Abstract
Fifty-six azithromycin-resistant (MICs, 2.0 to 4.0 μg/ml) Neisseria gonorrhoeae strains with cross-resistance to erythromycin (MICs, 2.0 to 64.0 μg/ml), isolated in Canada between 1997 and 1999, were characterized, and their mechanisms of azithromycin resistance were determined. Most (58.9%) of them belonged to auxotype-serotype class NR/IB-03, with a 2.6-mDa plasmid. Based on resistance to crystal violet (MICs ≥ 1 μg/ml), 96.4% of these macrolide-resistant strains appeared to have increased efflux. Nine of the eleven strains selected for further characterization were found to have a promoter region mtrR mutation, a single-base-pair (A) deletion in the 13-bp inverted repeat, which is believed to cause overexpression of the mtrCDE-encoded efflux pump. The two remaining macrolide-resistant strains (erythromycin MIC, 64.0 μg/ml; azithromycin MIC, 4.0 μg/ml), which did not have the mutation in the mtrR promoter region, were found to have a C2611T mutation (Escherichia coli numbering) in the peptidyltransferase loop in domain V of the 23S rRNA alleles. Although mutations in domain V of 23S rRNA alleles had been reported in other bacteria, including E. coli, Streptococcus pneumoniae, and Helicobacter pylori, this is the first observation of these mutations associated with macrolide resistance in N. gonorrhoeae.
Erythromycin is sometimes used to treat gonorrhea if a patient has a cephalosporin allergy or a history of immediate and/or anaphylactic reaction to penicillins (23). In some parts of the world, azithromycin, a 15-membered macrolide, is used to treat gonorrhea (1 g orally in a single dose) (29), but this dosage is not recommended for such use in Canada (10). In Canada, azithromycin (1 g dose) is used either to treat Chlamydia trachomatis infections or in conjunction with expanded spectrum cephalosporins or quinolones to treat coinfections of gonococcus and C. trachomatis (10). The increased use of azithromycin in treating other diseases may increase the selective pressure for macrolide-resistant gonococcus.
Macrolides such as erythromycin and azithromycin act by binding to the 50S subunit of bacterial ribosomes and restrain protein synthesis by inhibiting the elongation of peptide chains (11). The mechanisms of resistance to macrolides include efflux of these antibiotics and modification of the ribosomal target by modification of enzymes or mutations to reduce the affinity of the antibiotics for ribosomes.
A number of bacterial efflux systems which confer macrolide resistance have been described (18). The first efflux pump described for Neisseria gonorrhoeae was the MtrC-MtrD-MtrE system, encoded by the mtrRCDE operon, which exports hydrophobic agents including dyes such as crystal violet (29) and macrolides such as azithromycin and erythromycin (20). A fourth Mtr protein (MtrF) may also be associated with this efflux system (20). The MtrR protein regulates the expression of MtrCDE, and increased expression of the MtrCDE pump is due to either the lack of the repressor (MtrR) protein (through deletion or insertional inactivation of mtrR) or a mutation of the promoter region of this repressor gene (20). More recently, another macrolide efflux pump, encoded by the mef gene, which had originally been described for some of the gram-positive organisms (18), has now been found in clinical strains of N. gonorrhoeae (13).
Mechanisms of resistance related to modification of the ribosomal target by methylases have been described for Streptococcus pneumoniae (21), Staphylococcus aureus (7), and N. gonorrhoeae (19). There are reports of the emergence of azithromycin-resistant gonococci which show cross-resistance with erythromycin (19). One study showed that strains from Uruguay and Seattle, Washington, had acquired from other bacterial species one or more of the 23S rRNA methylases [erm(B), erm(C) and erm(F)] which modified the antimicrobial targets (19).
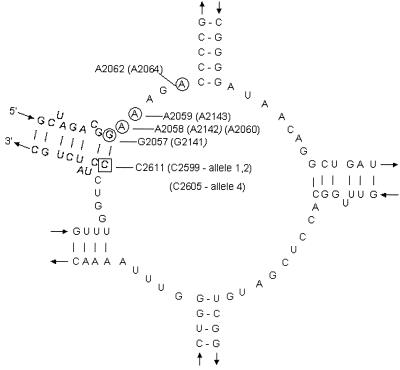
Another ribosomal modification, which involves mutations in the peptidyltransferase loop in domain V of 23S rRNA, also confers resistance to macrolides (Fig. 1). This mechanism has been described in a variety of organisms: Escherichia coli (27), Streptococcus pneumoniae (2), and Helicobacter pylori (24, 26). Mutants selected for resistance to macrolides have a substitution at nucleotide position 2058 (E. coli coordinates; GenBank accession number AJ007584) or at nearby nucleotides, suggesting their involvement in the binding of these macrolides to domain V of 23S rRNA (25, 27). Point mutations in ribosomal proteins L4 (rplD) and L22 (rplV) conferring resistance to macrolides have been described for E. coli (5, 17, 28). More recently, mutations of ribosomal protein L4 were also described for macrolide-resistant S. pneumoniae (22). Proteins L4 and L22 primarily bind to domain I of 23S rRNA, but macrolide resistance mutations in these proteins cause a change in the conformation in domains II, III, and V affecting the action of the macrolides for domain V of 23S rRNA (5).
FIG. 1.
Secondary structure of the peptidyltransferase loop in domain V of 23S rRNA. Nucleotide numbering is that of E. coli 23S rRNA (24); the corresponding N. gonorrhoeae, S. pneumoniae (2), and H. pylori (26) numbers are in parentheses. The C→T mutation at position 2611 (position 2599 in alleles 1 and 2 and position 2605 in allele 4) in N. gonorrhoeae strains 20869 and 20870, also previously reported for E. coli, is indicated by a square. The other mutations that confer macrolide drug resistance are circled: G2057 (E. coli numbering) and G2141 (H. pylori numbering); A2058 (E. coli numbering), A2142 (H. pylori numbering), and A2060 (S. pneumoniae numbering); A2143 (H. pylori numbering); and A2064 (S. pneumoniae numbering).
In 1997, we detected in Canada the emergence of azithro-mycin-resistant N. gonorrhoeae strains (MIC ≥ 2.0 μg/ml) which also have resistance to erythromycin (MIC ≥ 2.0 μg/ml). In this study, we determined the phenotypic characteristics of 56 azithromycin-resistant N. gonorrhoeae strains which were isolated in Canada between 1997 and 1999 and investigated their mechanisms of macrolide resistance.
MATERIALS AND METHODS
Bacterial strains.
Between 1997 and 1999, a total of 56 erythromycin- and azithromycin-resistant N. gonorrhoeae isolates were collected as part of the National Surveillance of N. gonorrhoeae Program in Canada. The strains used in this study originated from British Columbia, Alberta, Ontario, Québec, and Nova Scotia. N. gonorrhoeae strains were subcultured on GC medium base (GCMB) (Difco Laboratories, Detroit, Mich.) containing 0.2% BioX (QueLab, Montreal, Québec, Canada) and incubated for 24 h at 35°C in a 5% CO2 atmosphere with or without antibiotics and maintained in brain heart infusion (BHI) medium (Difco Laboratories) containing 20% glycerol and stored at −80°C. All other nonfastidious organisms were grown aerobically on BHI medium.
Antimicrobial susceptibility testing and strain characterization.
Antimicrobial susceptibilities of N. gonorrhoeae to azithromycin (compliments of Pfizer, Pointe-Claire/Dorval, Québec, Canada), cefixime (Wyeth-Ayerst Laboratories, Mason, Mich.), ceftriaxone (Roche, Laval, Québec, Canada), ciprofloxacin (compliments of Bayer, Etobicoke, Ontario, Canada), erythromycin (compliments of Lilly, Indianapolis, Ind.), penicillin (compliments of Novopharm, Scarborough, Ontario), spectinomycin (compliments of Pharmacia & Upjohn, Kalamazoo, Mich.), and tetracycline (Sigma-Aldrich Canada Ltd., Oakville) were performed according to the agar dilution method described by the National Committee for Clinical Laboratory Standards (NCCLS) (15), but the GC medium base was modified to contain 1% Kellogg's supplement (0.05 g of ferric nitrate, 1.0 g of cocarboxylase, 5.0 g of glutamine, 200.0 g of glucose in 1 liter of H2O) (8). The MICs of all antibiotics except azithromycin and erythromycin (for which there are no NCCLS MIC breakpoints) were interpreted according to the NCCLS guidelines (15). For the purposes of this study, the azithromycin and erythromycin MICs at which strains exhibited resistance were defined as ≥2.0 μg/ml (4, 21). N. gonorrhoeae reference cultures ATCC 49226, WHO III, WHO V, and WHO VII were used as quality control strains. Susceptibility to crystal violet was used as an indicator of efflux, and crystal violet susceptibility assays were performed as previously described (14).
Auxotyping of the N. gonorrhoeae strains was performed as previously described (6) on a chemically defined medium to determine their nutritional requirements for leucine (L), ornithine (O), citrulline (C), proline (P), arginine (A), hypoxanthine (H), uracil (U), and methionine (M). Prototype strains that did not require amino acid supplements for growth were designated NR (no requirements). Serotyping of the N. gonorrhoeae isolates was performed as described by Knapp et al. (9). Plasmid profiles of the N. gonorrhoeae isolates were performed as previously described (3).
PCR and DNA sequencing of the macrolide resistance targets.
PCR was used to determine the presence or absence of methylase genes and to determine mutations in the mtr efflux pump and ribosomal genes. Using PCR with primers and conditions as previously described, clinical isolates were tested for the presence of one or more of the methylase genes encoded by erm(A), erm(B), erm(C), erm(F) (19), and an efflux pump mef(A) (21). The following reference strains were used as positive controls for PCR: for erm(A), Staphylococcus aureus RN1389; for erm(B), Streptococcus pyogenes AC1; for erm(C), S. aureus RN4220; for erm(F), E. coli V831; and for mef(A), S. pyogenes 02C1064.
In an attempt to identify point mutations, sequences of genes related to macrolide resistance were determined for the mtrR promoter region, domain V of the 23S rRNA, and rplD and rplV (encoding ribosomal proteins L4 and L22, respectively) (2). The mtrR promoter region of the mtrR efflux pump operon was amplified using the published primers RPMAL#2 (5′ -ACTGAAGCTTATTTCCGGCGCAGGCAGGG-3′) and KH9#3 (5′-GACGACAGTGCCAATGCAACG-3′) (12) and sequenced in an attempt to identify mutations in this region.
To identify mutations within domain V (of the peptidyltransferase loop) for each of the four copies of the 23S rRNA gene, a two-step PCR method was developed. The first step used the gonrRNA-F primer, paired with a specific primer for each of the 23S rRNA alleles listed in Table 1, to amplify specific regions of 23S rRNA in eight strains. PCR conditions were 1 min of denaturation at 94°C, 1.5 min at 66°C (for alleles 2 and 3 of domain V) or 68°C (for alleles 1 and 4 of domain V) for annealing, and 2.5 min at 72°C for elongation for 30 cycles. The amplicons obtained were then used as templates in a second PCR using gonrRNA-F and gonrRNA-R2 primers to amplify the peptidyltransferase loop for sequencing determination. The conditions for this PCR step were 94°C at 1 min for denaturing, 59°C for 1 min for annealing, and 72°C at 1 min for elongation for 35 cycles.
TABLE 1.
Oligonucleotides used in the amplification of the four alleles of the peptidyltransferase region in domain V of the 23S rRNA
| Designation | PCR primer sequence (5′ to 3′) | Posi- tiona | Product size (bp) |
|---|---|---|---|
| gonrRNA-F | ACGAATGGCGTAACGATGGCCACA | 1954 | 712 |
| gonrRNA-R2 | TTCGTCCACTCCGGTCCTCTCGTA | 2665 | |
| rRNA (allele 1) | TCAGAATGCCACAGCTTACAAACT | +1116 | 2,054 |
| rRNA (allele 2) | GCGACCATACCAAACACCCACAGG | +1302 | 2,240 |
| rRNA (allele 3) | GATCCCGTTGCAGTGAAGAAAGTC | +1279 | 2,217 |
| rRNA (allele 4) | AACAGACTTACTATCCCATTCAGC | +909 | 1,847 |
Primer positions were determined from the N. gonorrhoeae FA1090 contig (Gonococcal Genome Sequencing Project, University of Oklahoma); allele-specific primers are downstream of the 23S rRNA.
PCR products were also sequenced in an attempt to identify point mutations in rplD and rplV genes in ribosomal proteins L4 and L22, respectively (2). All PCR amplicons were purified (Promega PCR Prep Kit; Fisher, Nepean, Ontario, Canada), and both strands were sequenced using an ABI Prism 377 DNA sequencing system (Applied Biosystems, Foster City, Calif.). DNA sequences were aligned using BLAST and GenBank (1) programs to identify the mutations of these genes.
Transformation studies.
Total genomic DNA from both clinical strain 20870 and strain 19416 was introduced into piliated N. gonorrhoeae F62 (MICs of erythromycin and azithromycin, 0.25 and 0.125 μg/ml, respectively) by transformation. As a control, a ciprofloxacin-resistant strain with a ciprofloxacin MIC of 8.0 μg/ml and with identified mutations of gyrA and parC (16) was transformed into F62. Briefly, N. gonorrhoeae F62 was suspended in 1 ml of 10 mM Tris-HCl-10 mM EDTA disodium salt (pH 8.0) to an optical density of 0.60 to 0.63 at 600 nm, inoculated on GCMB, and incubated for 1 h at 37°C in an atmosphere enriched with 7% CO2. Chromosomal or amplified DNA (1 μg) was then applied to the inoculated plate. After a 24-h incubation period, individual colonies from the GCMB plates were transferred to GCMB containing erythromycin (0.5, 1.0, and 2.0 μg/ml). Potential transformants were again subcultured onto GCMB containing erythromycin (0.5, 1.0, and 2.0 μg/ml) prior to MIC determination and examination of mutations in the 23S rRNA and the promoter region of mtrR.
In vitro conversion of resistant strains to susceptibility.
The following procedure was performed to obtain spontaneous susceptible revertants from macrolide-resistant strains. Single colonies of the macrolide-resistant cultures 20869 and 20870 (erythromycin MIC, 64.0 μg/ml; azithromycin MIC, 4.0 μg/ml) were subcultured daily for 7 days onto nonselective GCMB plates. Fifty isolated colonies were randomly selected and replicated onto GCMB plates with and without erythromycin (8.0 μg/ml), and another single colony was also subcultured onto GCMB to obtain isolated colonies. This step was repeated until colonies on replicated plates grew on GCMB but not on erythromycin plates (8.0 μg/ml) after 3 days of culturing. Mutants with decreased susceptibility were selected for MIC determination. Sequencing of the mtrR promoter region flanking mtrC and mtrR and the domain V region of the four alleles of 23S rRNA (rrl gene) of the resistant mutants and of the susceptible parent strains was also performed, and the results were compared.
In vitro mutation to erythromycin resistance.
To generate macrolide-resistant mutants, the following procedure was performed. Three susceptible N. gonorrhoeae strains (21855, 21305, and 22831) (erythromycin and azithromycin MICs, 0.5 to 1 and 0.125, respectively) were selected from our culture collection and subcultured onto nonselective GCMB plates. After a 24-h incubation period, the growth from a single colony of each strain was swabbed onto the entire surface of GCMB plates containing a gradient of erythromycin of 0 to 4.0 μg/ml. After a 24-h incubation, the plates were examined for growth and isolated colonies on the highest concentration of antibiotics were selected and streaked onto another GCMB gradient plate with a concentration of erythromycin from 0 to 4.0 μg/ml. After the selection step was repeated seven times, a gradient plate with a higher concentration of erythromycin (0 to 12.0 μg/ml) was used. Colonies in the region of the highest concentration of erythromycin were selected and subcultured onto three plates, namely, a GCMB gradient plate (0 to 12.0 μg of erythromycin/ml), a GMCB plate containing erythromycin at 8.0 μg/ml, and a GCMB plate containing erythromycin at 16.0 μg/ml. Colonies that grew on GCMB containing erythromycin (8.0 or 16.0 μg/ml) were subcultured twice prior to MIC determination. Sequencing of the mtrR promoter region between mtrC and mtrR and the domain V region of the four alleles of 23S rRNA (rrl gene) of the resistant mutants and of the susceptible parent strains was also performed, and the resulting sequences were compared.
Nucleotide sequence accession numbers.
The nucleotide sequences of the four alleles of 23S rRNA for strains 20869 and 20870 have been deposited in GenBank under accession numbers AF450074 to AF450081.
RESULTS AND DISCUSSION
Antibiotic susceptibilities and characterization of N. gonorrhoeae isolates.
Our laboratory began monitoring azithromycin resistance in 1996, and the emergence of resistant strains was first observed in 1997 in Canada. During 1997 to 1999, 56 azithromycin-resistant N. gonorrhoeae strains (MICs, 2.0 to 4.0 μg/ml) were isolated in five provinces in Canada (Québec, British Columbia, Ontario, Alberta, and Nova Scotia). In 1997, 95% (19 of 20) of the azithromycin-resistant strains were isolated from Québec and only 1 other strain was isolated from British Columbia. In 1998, 52% (13 of 25) of the azithromycin-resistant strains were from British Columbia, 5 from Ontario, 3 from Alberta, 2 from Nova Scotia, and 2 from Québec. Of the 11 strains isolated in 1999, 8 were from British Columbia and 3 were from Québec. The 56 strains were classified into 16 types by auxotype and serotype (A/S). The most common classes were type NR/IB-01 (33 strains [58.9%]) and type NR/IB-03 (6 strains [10.7%]). Three A/S classes had two strains each, NR/IA-04, H/IB-01, and H/IB-03. Eleven A/S classes (NR/IB-06, NR/IB-10, NR/IB-14, NR/IB-17, NR/IB-23, NR/IB-32, NR/IB-26, H/IB-20, P/IB-03, P/IB-02, and P/serotype [not typeable]) each had a unique strain. All the strains contained a 2.6-mDa plasmid, except for one P-requiring strain. All azithromycin-resistant strains (azithromycin MIC ≥ 2.0 μg/ml) were also resistant to erythromycin (erythromycin MIC ≥ 2.0 μg/ml). Some of these strains (58.9%) also had chromosome-mediated resistance to both tetracycline and penicillin, and the remaining 41.1% were resistant to tetracycline (tetracycline MIC ≥ 2.0 μg/ml). A total of 96.4% of the azithromycin-resistant strains showed resistance to crystal violet (crystal violet MICs ≥ 1.0 μg/ml), indicating that their cell envelopes may have had reduced permeability (14).
Detection of mechanisms of macrolide resistance.
Several targets were used to determine different mechanisms of resistance in the azithromycin-resistant N. gonorrhoeae. Forty-six selected strains, including strains from all A/S classes and strains from different geographical locations, were all negative by PCR for rRNA methylases using erm(A), erm(B), erm(C), or erm(F) (data not shown), indicating that our azithromycin-resistant strains did not have the same mechanisms of macrolide resistance as those found in Seattle and Uruguay (19). PCR amplification of a subset of 14 strains selected randomly also did not show the presence of the mef(A) gene, a macrolide transporter (data not shown). PCRs using genomic DNA of control strains all showed positive results, as expected. In addition, the structural genes rplD (ribosomal protein L4) and rplV (ribosomal protein L22) were examined for point mutations, using nine and eight strains, respectively (Table 2). The DNA sequences of the rplD and rplV genes were similar to those determined in a susceptible N. gonorrhoeae strain, FA1090 (Gonococcal Genome Sequencing Project, University of Oklahoma [http://dna1.chem.ou.edu/gono.html]). We concluded that macrolide resistance in these strains was not due to mutations of rplD or rplV or to the acquisition of erm genes.
TABLE 2.
Characterization of 14 clinical isolates of N. gonorrhoeae for which erythromycin and azithromycin MICs are elevated
| Strain no. | A/Sa class | ERY MIC (μg/ml)b | AZM MIC (μg/ml)c | 23S rRNA mutation at position 2611 (E. coli numbering system) | mtr mutationd | rplD mutation (L4 protein) | rplV mutation (L22 protein) |
|---|---|---|---|---|---|---|---|
| 20869 | NR/IB-01 | 32.0-64.0 | 4.0 | C→T, alleles 1, 2, and 4e | No mutation | No mutation | NTf |
| 20870 | NR/IB-01 | 32.0-64.0 | 4.0 | C→T, alleles 1, 2, and 4e | No mutation | No mutation | NT |
| 18649 | NR/IB-01 | 8.0 | 2.0 | No mutation | A deletion | NT | NT |
| 19255 | H/IB-01 | 2.0 | 2.0 | No mutation | A deletion | NT | NT |
| 19283 | NR/IB-01 | 4.0 | 2.0 | No mutation | A deletion | NT | NT |
| 19378 | NR/IB-03 | 4.0 | 2.0 | No mutation | A deletion | No mutation | NT |
| 19398 | NR/IB-17 | 4.0 | 2.0 | No mutation | A deletion | No mutation | NT |
| 19416 | H/IB-03 | 8.0 | 2.0 | No mutation | A deletion | No mutation | NT |
| 19418 | NR/IB-01 | 8.0 | 2.0 | No mutation | A deletion | No mutation | NT |
| 19446 | NR/IB-03 | 8.0 | 2.0 | No mutation | A deletion | NT | NT |
| 19486 | NR/IB-32 | 8.0 | 2.0 | No mutation | A deletion | No mutation | NT |
| 19491 | NR/IB-03 | 8.0 | 2.0 | No mutation | A deletion | No mutation | NT |
| 19561 | NR/IB-01 | 8.0 | 2.0 | No mutation | A deletion | NT | NT |
| 21631 | NR/IB-01 | 16.0 | 2.0 | No mutation | A deletion | No mutation | NT |
A/S, auxotype-serotype; all carried a 2.6-mDa plasmid.
ERY, erythromycin.
AZM, azithromycin.
The mtr mutation is the single-base-pair (A) deletion in the 13-bp inverted repeat in the mtrR-mtrC promoter region.
GenBank accession numbers: AF450074-AF45008; C2599T for alleles 1, 2 and C2605T for allele 4 (N. gonorrhoeae numbering).
NT, not tested.
It has been well established that overexpression of the mtr efflux pump can play a role in increasing resistance to macrolides (20, 29). DNA sequencing of the mtrR promoter region in the 14 azithromycin- and erythromycin-resistant strains (Table 2) revealed that 12 of these strains had a single-base-pair deletion within a 13-bp inverted repeat within the mtrR promoter (29) (Table 2). The deletion which activates the overexpression of the mtr pump (20) in these 12 strains was the same mutation as that reported for azithromycin-resistant N. gonorrhoeae strains in another study (29). The other two azithromycin-resistant strains (20869 and 20870) that did not have a mutation in the mtr promoter region therefore have other resistance mechanisms not previously described for N. gonorrhoeae.
There are other mechanisms of macrolide resistance, including mutations in the 23S rRNA as found in other bacteria. Therefore, we explored the possibility that changes occur in the 23S rRNA DNA sequence in strains 20869 and 20870 that affect their susceptibility to macrolides. The four copies of the 23S rRNA rrl gene present in N. gonorrhoeae were amplified individually for domain V in 14 azithromycin-resistant strains (Table 2). Twelve of these strains did not have mutations in the DNA sequence of the 23S rRNA peptidyltransferase region (Table 2). These 12 strains did have the deletion in the mtr promoter region. In the remaining two strains, 20869 and 20870 (erythromycin MIC, 64.0 μg/ml; azithromycin MIC, 4.0 μg/ml), alleles 1, 2, and 4 were found to contain a C2611T (E. coli numbering) mutation (Fig. 1). These mutations correspond to position C2599T for alleles 1 and 2 and position C2605T for allele 4 (N. gonorrhoeae numbering). Allele 4 was found to contain base insertions throughout domain V which account for the change in nucleotide position number. We believe that this mutation is responsible for macrolide resistance in these two strains. The secondary structure of domain V of the 23S rRNA gene shown in Fig. 1, based on an illustration by Depardieu and Courvalin (2), revealed a number of point mutations previously identified in other strains which confer resistance to macrolides (25). These include the following positions and mutations: positions A2058 and A2062 (E. coli numbering) in S. pneumoniae (2); positions G2057, A2058, and A2059 (E. coli numbering) in H. pylori (26); and positions G2057, A2058, and C2611 in E. coli. These mutations most likely affect the secondary structure of the peptidyltransferase loop in domain V, due to the pairing of different nucleotides. In the N. gonorrhoeae strains 20869 and 20870, the C→T mutation is located in this same region.
Further investigation of strains 20869 and 20870 revealed that they did not have the mutation in the mtrR promoter region causing the mtrR overexpression nor did they have mutations in the ribosomal protein L4 or L22. No mutations were observed in allele 3 of these two strains.
Transformation studies.
Transformation studies were used to confirm that mutations in the rrl gene within the peptidyltransferase loop of domain V of the 23S rRNA caused resistance to macrolides in N. gonorrhoeae. Transformation of total genomic DNA from macrolide-resistant strain 19416 (erythromycin MIC, 8.0 μg/ml; azithromycin MIC, 2.0 μg/ml) to N. gonorrhoeae F62 (an erythromycin- and azithromycin-susceptible strain) yielded transformants for which macrolide MICs were increased (erythromycin MIC of 4.0 μg/ml). Sequencing of the mtrR promoter region revealed that the transformants had the same deletion (A) in mtr found in the donor strain and did not have mutations in 23S rRNA. Transformation of total genomic DNA from macrolide-resistant strain 20870 (erythromycin MIC, 64.0 μg/ml; azithromycin MIC, 4.0 μg/ml) to F62 yielded resistant transformants (erythromycin MIC, 4.0 μg/ml; azithromycin MIC, 2.0 μg/ml). However, when the regions encoding domain V of the 4 alleles of the 23S rRNA rrl gene in the transformants were amplified, their DNA sequences showed that these strains did not have mutations in the 23S rRNA peptidyltransferase region or the deletion mutation in the mtrR promoter region. The increased macrolide MICs for the transformants remained unexplained in this experiment. The ciprofloxacin-resistant strain (MIC of 8.0 μg/ml) used as a positive control to transform F62 yielded transformants with a ciprofloxacin MIC of 2.0 μg/ml. Subsequent sequencing of the gyrA and parC genes of the transformants revealed that the mutations in the transformants were identical to the mutations of the donor strain (gyrA mutations of 91PHE and 95GLY and parC mutations of 86ASN).
Analysis of erythromycin- and azithromycin-resistant and -susceptible spontaneous mutants.
To confirm that the mutations of rrl were associated with macrolide resistance, we attempted to derive mutants with reduced susceptibility from two macrolide-resistant clinical isolates (20869 and 20870) that had mutations in alleles 1, 2, and 4 of the 23S rRNA (Table 3). Strains LK368 and LK370 were derived from 20869 and 20870, respectively, after subculturing 35 times on GCMB plates. The MICs of erythromycin were determined to be 4.0 μg/ml for LK368 and 32.0 to 64.0 μg/ml for LK370, and the MICs of azithromycin were 1.0 μg/ml for LK368 and 2.0 μg/ml for LK370. Strains LK368 and LK370 were then subcultured an additional 12 times on GCMB plates and yielded two derivatives, LK369 and LK371, respectively (Table 3). The MICs of erythromycin were determined to be 2.0 μg/ml for LK369 and 16.0 to 32.0 μg/ml for LK371, and the MICs of azithromycin were 0.5 μg/ml for LK369 and 2.0 to 4.0 μg/ml for LK371 (Table 3). The partial DNA sequence of 23S rRNA from these derivatives showed that allele 1 had a T→C reversion at nucleotide 2611 (E. coli numbering) for two strains, LK368 and LK369, and allele 2 had a T→C reversion at nucleotide 2611 (E. coli numbering) for one strain only (LK369). The sequences for alleles 3 and 4 remained identical to those of the parent strains. DNA sequencing of the mtrR repressor promoter region revealed that the deletion mutation was not present (Table 3). The two derivatives (LK370 and LK371) for which the erythromycin MIC values remained in the range of 16.0 to 64.0 μg/ml and the azithromycin MICs remained in the range of 2.0 to 4.0 μg/ml also retained the same DNA sequences for all four alleles of the 23S rRNA as the those of the parent strain. These derivatives (LK370, LK371, LK368, and LK369) did not show mutations in the mtrR promoter region (Table 3). The resistant- to susceptible-phenotype revertants showed the reversion of 23S rRNA sequence to wild type and no DNA sequence changes in the mtrR promoter.
TABLE 3.
Characterization of spontaneous N. gonorrhoeae mutants: group I, susceptible spontaneous mutants from erythromycin and azithromycin-resistant N. gonorrhoeae strains; group II, erythromycin- and azithromycin-resistant N. gonorrhoeae mutants from susceptible N. gonorrhoeae strains
| Group | Strain(s) | Origin | ERY MIC (μg/ml)a | AZM MIC (μg/ml)b | 23S rRNA mutation at position 2611 (E. coli numbering system)c
|
mtr mutationd | |||
|---|---|---|---|---|---|---|---|---|---|
| Allele 1 | Allele 2 | Allele 3 | Allele 4 | ||||||
| I | 20869 | Clinical isolate | 32.0-64.0 | 4.0 | T | T | C | T | No mutation |
| 20870 | Clinical isolate | 32.0-64.0 | 4.0 | T | T | C | T | No mutation | |
| LK368 | Spontaneous mutant of 20869 | 4.0 | 1.0 | C | T | C | T | No mutation | |
| LK369 | Spontaneous mutant of LK368 | 2.0 | 0.5 | C | C | C | T | No mutation | |
| II | 22831 | Clinical isolate | 0.5 | 0.125 | C | C | C | C | No mutation |
| 21305 | Clinical isolate | 0.5 | 0.125 | C | C | C | C | Insertion T | |
| 21855 | Clinical isolate | 1.0 | 0.125 | C | C | C | C | No mutation | |
| LK361, LK362 | Spontaneous mutant of 22831 | 8.0 | 2.0 | C | C | C | C | Deletion A | |
| LK363, LK364 | Spontaneous mutant of 21305 | 32.0 | 4.0 | T | T | T | T | Insertion T | |
| LK359, LK360 | Spontaneous mutant of 21855 | 64.0 | 8.0 | T | T | T | T | No mutation | |
| LK366 | Spontaneous mutant of 22831 | 64.0 | 32.0 | T | T | T | C | Deletion A | |
| LK367 | Spontaneous mutant of 22831 | 64.0 | 32.0 | T | T | C | T | Deletion A | |
ERY, erythromycin.
AZM, azithromycin.
C2599T for alleles 1 and 2 and C2605T for allele 4 (N. gonorrhoeae numbering).
mtr mutation is the single-base-pair (A) deletion in the 13-bp inverted repeat in the mtrR-mtrC promoter region.
We also attempted to generate spontaneous macrolide-resistant mutants from susceptible strains and compared the DNA sequences for the domain V region of the rrl gene. Three macrolide-susceptible N. gonorrhoeae strains, 22831, 21305, and 21855 (erythromycin MICs, 0.5 to 1.0 μg/ml; azithromycin MIC, 0.125 μg/ml), were selected from our culture collection (Table 3). The sequences of domain V of rrl of the three strains (22831, 21305, and 21855) were the same as that of the susceptible N. gonorrhoeae reference strain FA1090 (Gonococcal Genome Sequencing Project, University of Oklahoma). Initially, six spontaneously resistant mutants were obtained after subculturing the three parent strains 14 times. Strains LK359 and LK360 were obtained from parent strain 21855, strains LK361 and LK362 were from parent strain 22831, and strains LK363 and LK364 were from parent strain 21305. After subculturing an additional nine times, mutants LK366 and LK367 were obtained from parent strain 22831. The MICs and the DNA sequences of alleles 1, 2, 3, and 4 of the 23S rRNA of the spontaneous mutants and those of their parent strains were determined and compared (Table 3). No substitution mutations were identified in any of the four alleles of the 23S rRNA of LK361 and LK362 (erythromycin MIC, 8.0 μg/ml; azithromycin MIC, 2.0 μg/ml), but the alleles did have the deletion (A) in the mtrR promoter region, causing the formation of mutants with overexpressed mtr efflux pumps (29). LK363 and LK364 (erythromycin MIC, 32.0 μg/ml; azithromycin MIC, 4.0 μg/ml) had the C→T substitution in all four alleles of the 23S rRNA. They did not have the deletion in the mtrR promoter region, but they did have an insertion (T). The nucleotide insertion (T) in the mtrR promoter region was also identified in one of the parent strains, and this mutation did not seem to confer resistance. The MICs of erythromycin and azithromycin were found to be 64.0 μg/ml and 8.0 μg/ml, respectively, for strains LK359 and LK360. These strains all had the C→T substitution in all four alleles of the 23S rRNA and did not have the deletion in the mtrR promoter region. Mutant LK366 had the C2611T mutation (E. coli numbering) in alleles 1, 2, and 3 of the 23S rRNA. LK367 had the C2611T mutation (E. coli numbering) in alleles 1, 2, and 4 of the 23S rRNA (Table 3). In comparison with those for the other mutants, the MICs of erythromycin (64.0 μg/ml) and azithromycin (32.0 μg/ml) were found to be the highest for LK366 and LK367. These strains (LK366 and LK367) had mutations in three of the four alleles of rrl, while strains LK359 and LK360, for which azithromycin had a lower MIC (8.0 μg/ml), had mutations in all four alleles of rrl. Therefore, the higher azithromycin MIC does not correlate with the number of alleles with mutations. Both LK366 and LK367 had one deletion (A) in the mtrR promoter region which LK359 and LK360 did not have. This deletion may account for the finding of higher azithromycin MICs for LK366 and LK367 than for LK359 and LK360, which indicates the additive effects of multilocus mutations. On the other hand, other unknown mechanisms could also attribute to higher azithromycin resistance in LK366 and LK367.
In conclusion, we have described a novel C2611T (E. coli numbering) mutation in alleles 1, 2, and 4 in 23S rRNA that confers macrolide resistance in two clinical N. gonorrhoeae. The mtr efflux pump contributes to erythromycin resistance in strains for which erythromycin MICs range from 2.0 to 8.0 μg/ml and azithromycin MICs range from 0.5 to 2.0 μg/ml. Higher levels of macrolide resistance (erythromycin MIC, 16.0 μg/ml; azithromycin MIC, 4.0 to 8.0 μg/ml) are associated with mutations of one or more alleles of domain V of rrl. Neither clinical isolates nor spontaneous mutants in this study showed that rplD (ribosomal protein L4) or rplV (ribosomal protein L22) contributed to resistance. The spontaneous mutants derived by in vitro mutation to erythromycin resistance revealed that high-level macrolide resistance (erythromycin MIC, 64.0 μg/ml; azithromycin MIC, 32.0 μg/ml) may require three of the four mutations in rrl as well as the mutation in the mtr loci, showing the additive effects of mutations on resistance levels. Although we observed additive mechanisms of macrolide resistance with in vitro mutants, no clinical isolates with both mechanisms have yet been identified. This study provided further evidence that N. gonorrhoeae is evolving continuously in response to antibiotic selection pressure. Characterization of mechanisms of resistance in these organisms will provide information necessary for the development of new therapies.
Acknowledgments
We thank Allan Lau and Shane Jones for their technical assistance and the DNA Core Facility, NML, for the synthesis of oligonucleotides and for performing the DNA sequencing. We thank Mei Ling Lam for preparing Fig. 1. This project would not be possible without the contributions of our provincial partners (British Columbia Centre for Disease Control, Public Health Laboratory for Alberta, Ontario Ministry of Health, Laboratorie de santé publique du Québec, and Queen Elizabeth II Health Science Centre, Nova Scotia) in the monitoring of antimicrobial resistance in N. gonorrhoeae and also in supplying strains for our research program. We thank K. Jones, Virginia Commonwealth University, Richmond, Va., for E. coli V831 and J. Sutcliffe, Pfizer, Groton, Conn., for the following control strains: Staphylococcus aureus RN1389, Streptococcus pyogenes Ac1, S. aureus RN4221, and Streptococcus pyogenes 02C1064.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dillon, J. R., A. Nasim, and E. R. Nestmann. 1985. Recombinant DNA methodology. John Wiley & Sons, New York, N.Y.
- 4.Ehret, J. M., L. J. Nims, and F. N. Judson. 1996. A clinical isolate of Neisseria gonorrhoeae with in vitro resistance to erythromycin and decreased susceptibility to azithromycin. Sex. Transm. Dis. 23:270-272. [DOI] [PubMed] [Google Scholar]
- 5.Gregory, S. T., and A. E. Dahlberg. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J. Mol. Biol. 289:827-834. [DOI] [PubMed] [Google Scholar]
- 6.Hendry, A. T., and I. O. Stewart. 1979. Auxanographic grouping and typing of Neisseria gonorrhoeae. Can. J. Microbiol. 25:512-521. [DOI] [PubMed] [Google Scholar]
- 7.Jensen, L. B., N. Frimodt-Moller, and F. M. Aarestrup. 1999. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 170:151-158. [DOI] [PubMed] [Google Scholar]
- 8.Kellogg, D. S., W. L. Peacock, W. E. Deacon, L. Brown, and C. I. Pirkle. 1963. Neisseria gonorrhoeae I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knapp, J. S., M. R. Tam, R. C. Nowinski, K. K. Holmes, and E. G. Sandstrom. 1984. Serological classification of Neisseria gonorrhoeae with use of monoclonal antibodies to gonococcal outer membrane protein I. J. Infect. Dis. 150:44-48. [DOI] [PubMed] [Google Scholar]
- 10.Laboratory Centre for Disease Control Expert Working Group on Canadian Guidelines for Sexually Transmitted Diseases. 1998. Canadian STD guidelines. Health Canada, Ottawa, Ontario.
- 11.Leclercq, R., and P. Courvalin. 1998. Streptogramins: an answer to antibiotic resistance in gram-positive bacteria. Lancet 352:591-592. [DOI] [PubMed] [Google Scholar]
- 12.Lucas, C. E., J. T. Balthazar, K. E. Hagman, and W. M. Shafer. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 179:4123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luna, V. A., S. Cousin, Jr., W. L. Whittington, and M. C. Roberts. 2000. Identification of the conjugative mef gene in clinical Acinetobacter junii and Neisseria gonorrhoeae isolates. Antimicrob. Agents Chemother. 44:2503-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morse, S. A., P. G. Lysko, L. McFarland, J. S. Knapp, E. Sandstrom, C. Critchlow, and K. K. Holmes. 1982. Gonococcal strains from homosexual men have outer membranes with reduced permeability to hydrophobic molecules. Infect. Immun. 37:432-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2002. Performance standard for antimicrobial susceptibility testing: twelfth informational supplement M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 16.Ng, L.-K., P. Sawatzky, I. E. Martin, and S. Booth.. Characterization of ciprofloxacin resistance in Neisseria gonorrhoeae isolates in Canada. Sex. Transm. Dis., in press. [DOI] [PubMed]
- 17.Pardo, D., and R. Rosset. 1977. Properties of ribosomes from erythromycin resistant mutants of Escherichia coli. Mol. Gen. Genet. 156:267-271. [DOI] [PubMed] [Google Scholar]
- 18.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts, M. C., W. O. Chung, D. Roe, M. Xia, C. Marquez, G. Borthagaray, W. L. Whittington, and K. K. Holmes. 1999. Erythromycin-resistant Neisseria gonorrhoeae and oral commensal Neisseria spp. carry known rRNA methylase genes. Antimicrob. Agents Chemother. 43:1367-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shafer, W. M., W. L. Veal, E.-H. Lee, L. Zarantonelli, J. T. Balthazar, and C. Rouquette. 2001. Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis. J. Mol. Microbiol. Biotechnol. 3:219-224. [PubMed] [Google Scholar]
- 21.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tapsall, J. W., T. R. Shultz, E. A. Limnios, B. Donovan, G. Lum, and B. P. Mulhall. 1998. Failure of azithromycin therapy in gonorrhea and discorrelation with laboratory test parameters. Sex. Transm. Dis. 25:505-508. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, D. E., Z. Ge, D. Purych, T. Lo, and K. Hiratsuka. 1997. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob. Agents Chemother. 41:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, G. E., and D. E. Taylor. 1998. Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics. Antimicrob. Agents Chemother. 42:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wittmann, H. G., G. Stöffler, D. Apirion, L. Rosen, K. Tanaka, M. Tamaki, R. Takata, S. Dekio, and E. Otaka. 1973. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol. Gen. Genet. 127:175-189. [DOI] [PubMed] [Google Scholar]
- 29.Zarantonelli, L., G. Borthagaray, E.-H. Lee, and W. M. Shafer. 1999. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob. Agents Chemother. 43:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]