+ or produced by etoposide, the fluoroquinolone CP-115,953, or mitoxantrone. By contrast, Thr744Ala substitution had minimal effect on Ca2+- and drug-stimulated DNA cleavage sites, indicating the selectivity of single amino acid substitutions within the α4-helix on type II topoisomerase-mediated DNA cleavage. The equivalent mutation in the gene for Escherichia coli gyrase causing Ser83Trp also changed the DNA cleavage pattern generated by Ca2+ or quinolones. Finally, Thr744Pro substitution in the yeast type II topoisomerase rendered the enzyme sensitive to antibacterial quinolones. This study shows that the α4-helix within the conserved CAP homology domain of type II topoisomerases is critical for selecting the sites of DNA cleavage. It also demonstrates that selective amino acid residues in the α4-helix are important in determining the activity and possibly the binding of quinolones to the topoisomerase II-DNA complexes.
DNA topoisomerases are enzymes that catalyze changes in the topology of DNA via a mechanism involving the transient breakage and rejoining of phosphodiester bonds in the DNA backbone (9, 59). Studies in both prokaryotic and eukaryotic cells have demonstrated the importance of topoisomerases in transcription, DNA replication, and chromosome segregation (41, 58). Type II topoisomerases (Top2p) are conserved among eukaryotes. Gyrase and topoisomerase IV are bacterial Top2 enzymes with significant sequence similarity to eukaryotic Top2 proteins (4, 39). Eukaryotic Top2 enzymes are homodimeric and cleave one DNA duplex (the G, or gate, segment) using two tyrosine residues (one from each Top2p monomer) to attack phosphodiester bonds on both DNA strands. In this process, each enzyme monomer becomes covalently attached to the 5′ end of the cleaved DNA by a phosphotyrosine linkage. Top2p then opens the DNA, bridging the gap in the G segment. A second DNA duplex (the T, or transported, segment) passes through the gap. Thereafter, the G segment is resealed, and the T segment is liberated from the enzyme (4, 53, 59).
Under physiological conditions, the covalent Top2p-DNA complexes (referred to as cleavage complexes) are normally short-lived intermediates in the catalytic cycle of the enzyme. Top2p requires the presence of a divalent cation for catalytic activity, which is Mg2+ under physiological conditions. However, when magnesium is replaced by Ca2+, a higher level of Top2p is trapped in a Ca2+-promoted covalent enzyme-DNA complex, and DNA double- and single-stranded DNA cleavage can be detected (44, 45).
Beyond its vital cellular functions, Top2p is the primary cytotoxic target for some of the most active drugs used for the treatment of human cancers (see below) (3, 10, 11, 29, 48). In addition, inhibitors of bacterial type II topoisomerases, such as quinolones, are among the most widely prescribed antibacterial drugs (24, 36). Top2p inhibitors can be divided in two groups: Top2p catalytic inhibitors and Top2p poisons (48). Top2p catalytic inhibitors (“suppressors”) are defined as drugs that inhibit enzyme activity but do not stabilize DNA cleavage complexes. Proflavine and 9-aminoacridine belong to this category (2, 54). Top2p poisons inhibit the enzyme by increasing the steady-state levels of DNA cleavage complexes (18, 33, 48). Hence, they convert Top2p into a physiological toxin that creates protein-linked DNA breaks in the genome of treated cells (10, 18, 34, 48). Top2p poisons can, in turn, be subdivided in two groups: DNA intercalators—which include doxorubicin, mitoxantrone, amsacrine, and ellipticines—and nonintercalators, whose main representatives are the demethylepipodophyllotoxins etoposide (VP-16), teniposide (VM-26), and the quinolones (37, 52), such as ciprofloxacin and norfloxacin, which are specific for bacterial Top2 enzymes (24, 36).
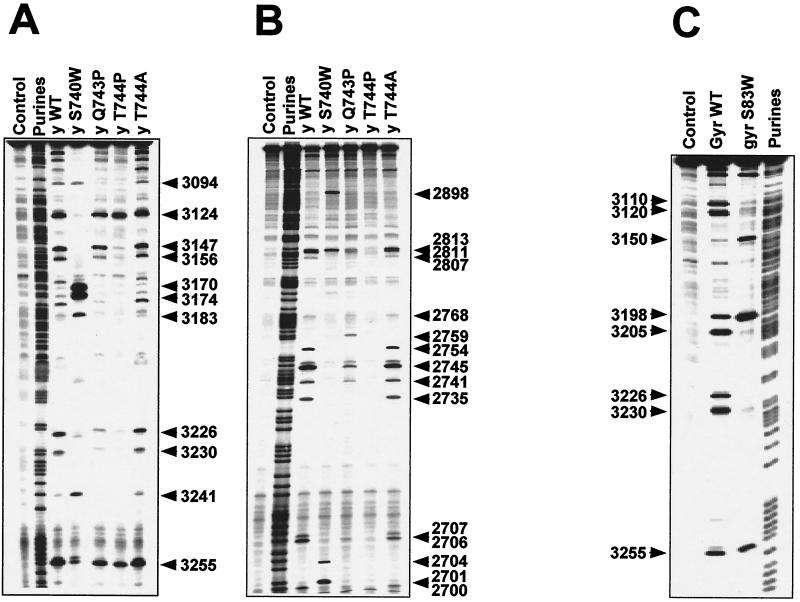
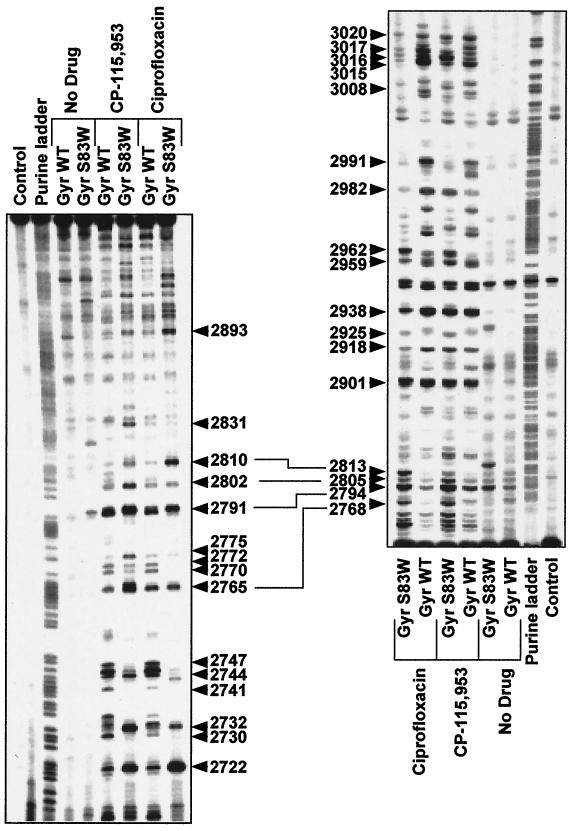
Previous studies showed that substitution of Gly747 to Glu of Saccharomyces cerevisiae Top2p (Fig. 1C) and B show enzyme-specific differences. Cleavage was reduced at several sites for Top2pS740W and Top2pQ743P compared to Top2pWT. For example, cleavage at positions 3124, 3147, 3156, 3226, 2706 or 2707, 2735, 2741, 2745, and 2754 was reduced for Top2pS740W and at that at positions 3094, 3170, 2754, and 2735 was reduced for Top2pQ743P. Most reduction in Ca2+-promoted cleavage was seen for Top2pT744P (Fig. 2B), which is in contrast to the intensity of the drug-induced cleavage sites generated by Top2pT744P (Fig. 3). Cleavage was enhanced shows a comparison of the DNA cleavage patterns observed in the presence of two different quinolones, CP-115,953 and ciprofloxacin. Because the GyrAS83W mutant is quinolone resistant (13, 42, 58, 60) and therefore bound to give less cleavage than gyraseWT, we chose drug concentrations (100 μM) that are approximately three times the 50% inhibitory concentration for supercoiling (data not shown). As for yeast Top2p, the S83W substitution changed the enzyme cleavage pattern. Several cleavage sites were reduced in intensity for GyrAS83W in the presence of CP-115,953 and ciprofloxacin (e.g., 2747, 2744, 2772, 2770, 2991, 3020, 3017, and 3020), whereas other sites were enhanced (e.g., 2722, 2893, 2768, 2794, 2805, and 2962). These results demonstrate that the GyrAS83W mutation markedly affects the DNA sequence specificity of DNA gyrase.
FIG. 1.
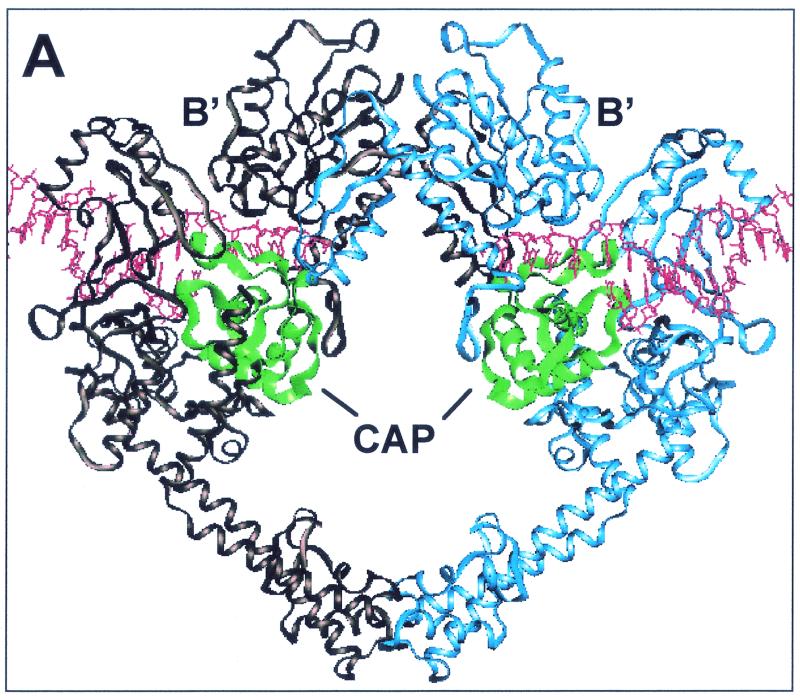
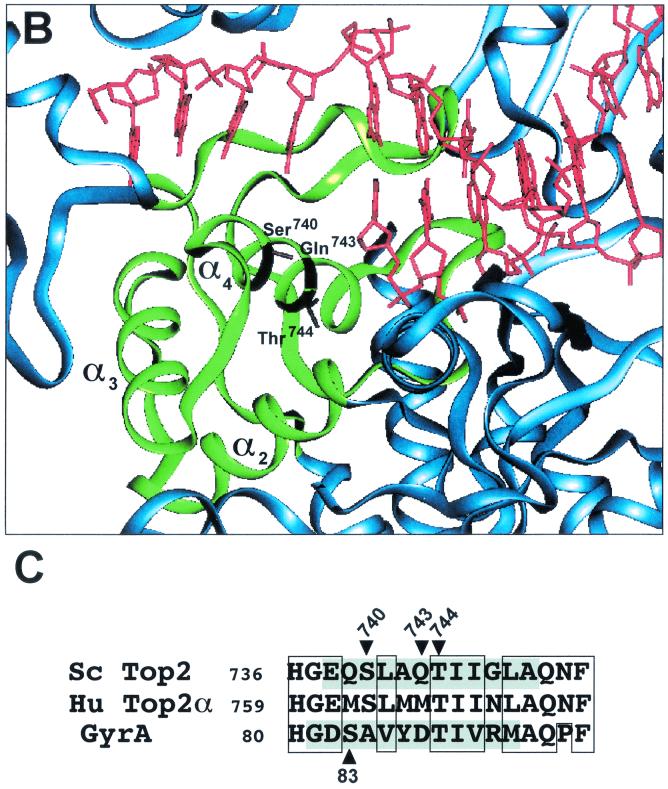
CAP homology domain and positions of Ser740, Gln743, and Thr744 in yeast Top2p. (A) Illustration of dimeric S. cerevisiae topoisomerase II structure (Protein Data Bank accession number 1bgw). Drawings were generated using the program QUANTA (version 97). The helical ribbon representation shows the 92-kDa fragment of the yeast enzyme with a DNA fragment modeled into each of the putative DNA-binding sites (6). The CAP homology domain of each protomer is highlighted in green. (B) Close view of the putative DNA-binding region, presenting the proposed proximity of the α4-helix within the CAP homology domain, including Ser 740, Gln 743, and Thr744 (in a stick model), to DNA. (C) Alignment of protein sequence for the yeast (Sc Top2p), human Top2pα (Hu Top2pα) and E. coli gyrase. The mutated residues studied in the present report are indicated by arrowheads. Shaded residues correspond to the α4-helix (5). No shading is shown for human Top2p because of lack of structural data. Boxed regions indicate similarity between the amino acid residues. Ser763 in the human Top2pα sequence corresponds to Ser740 in yeast Top2p.
FIG. 2.
Comparison of Ca2+-promoted DNA cleavage patterns of yeast Top2pWT, Top2pS740W, Top2pQ743P, Top2pT744P, and Top2pT744A as well as gyraseWT and GyrAS83W. DNA fragments from the human c-myc first intron (A) and from the junction between the c-myc first intron and first exon (B) were prepared by PCR. For each fragment, 5′-end-labeling was performed with 32P. Yeast Top2p reactions were performed at 37°C for 30 min (A and B), and gyrase reactions were performed at 25°C for 2 h (C). In all reactions, the CaCl2 concentration was 5 mM. Lanes: Control, without Top2 enzyme; Purine, ladder obtained after formic acid reaction; y WT, yeast wild-type Top2p; y S74 0W, yeast Top2pS740W; y Q743P, yeast Top2pQ743P; y T744P, yeast Top2pT744P; y T744A, yeast Top2pT744A; gyrWT, gyraseWT from E. coli; GyrAS83W, mutant Ser83Trp gyrase. Numbers correspond to genomic positions of the nucleotide covalently linked to the enzyme via the 5′ phosphate.
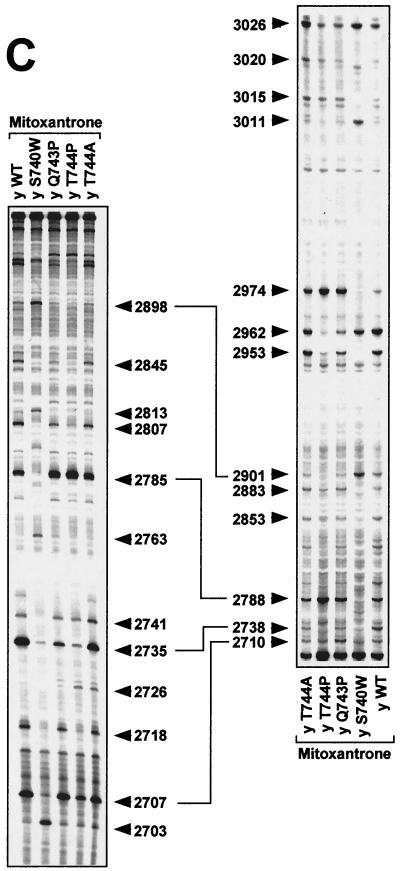
FIG. 3.
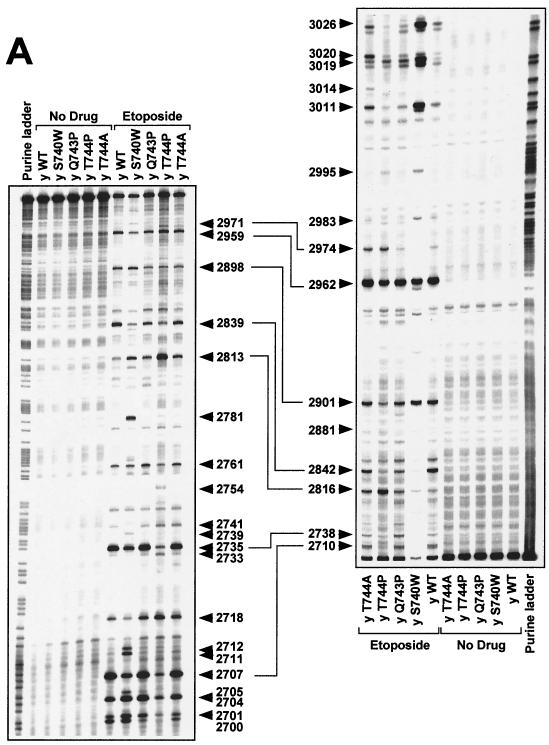
DNA cleavage patterns generated by yeast Top2pWT, Top2pS740W, Top2pQ743P, Top2pT744P, and Top2pT744A enzymes in the presence of etoposide, the fluoroquinolone CP-115,953, and the intercalator mitoxantrone. DNA fragments from the junction between the c-myc first intron and first exon between positions 2671 and 3072 were prepared by PCR using one primer labeled with 32P at the 5′ terminus. The left-hand panels show labeling of the upper DNA strand at position 2671. The right-hand panels show that labeling of the lower DNA strand was at position 3072. Top2 enzymes are indicated above each lane. Lanes: y WT, yeast wild-type Top2p; y S740W, yeast Top2pS740W; y Q743P, yeast Top2pQ743P; y T744P, yeast Top2pT744P; y T744A, yeast Top2pT744A. Concentrations were 100 μM for etoposide (A) and CP-115,953 (B) and 1 μM for mitoxantrone (C). Reactions were performed at 37°C for 30 min in the presence of 5 mM MgCl2. Purine ladders were obtained after formic acid reaction. Numbers correspond to genomic positions of the nucleotide covalently linked to Top2p via the 5′ phosphate. Double-headed arrows correspond to DNA cleavage sites with a 4-bp stagger that represent potential DNA double-strand breaks.
Comparison of yeast Top2pT744Pand E. coli gyrase DNA cleavage sites generated in the presence of quinolones.
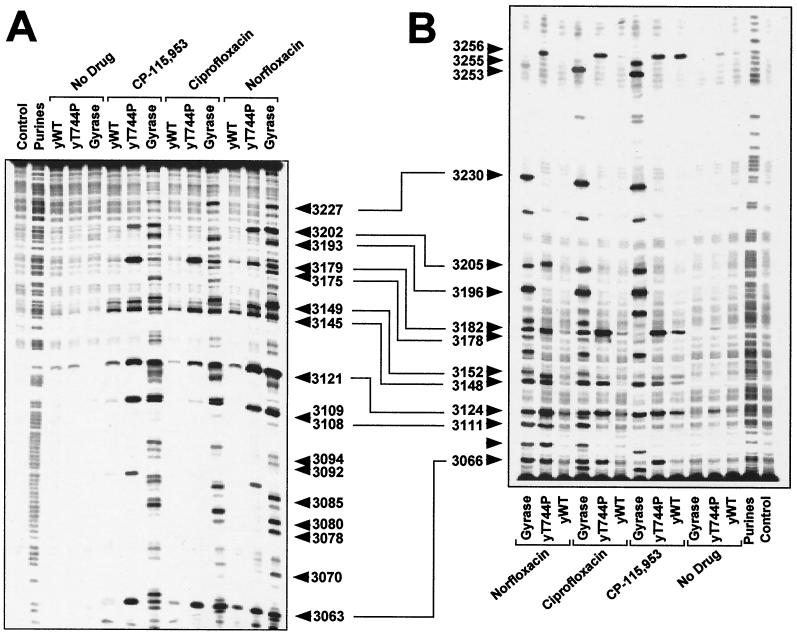
In contrast to the mutation causing Ser740Trp, which confers quinolone resistance, the Thr744Pro substitution causes hypersensitivity to CP-115,953 (14) (Fig. 3). Figure 5 compares DNA cleavage produced by CP-115,953, ciprofloxacin, and norfloxacin for GyrAWT, Top2pWT, and Top2pT744P. Remarkably, prominent cleavage was observed for Top2pT744P in the presence of ciprofloxacin and norfloxacin, whose antibacterial activities are due to specific inhibition of bacterial Top2p and have no effect on mammalian Top2p.
FIG. 5.
Generation of DNA cleavage by the yeast Top2pT744P mutant in the presence of antibacterial quinolones. A 254-bp DNA fragment from the c-myc first intron was prepared. (A) Labeling of the upper DNA strand at position 3035. (B) Labeling of the lower DNA strand at position 3288. Drugs (100 μM each) and enzymes are indicated above the lanes. Lanes: yWT, yeast wild-type Top2p; Y T744P, Top2pT744P. Numbers correspond to genomic positions of the nucleotide covalently linked to Top 2p. Double-headed arrows correspond to DNA cleavage sites with a 4-bp stagger that represent potential DNA double-strand breaks.
A number of cleavage sites stimulated by Top2pT744P in the presence of quinolones matched the E. coli DNA gyrase sites (e.g., at positions 3202, 3179, 3145, 3121, 3108, 3066, and 3111). However, other sites were different for DNA gyrase. Enhanced sensitivity to antibacterial quinolones by the Thr744Pro substitution in yeast Top2p was also detected in other DNA fragments (data not shown).
Base preference of the CP-115,953-stabilized cleavage complexes for mutant Top2T744P enzyme.
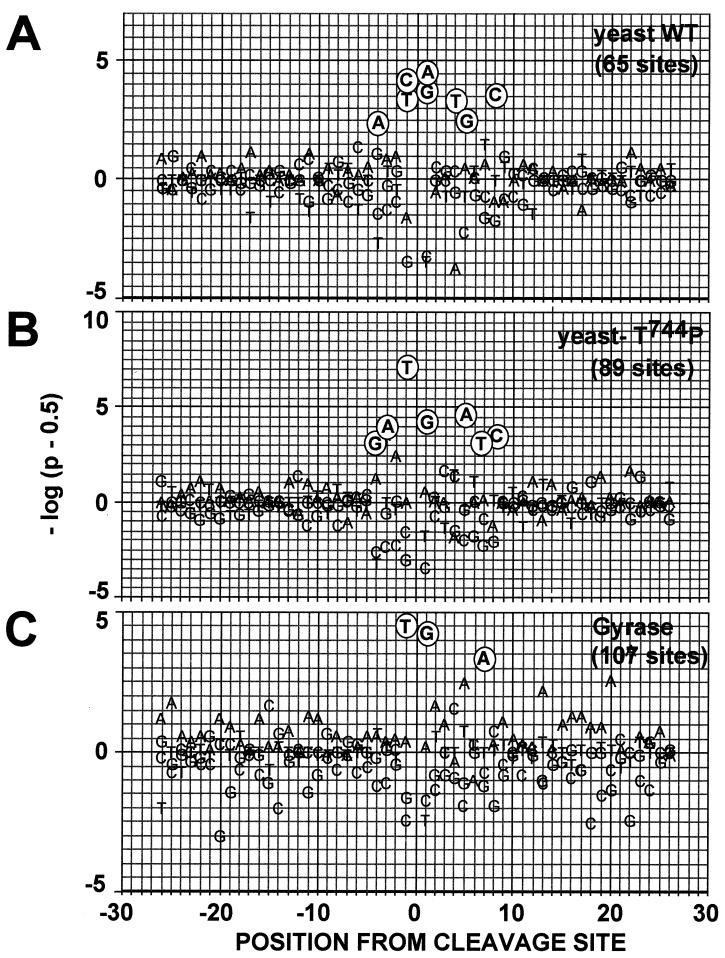
Because we observed that the Thr744Pro substitution enhanced sensitivity to quinolones (including the fluoroquinolone CP-115,953) and produced common DNA cleavage sites with E. coli DNA gyrase, we examined whether this particular amino acid substitution affected the DNA base preference of yeast Top2p in the presence of CP-115,953. Cleavage sites for the three c-myc DNA fragments and one c-jun fragment (see Materials and Methods) (Fig. 6) were analyzed for both DNA strands. For the wild-type yeast Top2p, CP-115,953 preferentially stabilized sites with C or T at position −1 (C−1 + T−1: 49 out of 65 sites) and A or G at position +1 (A+1 + G+1: 54 out of 65 sites). Complementary preferences (although slightly weaker) were seen for T+4 and G+5.
FIG. 6.
Probability of the observed base frequency deviations at Top2p cleavage sites for the yeast wild-type enzyme, for Top2pT744P and for E. coli gyrase in the presence of CP-115,953. Position 0 corresponds to the cleavage site, and positions −1 and +1 to the bases immediately 3′ and 5′ from the cleavage site, respectively. The panels present the probability of the observed base frequency deviations from expectation for the indicated enzyme. On the y axis, P is the probability of observing that deviation or more, either as excess (above baseline) or deficiency (below baseline), relative to the expected frequency of each individual base (48). Cleavage sites for the yeast wild-type Top2p (A), the yeast Top2pT744P (B), and E. coli gyrase (C) were analyzed after treatment with 100 μM CP-115,953.
Top2pT744P also demonstrated a preference for T at position −1, which was higher than that for the wild-type enzyme (41 out of 89 sites), in combination with preference for G at position +1 (44 out of 89 sites). However, preferences for C at position −1 and A at position +1 were not detectable in the mutant protein (Thr744Pro). Moreover, complementary preference also changed to A+5. Because bacterial Top2 enzymes are also sensitive to CP-115,953 (Fig. 5), we analyzed the base preferences for DNA gyrase. The observed preferences for T at position −1 (41 out of 107 sites) and G at position +1 (50 out of 107 sites) are in agreement with previous reports obtained with the antibacterial fluoroquinolones (12, 17, 21). These data indicate that enhanced quinolone sensitivity of the mutation causing Thr744Pro is associated with a shift in base sequence preferences, resembling that of DNA gyrase.
DISCUSSION
The A′ domains of eukaryotic Top2 enzymes (homologous to the amino terminus of the A subunit of gyrase) comprise the primary DNA binding sites and include the catalytic tyrosine Y782 for the yeast S. cerevisiae (59), which is homologous to Y122 for E. coli gyrase (6, 25, 39). DNA footprinting analyses indicate that for eukaryotic Top2p and bacterial topoisomerase IV, ∼15 to 35 bp of DNA is tightly bound to the enzyme (30, 47). In the case of gyrase, a more extensive stretch of DNA (∼130 bp) is protected by the enzyme (43). However, the central section of this region around the DNA cleavage site is very similar in size to that bound by Top2p and topoisomerase IV (12, 21, 39). The length of this DNA segment is consistent with the combined length of the A′ grooves present in the Top2p and GyrA structures (6, 39). Within the A′ domain of Top2p, a 29-kDa fragment containing the active-site tyrosine and the helix-turn-helix motif from the CAP homology domain can be cross-linked to DNA (27), and protein footprinting suggest that the binding of DNA protects lysines from chemical modification within the same region (31). CAP homology domains contain typically a three-alpha-helix bundle backed by β-sheets. Two adjacent alpha-helices, usually the second helix (referred to as α3-helix in yeast Top2p according to reference 6)and the third helix (referred to as α4-helix of the bundle in yeast Top2p according to reference 6), are connected by a short turn. This motif known as the helix-turn-helix is responsible for many of the critical contacts between CAP-like proteins and DNA. The second helix of this motif generally inserts into the major groove of DNA with the turn contacting the phosphodiester backbone (5, 23, 40) (Fig. 1).
Our observations suggest that specific substitutions in the α4-helix (Thr744Pro, Gln743Pro, and Ser740Trp of eukaryotic Top2p), which corresponds to the third helix of the CAP homology domain, can change the enzyme-DNA interactions, as reflected by differences in DNA cleavage patterns induced in the presence of Ca2+ or in the presence of intercalating and nonintercalating drugs. We also observed differences in base sequence preferences. We interpret these results as an indication that the α4-helix of the Top2p CAP homology domain is critical for DNA sequence recognition prior to cleavage of the G strand, which is consistent with the proposed binding of DNA to yeast Top2p with the α4-helix close to the catalytic tyrosine and to the ends of the cleaved DNA (6). Figure 1 presents the positions of Ser740, Gln743, and Thr744 in the yeast Top2 protein (4, 6). These conserved residues (Fig. 1C) tend to be buried inside the protein, while the nonconserved residues Ser740 and Gln743 are more solvent accessible (22) (Fig. 1B). The nonconserved amino acid residues might contribute to the observed differences in DNA sequence selectivity and drug sensitivity between the bacterial, the yeast, and the human Top2 enzymes (22, 55).
Other regions of Top2p must interact with different DNA segments, such as the T segment for the strand passage reaction. The B′ domain (homologous to B subunit of gyrase) is also essential for DNA binding and cleavage (7, 19, 20, 31, 35). Recently it was proposed that DNA breakage and rejoining by Top2p involve the coordinated action of the CAP homology domain in the A′ fragment containing the active-site tyrosine together with the Rossmann fold in the B′ fragment (5, 16, 35; J. G. Heddle, and A. Maxwell, submitted for publication). The Rossmann fold, which is important for DNA binding and cleavage reactions, contains a number of highly conserved acidic residues that probably bind divalent metals (5, 16, 35). Structural and biochemical studies suggest that the active-site tyrosine in the DNA-binding domain of one protomer cooperates with several residues in the Rossmann fold of the other protomer (16, 35). Our data suggest that not only the active-site tyrosine and the Rossmann fold domain but also the α4-helix of the CAP domain are involved in the concerted molecular actions, including DNA interactions leading to sequence-specific cleavage of the G-strand duplex DNA.
We have shown that the Ca2+- and drug-generated DNA cleavage patterns of Top2pT744P and Top2p Q743P are different from those of Top2pT744A and Top2pWT and that Top2pT744A is comparable to Top2pWT. Since the Top2p Q743P substitution does not lead to enhanced drug sensitivity (14), the change in the observed pattern of Top2p cleavage sites cannot arise solely from global quantitative alterations. Also, since Top2pT744A does not have a pattern of cleavage that differs from the wild-type enzyme, Thr744 probably does not play a crucial role in determining cleavage site selection by the enzyme. Since detailed structural data for any of the mutant proteins are not yet available, we modeled the specific molecular changes that might result from the Thr744Ala/Pro and Gln743Pro substitutions in the yeast Top2p structure (6). Molecular dynamics simulations of the Thr744Pro mutant Top2p predicted structural changes within the CAP homology domain (data not shown; see also reference 14). Compared to wild-type Top2p and Top2pT744A, the Thr744Pro substitution was associated with a kink in the α4-helix and a bending angle of approximately 37o. The helical structure was preserved on both sides of the kink (data not shown). These simulations suggest that an alteration in the secondary structure of the α4-helix can alter protein-DNA interactions. It is important to note, however, that changes in other parts of the protein also occurred. Therefore, it is premature to conclude that all of the biochemical alterations we observed in the mutant proteins arise solely from changes in the α4-helix.
A key issue in understanding the mechanism of action of Top2p targeting drugs is the determination of where the drugs bind in the Top2p/DNA complex. Our present findings indicate that the same Top2p domain that presumably interacts with DNA is also critical for drug interaction in the Top2p/DNA/drug ternary complex. In particular, the solvent-exposed residues Ser740, Gln743, and to a lesser extent Thr744, which affect drug activity, are important, directly or indirectly, for drug binding to the Top2p/DNA complex (22).
An intriguing finding arising from this work is that a quinolone-hypersensitive yeast Top2p mutant, resulting in Thr744Pro substitution, changed the DNA cleavage pattern in the presence of quinolones so that it more closely resembled the DNA cleavage pattern of a quinolone-sensitive prokaryotic topoisomerase II. Quinolones act against eukaryotic topoisomerase II by accelerating the rate of DNA cleavage (51). Recent studies have shown that along with gyrase and topoisomerase IV of E. coli, quinolones can also inhibit religation (1, 28). A plausible model is that the quinolone-hypersensitive mutant allows quinolones to inhibit religation as well. By this hypothesis, the action of quinolones against Top2pThr744Pro more closely resembles the action of quinolones against prokaryotic topoisomerase II, demonstrating conservation of mechanisms of drug action between eukaryotic and prokaryotic type II topoisomerases. Thus, the quinolone-protein interaction domains between the prokaryotic and eukaryotic enzymes may be comparable, with only limited differences preventing antibacterial quinolones from acting against eukaryotic Top2p.
Preparation of end-labeled DNA fragments by PCR.
Three sets of labeled DNA fragments were prepared from the human c-myc gene by PCR. A 254-bp DNA fragment from the first intron was prepared between positions 3035 and 3288 (GenBank accession X00364) using the following oligonucleotides: 5′-GTAATCCAGAACTGGATCGG-3′ for the upper strand and 5′-ATGCGGTCCCTACTCCAAGG-3′ for the lower strand (annealing temperature, 56°C). A 401-bp DNA fragment from the junction between the first intron and the first exon was prepared between positions 2671 and 3072 using the following oligonucleotides: 5′-TGCCGCATCCACGAAACTTT-3′ for the upper strand and 5′-TTGACAAGTCACTTTACCCC-3′ for the lower strand (annealing temperature, 60°C). A 480-bp fragment from the first exon containing promoters P1and P2 was prepared between positions 2265 and 2745 using the following oligonucleotides: 5′-GATCCTCTCTCGCTAATCTCCGCCC-3′ for the upper strand and 5′-TCCTTGCTCGGGTGTTGTAAGTTCC-3′ for the lower strand (annealing temperature, 70°C). A 213-bp fragment from the human c-jun gene was prepared between positions 5′-TGTTGACAGCGGCGGAAAGCAGS-3′ for the upper strand and 5′-CGTCCTTCTTCTCTTGCGTGGCTCT-3′ for the lower strand (annealing temperature, 64°C). Single-end labeling of these DNA fragments was obtained by 5′-end labeling of the specific primer oligonucleotide. Ten picomoles of DNA was incubated for 60 min at 37°C with 10 U of T4 polynucleotide kinase and 10 pM [γ-32P]ATP (100 μCi) in kinase buffer (70 mM Tris-HCl, pH 7.6; 0.1 M KCl; 10 mM MgCl2; 5 mM dithiothreitol; bovine serum albumin [0.5 mg/ml]). Reactions were stopped by heat denaturation at 70°C for 15 min. After purification using Sephadex G-25 columns (Boehringer Mannheim), the labeled oligonucleotides were used for PCR. Approximately 0.1 μg of the c-myc DNA that had been restricted by SmaI and PvuII (fragment spanning positions 2265 to 2745) and XhoI and XbaI (fragment spanning positions 2671 to 3072 and fragment 3035 to 3288) was used as a template for the PCR. Ten picomoles of each oligonucleotide primer, one of them being 5′ labeled, was used in 22 temperature cycle reactions (each cycle with 94°C for 1 min, annealing for 1 min, and 72°C for 2 min). The last extension was for 10 min, and DNA was purified using PCR Select-II columns (5Prime-3Prime Inc., Boulder, Colo.).
Overexpression and purification of yeast topoisomerase II and E. coli DNA gyrase.
Wild-type yeast Top2p, Ser740Trp, Gln743Pro, Thr744Pro, and Thr744Ala proteins were overexpressed using YEpTOP2-PGAL1 or YEptop2-S*W-PGAL1 and yeast strain JEL1t1− (32)and purified to homogeneity as previously described (59). The detailed procedure has been described elsewhere (15). GyrA (gift of N. A. Gormley), GyrAS83W (gift of C. J. R. Willmott), and GyrB were purified as described previously (38). GyrAS83W was shown to have the expected level of quinolone-resistant supercoiling activity when complexed with wild-type gyrB.
Topoisomerase II-generated DNA cleavage reactions.
For the yeast Top2p experiments, DNA fragments (5 × 104 to 10 × 104 dpm/reaction) were equilibrated with drug or with the corresponding 1% dimethyl sulfoxide concentration in 10 mM Tris-HCl (pH 7.5)-50 mM KCl-5 mM MgCl2-2 mM dithiothreitol-0.1 mM Na2EDTA-1 mM ATP-bovine serum albumin (15 μg/ml) for 5 min before the addition of 8 U (80 ng) of purified Top2p in a 10-μl final reaction volume. Unless otherwise indicated, reactions were for 30 min at 37°C. Reactions were stopped by adding EDTA and sodium dodecyl sulfate (25 mM and 1% final concentrations, respectively) and were further digested with proteinase K (0.4-mg/ml final concentration for 30 min at 55°C). For the DNA gyrase experiments, similar conditions were used except that the DNA cleavage reactions were performed with 10 to 15 U of purified E. coli gyrase for 120 min at 25°C (60).
Electrophoresis and base sequence analysis.
For DNA sequence analysis, samples were precipitated with ethanol and resuspended in 5 μl of loading buffer (80% formamide, 10 mM NaOH, 1 mM EDTA, 0,1% xylene cyanol, 0.1% bromphenol blue). Samples were heated to 95°C for 5 min and thereafter loaded onto DNA sequencing gels (7% polyacrylamide; 19:1 acrylamide-bisacrylamide) containing 7 M urea in 1× Tris-borate-EDTA buffer. Electrophoresis was performed at 2,500 V (60 W) for 2 to 3 h. The gels were dried on Whatman no. 3MM paper sheets and visualized using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and ImageQuant software. The determination of preferred bases around Top2p cleavage sites was done as described previously (8, 49, 57).
Molecular dynamics simulation.
Modeling of the Thr744Pro substitution into published crystal structure data of a 92-kDa fragment of yeast Top2p (6) (PDB identification code 1bgw) was performed with the molecular modeling package QUANTA (version 96) run on both an SGI Crimson and an SGI Octane workstation. Energy calculations and related operations, such as energy minimizations and the molecular dynamics simulations, were carried out with QUANTA's associated molecular mechanics program CHARMm, version 23.1, run on a separate computation host, a Digital 2100 AlphaServer 4/275, using the MSI parameter set version 22.0.
In order to generate a low-energy starting geometry for the subsequent molecular dynamics simulation, each of the wild-type and mutant structures was energy minimized in 5,000 steps of Adopted-Basis Newton-Raphson minimization. Each of these minimized structures was subjected to a total of 300 ps of molecular dynamics simulation using the Verlet algorithm for integration of the equations of motion. Using a step size of 1 fs, the structures were heated from 0 to 310 K in approximately 10 ps (9,982 steps), then equilibrated at 310 K for 40 ps (40,000 steps) with an equilibration frequency of 200 steps, and finally subjected to a simulation at 310 K for 250 ps (250,000 steps). Snapshot frames containing the current coordinates were written out every 50 steps.
RESULTS
Alteration of Ca2+-promoted DNA cleavage by Ser740Trp, Gln743Pro, Thr744Pro, and Thr744Ala substitutions in yeast Top2p and by the Ser83Trp substitution in bacterial GyrA.
It is well known that DNA cleavage by type II DNA topoisomerases can be stimulated by a variety of drugs (2, 18, 33, 34, 36, 37, 52), but cleavage can also be stimulated when Ca2+ is substituted for Mg2+ in the enzyme reaction (45, 50). In this case it is likely that the cleavage specificity reflects the natural sequence specificity of the enzyme. The Ca2+-promoted, drug-independent DNA cleavage sites generated by yeast wild-type Top2p (Top2pWT) and four yeast mutant proteins (Top2pS740W, Top2pQ743P, Top2pT744P, and Top2pT744A) were mapped in two different c-myc DNA fragments. Figure 2A
To further investigate differences between Top2pS740W, Top2pQ743P, Top2pT744P, Top2pT744A, and Top2pWT, we compared drug-stimulated DNA cleavage patterns. Etoposide results are shown in Fig. 3A. Results for the fluoroquinolone CP-115,953 in are shown in Fig. 3B, and those for mitoxantrone are shown in Fig. 3C.
Top2pS740W (as well as the corresponding yeast mutant) has been previously characterized as being resistant to the fluoroquinolone CP-115,953 and hypersensitive to etoposide (26, 56). Compared with Top2pWT and the other mutant proteins, several cleavage sites stimulated by etoposide were markedly enhanced with Top2pS740W, e.g., at positions 2711 to 2712, 2781, 2983, 3011, 3019, and 3026 (Fig. 3A).
Proline substitutions at positions 744 and 743 also changed the distribution of Top2p-mediated DNA cleavages. These effects were more pronounced for Top2pT744P than for Top2pQ743P (Fig. 3). By contrast, Top2pT744A behaved very similarly to Top2pWT (Fig. 3).
Together, the data shown in Fig. 3 demonstrate that single mutations in the α4-helix of Top2p (Ser740Trp, Gln743Pro, and Thr744Pro) changed enzyme-specific DNA cleavage patterns produced by nonintercalating and intercalating drugs.
Differences in quinolone-generated DNA cleavage patterns by the mutant GyrAS83W and wild-type DNA gyrase.
We next tested the role of the corresponding α4-helix in the bacterial topoisomerase II, DNA gyrase. Figure 4
FIG. 4.
DNA cleavage patterns generated by gyraseWT and mutant GyrAS83W in the presence of the quinolones CP-115,953 and ciprofloxacin. The same DNA fragments presented in Fig. 3 were used. The left-hand panel shows labeling of the upper DNA strand; the right-hand panel shows labeling of the lower DNA strand. Drugs and enzymes are indicated above each lane. Concentrations were 100 μM for CP-115,953 and ciprofloxacin, respectively. Cleavage reactions were performed at 25°C for 2 h in the presence of 5 mM MgCl2. Purine ladders were obtained after formic acid reaction. Lanes: Control, no Top2p, no drug treatment; Gyr WT: gyraseWT from E. coli; GyrAS83W, mutant Ser83Trp gyrase. Numbers correspond to genomic positions of the nucleotide covalently linked to Top2p. Double-headed arrows correspond to DNA cleavage sites with a 4-bp stagger that represent potential DNA double-strand breaks.
Etoposide (VP16) was obtained from the Bristol-Myers Squibb Co., Wallingford, Conn. Mitoxantrone was from the Drug Synthesis and Chemistry Branch (National Cancer Institute, Bethesda, Md.). Azatoxin was provided by T. Macdonald, Department of Chemistry, University of Virginia, Charlottesville. CP-115,953 was a gift from P. R. McGuirk and T. D. Gootz of Pfizer Laboratories. Drug stock solutions were made in dimethyl sulfoxide at 10 mM. Further dilutions were made in distilled water immediately before use. Human c-myc inserted into pBR322, restriction enzymes, T4 polynucleotide kinase, polyacrylamide-bisacrylamide, and Taq DNA polymerase were purchased from Lofstrand Labs (Gaithersburg, Md); Life Technologies, Inc. (Gaithersburg, Md.); New England Biolabs (Beverly, Mass.), or Qiagen Inc. (Valencia, Calif.). [γ-32P]ATP (6,000 Ci/mmol) was purchased from DuPont NEN (Boston, Mass.). PCR oligonucleotide primers were obtained from GIBCO BRL (Gaithersburg, Md.).
Acknowledgments
Dirk Strumberg was supported by the Deutsche Forschungsgemeinschaft (grant Str 527/2-1), Bonn, Germany. John L. Nitiss was supported by grants from the National Cancer Institute (CA52814 and CA21765) and the American Lebanese Syrian Associated Charities (ALSAC). Jonathan G. Heddle was supported by the Medical Research Council (United Kingdom).
REFERENCES
- 1.Anderson, V. E., T. D. Gootz, and N. Osheroff. 1998. Topoisomerase IV catalysis and the mechanism of quinolone action. J. Biol. Chem. 273:17879-17885. [DOI] [PubMed] [Google Scholar]
- 2.Andoh, T., and R. Ishida. 1998. Catalytic inhibitors of DNA topoisomerase II. Biochim. Biophys. Acta 1400:155-171. [DOI] [PubMed] [Google Scholar]
- 3.Beck, W. T., M. K. Danks, J. S. Wolverton, R. Kim, and M. Chen. 1993. Drug resistance associated with altered DNA topoisomerase II. Adv. Enzyme Regul. 33:113-127. [DOI] [PubMed] [Google Scholar]
- 4.Berger, J. 1998. Type II DNA topoisomerases. Curr. Opin. Struct. Biol. 8:26-32. [DOI] [PubMed] [Google Scholar]
- 5.Berger, J. M., D. Fass, J. C. Wang, and S. C. Harrison. 1998. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc. Natl. Acad. Sci. USA 95:7876-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, J. M., S. J. Gamblin, S. C. Harrison, and J. C. Wang. 1996. Structure and mechanism of DNA topoisomerase II. Nature 379:225-232. [DOI] [PubMed] [Google Scholar]
- 7.Brown, P. O., C. L. Peebles, and N. R. Cozzarelli. 1979. A topoisomerase from Escherichia coli related to DNA gyrase. Proc. Natl. Acad. Sci. USA 76:6110-6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capranico, G., F. Zunino, K. W. Kohn, and Y. Pommier. 1990. Sequence-selective topoisomerase II inhibition by anthracycline derivatives in SV40 DNA: relationship with DNA binding affinity and cytotoxicity. Biochemistry 29:562-569. [DOI] [PubMed] [Google Scholar]
- 9.Champoux, J. J. 2001. DNA Topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 10.Chen, A. Y., and L. F. Liu. 1994. DNA topoisomerases: essential enzymes and lethal targets. Annu. Rev. Pharmacol. Toxicol. 34:191-218. [DOI] [PubMed] [Google Scholar]
- 11.Corbett, A. H., D. Hong, and N. Osheroff. 1993. Exploiting mechanistic differences between drug classes to define functional drug interaction domains on topoisomerase II. Evidence that several diverse DNA cleavage-enhancing agents share a common site of action on the enzyme. J. Biol. Chem. 268:14394-14398. [PubMed] [Google Scholar]
- 12.Cove, M. E., A. P. Tingey, and A. Maxwell. 1997. DNA gyrase can cleave short DNA fragments in the presence of quinolone drugs. Nucleic Acids Res. 25:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen, M. E., A. W. Wyke, R. Kuroda, and L. M. Fisher. 1989. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob. Agents Chemother. 33:886-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, J., J. Walker, and J. L. Nitiss. 2000. A mutation in yeast topoisomerase II that confers hypersensitivity to multiple classes of topoisomerase II poisons. J. Biol. Chem. 275:7980-7987. [DOI] [PubMed] [Google Scholar]
- 15.Elsea, S. H., Y. Hsiung, J. L. Nitiss, and N. Osheroff. 1995. A yeast type II topoisomerase selected for resistance to quinolones. Mutation of histidine 1012 to tyrosine confers resistance to nonintercalative drugs but hypersensitivity to ellipticine. J. Biol. Chem. 270:1913-1920. [DOI] [PubMed] [Google Scholar]
- 16.Fass, D., C. E. Bogden, and J. M. Berger. 1999. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Biol. 6:322-326. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, L. M., K. Mizuuchi, M. H. O'Dea, H. Ohmori, and M. Gellert. 1981. Site-specific interaction of DNA gyrase with DNA. Proc. Natl. Acad. Sci. USA 78:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froelich-Ammon, S. J., and N. Osheroff. 1995. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 270:2142. 9-32. [DOI] [PubMed] [Google Scholar]
- 19.Funatsuki, K., R. Tanaka, S. Inagaki, H. Konno, K. Katoh, and H. Nakamura. 1997. AcrB mutation located at carboxyl-terminal region of gyrase B subunit reduces DNA binding of DNA gyrase. J. Biol. Chem. 272:13302-13308. [DOI] [PubMed] [Google Scholar]
- 20.Gellert, M., L. M. Fisher, and M. H. O'Dea. 1979. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc. Natl. Acad. Sci. USA 76:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gmunder, H., K. Kuratli, and W. Keck. 1997. In the presence of subunit A inhibitors DNA gyrase cleaves DNA fragments as short as 20 bp at specific sites. Nucleic Acids Res. 25:604-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammonds, T. R., A. Maxwell, and J. R. Jenkins. 1998. Use of a rapid throughput in vivo screen to investigate inhibitors of eukaryotic topoisomerase II enzymes. Antimicrob. Agents Chemother. 42:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison, S. C. 1991. A structural taxonomy of DNA-binding domains. Nature 353:715-719. [DOI] [PubMed] [Google Scholar]
- 24.Hooper, D. C. 1998. Clinical applications of quinolones. Biochim. Biophys. Acta 1400:45-61. [DOI] [PubMed] [Google Scholar]
- 25.Horowitz, D. S., and J. C. Wang. 1987. Mapping the active site tyrosine of Escherichia coli DNA gyrase. J. Biol. Chem. 262:5339-5344. [PubMed] [Google Scholar]
- 26.Hsiung, Y., S. H. Elsea, N. Osheroff, and J. L. Nitiss. 1995. A mutation in yeast TOP2 homologous to a quinolone-resistant mutation in bacteria. Mutation of the amino acid homologous to Ser83 of Escherichia coli GyrA alters sensitivity to eukaryotic topoisomerase inhibitors. J. Biol. Chem. 270:20359-20364. [DOI] [PubMed] [Google Scholar]
- 27.Hung, F., D. Luo, D. M. Sauve, M. T. Muller, and M. Roberge. 1996. Characterization of topoisomerase II-DNA interaction and identification of a DNA-binding domain by ultraviolet laser crosslinking. FEBS Lett. 380:127-132. [DOI] [PubMed] [Google Scholar]
- 28.Kampranis, S. C. M., A. 1998. The DNA gyrase-quinolone complex: ATP hydrolysis and the mechanism of DNA cleavage. J. Biol. Chem. 273:22615-22626. [DOI] [PubMed] [Google Scholar]
- 29.Kingma, P. S., D. A. Burden, and N. Osheroff. 1999. Binding of etoposide to topoisomerase II in the absence of DNA: decreased affinity as a mechanism of drug resistance. Biochemistry 38:3457-3461. [DOI] [PubMed] [Google Scholar]
- 30.Lee, M. P., M. Sander, and T. Hsieh. 1989. Nuclease protection by Drosophila DNA topoisomerase II. Enzyme/DNA contacts at the strong topoisomerase II cleavage sites. J. Biol. Chem. 264:21779-21787. [PubMed] [Google Scholar]
- 31.Li, W., and J. C. Wang. 1997. Footprinting of yeast DNA topoisomerase II lysyl side chains involved in substrate binding and interdomainal interactions. J. Biol. Chem. 272:31190-31195. [DOI] [PubMed] [Google Scholar]
- 32.Lindsley, J. E., and J. C. Wang. 1991. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc. Natl. Acad. Sci. USA 88:10485-10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, L. 1994. DNA topoisomerases: topoisomerase-targeting drugs. Academic Press, New York, N.Y.
- 34.Liu, L. F., and P. D'Arpa. 1992. Topoisomerase-targeting antitumor drugs: mechanisms of cytotoxicity and resistance. Important Adv. Oncol: 79-89. [PubMed]
- 35.Liu, Q., and J. C. Wang. 1999. Similarity in the catalysis of DNA breakage and rejoining by type IA and IIA DNA topoisomerases. Proc. Natl. Acad. Sci. USA 96:881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxwell, A. 1999. DNA gyrase as a drug target. Biochem. Soc. Trans. 27:48-53. [DOI] [PubMed] [Google Scholar]
- 37.Maxwell, A. 1992. The molecular basis of quinolone action. J. Antimicrob. Chemother. 30:409-414. [DOI] [PubMed] [Google Scholar]
- 38.Maxwell, A. 1999. Overexpression and purification of bacterial DNA gyrase, p. 135-144. In M.-A. Bjornsti (ed.), Protocols for DNA topoisomerases I. DNA topology and enzyme purification. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 39.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, H. C. 1995. Structure and function of DNA-binding proteins. Curr. Opin. Genet. Dev. 5:180-189. [DOI] [PubMed] [Google Scholar]
- 41.Nitiss, J. L. 1998. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta 1400:63-81. [DOI] [PubMed] [Google Scholar]
- 42.Oram, M., and L. M. Fisher. 1991. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob. Agents Chemother. 35:387-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orphanides, G., and A. Maxwell. 1994. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 22:1567-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osheroff, N. 1987. Role of the divalent cation in topoisomerase II mediated reactions. Biochemistry 26:6402-6406. [DOI] [PubMed] [Google Scholar]
- 45.Osheroff, N., and E. L. Zechiedrich. 1987. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: trapping the covalent enzyme-DNA complex in an active form. Biochemistry 26:4303-4309. [DOI] [PubMed] [Google Scholar]
- 46.Peng, H., and K. J. Marians. 1995. The interaction of Escherichia coli topoisomerase IV with DNA. J. Biol. Chem. 270:25286-25290. [DOI] [PubMed] [Google Scholar]
- 47.Pommier, Y. 1997. DNA topoisomerase II inhibitors, p. 153-174. In B. A. Teicher (ed.), Cancer therapeutics: experimental and clinical agents. Humana Press, Totowa, N.J.
- 48.Pommier, Y., G. Capranico, A. Orr, and K. W. Kohn. 1991. Local base sequence preferences for DNA cleavage by mammalian topoisomerase II in the presence of amsacrine or teniposide. Nucleic Acids Res. 19:5973-5980. (Erratum, 19: 7003.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reece, R. J., and A. Maxwell. 1989. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264:19648-19653. [PubMed] [Google Scholar]
- 50.Robinson, M. J., B. A. Martin, T. D. Gootz, P. R. McGuirk, M. Moynihan, J. A. Sutcliffe, and N. Osheroff. 1991. Effects of quinolone derivatives on eukaryotic topoisomerase II. A novel mechanism for enhancement of enzyme-mediated DNA cleavage. J. Biol. Chem. 266:14585-14592. [PubMed] [Google Scholar]
- 51.Robinson, M. J., B. A. Martin, T. D. Gootz, P. R. McGuirk, and N. Osheroff. 1992. Effects of novel fluoroquinolones on the catalytic activities of eukaryotic topoisomerase II: influence of the C-8 fluorine group. Antimicrob. Agents Chemother. 36:751-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roca, J. 1995. The mechanisms of DNA topoisomerases. Trends Biochem Sci. 20:156-160. [DOI] [PubMed] [Google Scholar]
- 53.Strumberg, D., J. L. Nitiss, J. Dong, K. W. Kohn, and Y. Pommier. 1999. Molecular analysis of yeast and human type II topoisomerases. Enzyme-DNA and drug interactions. J. Biol. Chem. 274:28246-28255. [DOI] [PubMed] [Google Scholar]
- 54.Strumberg, D., J. L. Nitiss, A. Rose, M. C. Nicklaus, and Y. Pommier. 1999. Mutation of a conserved serine residue in a quinolone-resistant type II topoisomerase alters the enzyme-DNA and drug interactions. J. Biol. Chem. 274:7292-7301. [DOI] [PubMed] [Google Scholar]
- 55.Tanizawa, A., K. W. Kohn, and Y. Pommier. 1993. Induction of cleavage in topoisomerase I c-DNA by topoisomerase I enzymes from calf thymus and wheat germ in the presence and absence of camptothecin. Nucleic Acids Res. 21:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 57.Wang, J. C. 1998. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31:107-144. [DOI] [PubMed] [Google Scholar]
- 58.Willmott, C. J., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Worland, S. T., and J. C. Wang. 1989. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 264:4412-4416. [PubMed] [Google Scholar]
- 60.Yoshida, H., T. Kojima, J. Yamagishi, and S. Nakamura. 1988. Quinolone-resistant mutations of the GyrA gene of Escherichia coli. Mol. Gen. Genet. 211:1-7. [DOI] [PubMed] [Google Scholar]