Abstract
The Listeria monocytogenes two-component signal transduction system, LisRK, initially identified in strain LO28, plays a significant role in the virulence potential of this important food-borne pathogen. Here, it is shown that, in addition to its major contribution in responding to ethanol, pH, and hydrogen peroxide stresses, LisRK is involved in the ability of the cell to tolerate important antimicrobials used in food and in medicine, e.g., the lantibiotic nisin and the cephalosporin family of antibiotics. A ΔlisK mutant (lacking the LisK histidine kinase sensor component) displays significantly enhanced resistance to the lantibiotic nisin, a greatly enhanced sensitivity to the cephalosporins, and a large reduction in the expression of three genes thought to encode a penicillin-binding protein, another histidine kinase (other than LisK), and a protein of unknown function. Confirmation of the role of LisRK was obtained when the response regulator, LisR, was overexpressed using both constitutive and inducible (nisin-controlled expression) systems. Under these conditions we observed a reversion of the ΔlisK mutant to wild-type growth kinetics in the presence of nisin. It was also found that overexpression of LisR complemented the reduced expression of two of the aforementioned genes. These results demonstrate the important role of LisRK in the response of L. monocytogenes to a number of antimicrobial agents.
The gram-positive pathogen Listeria monocytogenes imposes a significant burden in terms of both human and economic costs. Listeria was responsible for 71% of all recalls of food products due to bacterial contamination in the United States between 1993 and 1998 (30) and, more importantly, is the cause of almost 30% of all deaths caused by food-borne pathogens in the United States every year (46). As a consequence, developing methodologies to control the survival and growth of Listeria in foods and during infection is a significant research goal. In this respect it is vital that a more complete understanding of how L. monocytogenes responds to the presence of those antimicrobial agents currently used to control this pathogen, both ex vivo (e.g., food grade inhibitors such as bacteriocins in foods) and in vivo (e.g., antibiotics), is developed. This information will be crucial in reducing the human and economic costs associated with Listeria and listeriosis.
Our understanding of the mode of action of nisin, the only lantibiotic approved by the U.S. Food and Drug Administration for use as a food grade inhibitor, is increasing (3). It is now known that nisin functions, at least in part, by the formation of pores in the bacterial cell membrane, with the interaction being largely dependent on the type of lipids present and, most importantly, the charge carried by those lipids (2, 11, 26). Pore formation is facilitated by the binding of nisin to lipid II, a membrane-bound peptidoglycan precursor, which is thought to function as a docking molecule rather than as a receptor. In addition, the binding of nisin to lipid II is responsible for a secondary mode of action, i.e., inhibition of cell wall synthesis (45). Thus nisin inhibits bacterial growth by a combination of these two mechanisms. Despite the extent of this knowledge the role of host genes in determining nisin sensitivity and resistance is less clear.
In vivo, i.e., in infection by Listeria, ampicillin, alone or in combination with gentamicin, remains the treatment of choice (43). However, the high level of innate resistance to cephalosporin antibiotics that Listeria possesses may be especially significant as members of this family of drugs are used most frequently for sepsis due to unknown causes. While cephalosporins were found to be efficient inhibitors of penicillin-binding protein 1 (PBP1), -2, and -4 in L. monocytogenes, which are completely blocked at concentrations well below the MIC, the innate resistance to cephalosporins is thought to be due to their lack of affinity for PBP3, the primary lethal target for β-lactams in the species (18, 44).
Here we report that LisRK, an L. monocytogenes two-component signal transduction system which we previously identified and which has been found to play a role in acid, ethanol, and oxidative stress and in murine virulence (5, 20), also plays a major role in the retardation of the growth of Listeria in the presence of nisin and in the innate cephalosporin resistance of this pathogen. In addition, for the first time, genes regulated by LisRK have been identified. It was found that overexpression of the gene encoding the response regulator component of the system, lisR, could complement a number of the phenotypic consequences of mutating lisK (the gene encoding the histidine kinase component) as well as restoring expression of two of the three regulated genes.
MATERIALS AND METHODS
Strains, plasmids and media.
L. monocytogenes LO28 (serotype 1/2c) is a clinical isolate obtained from P. Cossart, Institut Pasteur, Paris, France. LO28ΔlisK is a mutant from which a portion of the histidine kinase-encoding gene, lisK, has been deleted by splicing by overlap extension (SOEing) PCR (5). The culture medium used was tryptone soy agar or tryptone soy broth (Oxoid, Basingstoke, Hampshire, England) supplemented with 0.6% yeast extract (Difco) (TSA-YE or TSB-YE) or brain heart infusion (BHI) agar or broth (Oxoid). Plasmid pKSV7, used for SOEing PCR, was a kind gift from Kathryn Boor, Cornell University, Ithaca, N.Y. Plasmids pNZ8048 (8) and pNZ9530 (21) were gifts from Michiel Kleerebezem, NIZO, Ede, The Netherlands. Plasmid pNZ44 (29) was a gift from Stephen McGrath, University College Cork, Cork, Ireland.
Growth in the presence of nisin.
The rates of growth of L. monocytogenes LO28 and LO28ΔlisK in the presence of different levels of nisin (2% inoculum in TSB-YE containing 50, 100, 150, 200, or 300 μg of nisin powder [Sigma, St. Louis, Mo.]/ml) were compared by monitoring optical density at 600 nm (OD600) with a Spectra Max 340 spectrophotometer (Molecular Devices, Sunnyvale, Calif.) over a 20-h period.
Antibiotic assays.
Assays to determine the sensitivities of LO28 and LO28ΔlisK to a wide range of antibiotics were carried out by agar diffusion. Overnight cultures were diluted to 106 CFU/ml and swabbed onto TSA-YE. Commercially purchased disks (6 mm in diameter; Oxoid) containing 30 μg (unless otherwise stated) of the antibiotics to be studied were then placed on the surfaces of agar plates. Following overnight incubation of the plates at 37°C, the diameters of the zones of bacterial growth inhibition surrounding the filter disks were measured. The relative susceptibilities of different strains to the various antibiotics tested were correlated with the sizes of the zones of inhibition, with increased zone size reflecting increased susceptibility. Initially the antibiotics assayed were cefotaxime, cefuroxime, vancomycin, erythromycin, kanamycin, fosfomycin (50 μg), minocycline, polymyxin B (300 μg), streptomycin (25 μg), fusidic acid (10 μg), oxytetracycline, penicillin (10 μg), clindamycin (10 μg), spectinomycin (25 μg), ampicillin (25 μg), novobiocin, rifampin, nalidixic acid, gentamicin, colistin sulfate (25 μg), chloramphenicol (CAM), and tetracycline.
In addition to this assay, designed to compare antibiotic susceptibilities in general, further studies involved disks containing antibiotics of the cephalosporin family. These were cefuroxime, ceftazidime, cefaclor, cephalothin, cefoxitin, ceftriaxone, cefotetan, cefoperazone, cephradine, cephalexin, and cefotaxime (all 30 μg).
Implementation of the nisin-controlled expression (NICE) system in L. monocytogenes LO28ΔlisK.
A strategy to replace the hemolysin gene, hly, on the L. monocytogenes chromosome with the nisRK genes was devised. Primers were designed to amplify the chromosomal regions flanking the hly gene (primers HSOEA, -B, -C, and -D; Table 1). The resultant A-B and C-D fragments were spliced by overlap extension PCR (19) using primers HSOEA and -D to create a single A-D fragment, representing the region surrounding hly but with the gene precisely removed. This fragment was subsequently cloned in temperature-sensitive plasmid pKSV7 (41). Since primers B and C have built-in restriction sites, we were able to clone the nisRK operon (amplified, by using primers nisRF and nisKB, from pNZ9530 [21]) into the location formerly occupied by hly. This plasmid construct, pCPL-53, was electroporated into L. monocytogenes LO28ΔlisK, and transformants were selected on BHI agar with 10 μg of CAM/ml (BHI/CAM). Chromosomal integration of the plasmid at 41°C was selected by serial passage of a transformant in prewarmed BHI/CAM broth and streaking onto prewarmed BHI/CAM agar. Plasmid excision was accomplished by continuous passage in BHI at 30°C, and clones in which nisRK had replaced hly on the chromosome were identified by plating them onto blood agar plates at 30°C. Replica plating of nonhemolytic colonies onto BHI and BHI/CAM at 30°C identified cured derivatives. PCR and sequencing analysis of one such strain confirmed that the nisRK operon had replaced the hly gene. This strain was designated LO28ΔlisK-NICE.
TABLE 1.
PCR primers used in this study.
| Name | Sequence (5′-3′) |
|---|---|
| nisRF | AATGATCGATAAACAATCGGAGGTa |
| nisKB | ACTAGTGGATCCCCCGGG |
| NZlisRF | GGGTTAGGCCCCATGGATAGAATACTAb |
| NZlisRB | AATGGTCTAGACGTCATGTACGCATc |
| lisRRTPCRF | CAGCGGTTGCTAATGATG |
| lisRRTPCRB | GATAACAGATCACGTGCG |
| HSOEA | CAGGTAGAGCGGAATTCATTGd |
| HSOEB | TTCATCGATTTCACTCTCCTTCCGTGTGTGTTAAGCGGATCCATa,e |
| HSOEC | ATGGATCCGCTTAACACACACGf |
| HSOED | CCTCTAGAGAATCTGCTTTTACCGTCc |
| PBP 1 | GCGACAAGGCCGGGGAAC |
| PBP 2 | CGGCGATTAGTCGCTTTG |
| CP 1 | AGACGCCCAGAACCGACTCCA |
| CP 2 | AATCGTACTCACGACGAGG |
| HKP 1 | CGGTACGGTCGGTTACTAT |
| HKP 2 | TGTCGCGCTTTTTTCACTCT |
Base changes made to incorporate a ClaI site are in boldface.
Base changes made to incorporate an NcoI site are in boldface.
Base changes made to incorporate an XbaI site are in boldface.
Base changes made to incorporate an EcoRI site are in boldface.
Overhang complementary to HSOEB primer is underlined.
Base changes made to incorporate a BamHI site are in boldface.
Overexpression of lisR.
PCR primers NZlisRF (containing the lisR start codon) and NZlisRB (containing the lisR stop codon) with incorporated NcoI and XbaI sites, respectively, were used to amplify lisR from LO28. The resultant PCR product was digested with NcoI and XbaI restriction enzymes (Roche) and cloned into similarly digested pNZ8048, resulting in the generation of a translational fusion between the nisin-inducible nisA promoter on pNZ8048 and the lisR gene. This event was confirmed by sequence analysis. This strain was designated LO28ΔlisK-NICE(pNZ8048-lisR). pNZ8048 was also introduced into the LO28ΔlisK-NICE background to create LO28ΔlisK-NICE(pNZ8048). As an alternative to inducible overexpression, lisR (amplified by primer pair NZlisRF and NZlisRB) was also cloned into plasmid pNZ44, a derivative of pNZ8048 in which the Pnis promoter was replaced by a constitutive P44 promoter (29), resulting in the creation of LO28ΔlisK(pNZ44-lisR). LO28ΔlisK(pNZ44) was also created to serve as a control for subsequent experiments.
RT-PCR.
RNA isolation and reverse transcription-PCR (RT-PCR) were carried out as described previously (6). RNA was isolated from overnight cultures (constitutive overexpression system) or following nisin induction (inducible overexpression system). For induction with nisin cultures were grown to an OD600 of 0.2 and preinduced with 4.5 μg of nisin powder/ml for 1 h, followed by induction with 45 μg of nisin powder/ml (a concentration at which there is no difference in the nisin sensitivities of the two strains) for 30 min and then isolation of RNA. In all cases cDNA was amplified by PCR with specific primers and samples were taken at regular intervals and run on agarose gels. Primers for the 16S rRNA of L. monocytogenes LO28 were used as controls (36).
RESULTS AND DISCUSSION
Nisin resistance of the ΔlisK mutant.
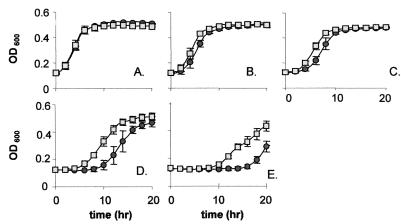
Because deletion of the L. monocytogenes histidine kinase-encoding gene, lisK, results in an altered response to environmental parameters (5), we examined the susceptibility of L. monocytogenes to the lantibiotic nisin. It was found that the levels of nisin required to inhibit growth of wild-type LO28 are high relative to those required to inhibit the growth of a number of other gram-positive bacteria (9). This is a feature of this strain which we have previously observed in relation to other bacteriocins such as lacticin 3147 and enterocin A (our unpublished data). Growth curves were carried out to determine whether deletion of lisK affected the response to nisin. In the absence of nisin the growth rates of the parent and mutant strains were identical, but in the presence of increasing levels of nisin differences became apparent, primarily manifested as a significantly longer lag phase in wild-type LO28 (Fig. 1). This is most obvious when one observes that the lag period for LO28 in the presence of 300 μg of nisin/ml is 6 h longer than that for LO28ΔlisK (Fig. 1E). There is no difference in the growth rates once the lag phase has been exited (e.g., both are 0.046 U/h during logarithmic growth in the presence of 200 μg of nisin/ml). Therefore, the LisK mutation results in a culture that is able to rapidly initiate growth in the presence of nisin, whereas the parent apparently needs a significant period in which to adapt to the presence of the inhibitor. We speculate that, during prior growth in the absence of nisin, the LisRK system plays a role in creating a particular cell envelope composition that renders the cell more susceptible to nisin (and also alters its response to ethanol, low pH, and hydrogen peroxide [5, 20]). In the absence of LisK, the cell must presumably fail to sense an as yet unknown environmental parameter, resulting in an altered envelope composition that is manifested as rapidly initiated growth in the nisin assay.
FIG. 1.
Growth of LO28ΔlisK in the presence of antimicrobial agents. Shown is growth of LO28 (circles) and LO28ΔlisK (squares) in TSB-YE with 0 (A), 50 (B), 100 (C), 200 (D), and 300 (E) μg of nisin powder/ml. Growth was determined by using a Spectra Max 340 spectrophotometer (Molecular Devices) over a 20-h period. Error bars, standard deviations from the means of quadruplicate experiments.
Antibiotic disk assays.
Antibiotic disks were used to determine if LO28 and LO28ΔlisK differed in their responses to other antimicrobial agents. An initial extensive study using a wide range of antibiotics (see Materials and Methods) revealed that only cefotaxime and cefuroxime differentiated significantly between the two strains (Table 2) though a slightly enhanced (though not statistically significant) sensitivity to ampicillin and penicillin was also observed. It was found that for the parent strain, LO28, the diameters of the zones surrounding the cefotaxime and cefuroxime disks were very small (16 and 15.4 mm, respectively). In contrast, for LO28ΔlisK the diameters extended to 26 and 26.4 mm, representing significant 63 and 71% increases in zone size for cefotaxime and cefuroxime, respectively. As a consequence of the dramatic nature of these findings, further antibiotic disk assays were performed using other antibiotics of the cephalosporin family (Table 2). While, as expected, it was observed that the LO28 background was, in general, more sensitive to narrow-spectrum cephalosporins (e.g., cephalothin, cefaclor, and cephradine), the most significant discovery was the greatly enhanced sensitivity of the LO28ΔlisK mutant in all cases (Table 2). Our observations are especially significant when one considers that, despite L. monocytogenes susceptibility to a wide range of antibiotics, it is resistant to the cephalosporins (18), a large and expanding family of drugs based on cephalosporin C which are frequently the initial choice for hospital treatment of bacterial infection resulting in fever due to unidentified organisms.
TABLE 2.
Cephalosporin resistance of LO28 and LO28ΔlisK
| Straina | Avg diam (mm) of zone of inhibition (SD)b for antibioticc:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ (III) | CTT (III) | CRO (III) | FOX (II) | LEX (I) | CXM (II) | CTX (III) | RAD (I) | CFP (III) | CEC (I) | CEF (I) | |
| L | 6.0d (0) | 9.6 (0.3) | 13.5 (0.8) | 14.6 (0.3) | 15.2 (0.5) | 15.4 (0.7) | 16.0 (1.0) | 17.8 (0.4) | 18.8 (0.5) | 23 (0.5) | 28.5 (1.0) |
| ΔK | 9.3 (1.0) | 12.3 (0.5) | 19.2* (0.5) | 19.5* (0.2) | 20.9** (0.4) | 26.4** (1.3) | 26.0** (1.3) | 20.3* (0.4) | 22.6* (1.0) | 24.4 (0.4) | 32.6* (1.4) |
| −R | 9.5 (0.6) | 12.3 (0.4) | 19.5 (0.4) | 19.3 (0.4) | 19.4 (0.8) | 26.5 (0.7) | 27.5 (0.7) | 20.9 (0.2) | 21.9 (0.1) | 24.3 (0.4) | 32.0 (0.5) |
| +R | 7.5 (0.7) | 9.25* (0.4) | 18.6 (0.5) | 15.5* (0.7) | 15.6* (0.6) | 23.8 (1.0) | 23.5* (0.7) | 17.9** (0.2) | 21.0* (0.2) | 21.5* (0.7) | 26.5* (0.7) |
L, LO28; ΔK, LO28ΔlisK; −R, LO28ΔlisK(pNZ44); +R, LO28ΔlisK(pNZ44-lisR).
Values are averages of triplicate experiments. Asterisks indicate significant differences (∗, P < 0.05; ∗∗, P < 0.01) between the strain and its control (ΔK versus L and +R versus −R).
Antibiotics are in order of increasing effectiveness against LO28. CAZ, ceftazidime; CTT, cefotetan; CRO ceftriaxone; FOX, cefoxitin; LEX, cephalexin; CXM, cefuroxime; CTX, cefotaxime; RAD, cephradine; CFP, cefoperazone; CEC, cefaclor; CEF, cephalothin. The amount of each was 30 μg. I, narrow spectrum; II, expanded spectrum; III, broad spectrum.
Represents the diameter of the disk, as no zone of inhibition was observed.
LisRK-regulated genes.
While the physiological changes present in nisin-resistant mutants have been examined on a number of occasions (25, 27, 28, 32), until the recent study by Gravesen et al. (14) it was not known which genes might be involved. In a high percentage of spontaneous nisin-resistant L. monocytogenes 412 mutants studied by this group increases in the levels of cDNA corresponding to three genes were uncovered by restriction fragment differential-display PCR. Of the putative proteins encoded by these genes one showed high homology to the glycosyltransferase domains of PBPs, another was a histidine kinase (though not LisK), and the third was a protein of unknown function. These findings were especially relevant as a large proportion of these spontaneous mutants also demonstrated enhanced resistance to cefuroxime (14), leading us to speculate that LisRK may regulate one or more of the same genes. To determine if this was the case, RT-PCR analysis was carried out to investigate whether transcription of these genes varies between LO28 and LO28ΔlisK. Significantly, it was found in all cases that the relative level of expression of these three genes was greatly reduced or eliminated in LO28ΔlisK (Fig. 2A). The gene encoding the putative PBP in strain 412 corresponds to the open reading frame designated lmo2229 in genome-sequenced strain L. monocytogenes EGD-e (13). This putative PBP shows highest homology to PBP2a of Streptococcus pneumoniae (16) and PBP1a of Bacillus subtilis (39), both of which are high-molecular-weight PBPs possessing both glycosyltransferase and transpeptidase domains. The gene encoding the histidine kinase corresponds to lmo1021 and shows homology with the yvqE gene of B. subtilis (23) and llkinD of Lactococcus lactis (34, 35) (genes encoding members of the NarQ/NarX subfamily). In each case the gene encoding the histidine kinase is located between a response regulator gene homolog (lmo1022, yvqC, or llrrD) and a gene encoding a protein of unknown function (lmo1020, yvqF, or tcdsorf1). While mutation of llkinD has not been achieved, it was found that the most significant trait associated with an llrrD mutant was an increased osmosensitivity. However, unlike lisK mutants, this mutant did not exhibit enhanced sensitivity to oxidative stress (35). The third gene fragment, originally designated fragment C, corresponds to lmo2487 and is homologous to B. subtilis yvlB, the predicted protein of which has not been assigned a function. While we have yet to ascertain what role, if any, these three genes play in the phenotypes associated with the lisK mutation, the variations in the levels of transcript and the observation that at least one other histidine kinase plays a role suggest that LisRK is involved in a complex regulatory pathway. The apparent inconsistency between our findings and those of Gravesen et al. (14) with regard to the relative increase or decrease in transcription of these genes may reflect the growth phases during which cells were studied, i.e., late exponential phase (OD600 = 0.6) and late stationary phase (overnight growth), respectively. Growth phase-dependent variations have previously been reported with respect to the acid resistance of the LO28ΔlisK mutant. We can, however, definitively state that alterations in the levels of these proteins are associated with nisin resistance and cephalosporin sensitivity.
FIG. 2.
(A and B) RT-PCRs to compare levels of transcripts for genes whose products show homology to histidine kinase (HK), a protein of unknown function (gene C), and a PBP in strains LO28 (L) and LO28ΔlisK (ΔK) (A) and LO28ΔlisK(pNZ44lisR) (+R) and LO28ΔlisK(pNZ44) (−R) (B). (C) Confirmation of the overexpression of lisR by RT-PCR using lisR-specific PCR primers to amplify cDNA templates of equal concentrations generated from LO28ΔlisKNICE(pNZ8048) or LO28ΔlisKNICE(pNZ8048-lisR) RNA following nisin induction. In all cases control PCR primers were used to ensure the complete removal of DNA from RNA preparations prior to reverse transcription and to ensure that levels of cDNA for samples that were to be compared were equal.
In addition to this now-established link between nisin resistance and cephalosporin sensitivity it has also been found that a number of spontaneous L. innocua 4202 mutants resistant to bacteriocin AS-48 displayed enhanced sensitivity to cephradine (37) while lacticin 3147-resistant L. monocytogenes LO28 mutants were more sensitive to cephalexin, cefaclor, and cephradine (22). The recurring link between bacteriocin resistance and cephalosporin sensitivity is thus significant and merits further study. Intriguing are suggestions that the S. pneumoniae CiaRH two-component system, which is closely related to LisRK and which plays a role in cefotaxime resistance as well as in other phenotypes attributable to the cell envelope (12, 15, 47), also controls levels of undecaprenol (bactoprenol), a component of lipid II (N-acetylglucosamine-beta-1,4-MurNAc-pentapeptide-pyrophosphoryl-undecaprenol) (17) as well as monitoring the integrity of the cell wall in general (48). This is especially relevant as lipid II serves as the docking molecule for nisin binding prior to pore formation and is the target for its secondary mode of action, inhibition of cell wall synthesis. Though the nisin resistance of CiaRH mutants has not been reported, it is tempting to suggest, on the basis of the involvement of lipid II, that their resistance would be modified and thus that further examination of this pathway in the appropriate mutants may reveal the mechanism responsible for the correlation between nisin resistance and cephalosporin sensitivity.
Implementation of the NICE system in L. monocytogenes LO28ΔlisK.
To confirm that the observed phenotypic and transcriptional changes associated with the deletion in lisK were linked to its interaction with the cognate LisR regulator (and not due to cross talk or a secondary undetected mutation in LO28ΔlisK), we designed a strategy to overexpress LisR in the LO28ΔlisK background. The rationale for this approach stems from the observation that in certain circumstances overexpression of a response regulator alone may mimic activation thereof (33). This phenomenon may be explained by the observation that a number of response regulators are capable of binding their target DNA when nonphosphorylated, though less efficiently than when phosphorylated (1, 7, 24). It may also be possible that the high stoichiometry of LisR allows some level of phosphorylation as a result of either cross talk or the possible contribution of another histidine kinase that specifically interacts with LisR (the existence of which has not been ruled out). Therefore, a system to allow overexpression of LisR was necessary to investigate whether excess LisR could complement the absence of LisK. In addition to the need for a very strong promoter it was desirable that the system also be inducible, in the hope that it might ultimately facilitate the identification of additional promoters controlled by LisRK by using a strategy analogous to that used by Soncini et al. to identify PhoPQ-regulated genes in Salmonella enterica serovar Typhimurium (42). Due to the paucity of controlled expression systems in Listeria and the extent to which it can overexpress proteins, the lactococcal NICE system was employed (8). The NICE system has the advantage of being extremely responsive to an external stimulus (nisin) and has been successfully used in a number of other gram-positive genera, including Leuconostoc and Lactobacillus (21, 38) and Streptococcus, Enterococcus, and Bacillus (9). It depends on the presence of a two-component signal transduction system (NisRK) which senses nisin in the external environment and stimulates transcription from the Pnis promoter. Although nisin is both the induction factor for the NICE system and one of the inhibitors under investigation in this study, because of the relative differences in concentration required for induction and for inhibition, we were able to utilize nisin as both inducer and challenge to determine whether overexpression of LisR can overcome the ΔlisK lesion.
Original applications of the NICE system used two plasmids, one with a nisA promoter and the relevant gene to be overexpressed and the second on which the nisRK regulatory genes were located in trans (9, 21). More-recent innovations involve either placing the nisRK genes on the host chromosome, allowing the subsequent use of a one-plasmid system (38), or placing both nisRK and the nisA promoter on a single plasmid (4). We used the former approach and introduced the nisRK genes onto the listerial chromosome, replacing the hemolysin (hly) gene and placing nisRK under the control of the hly promoter. This replacement did not impact on the level of resistance of the strain. This particular gene replacement strategy was chosen because transcription of hly in strain LO28 in vitro is constitutively high but can be further increased (by the addition of charcoal to the culture medium) or decreased (by a reduction in growth temperature from 37 to 20°C) if required (40). Second, this has the advantage of creating a much less virulent host with which to perform physiological assays. The strain resulting from this procedure, the construction of which is described in Materials and Methods, was designated LO28ΔlisK-NICE (Fig. 2B).
To overexpress LisR, the corresponding gene was cloned into pNZ8048 behind the Pnis promoter. The resulting plasmid, designated pNZ8048-lisR, was introduced into LO28ΔlisK-NICE. Plasmid pNZ8048 lacking an insert was also introduced into the same background for use as a negative control in subsequent experiments. To confirm that nisin induced overexpression of a lisR transcript, an RT-PCR comparing the relative quantities of lisR mRNA in the two backgrounds was performed (Fig. 2C). After 15 cycles it was apparent that the levels of lisR mRNA being produced, following induction by nisin (45 μg/ml), were much greater when pNZ8048-lisR, rather than pNZ8048, was present.
Complementation of the ΔlisK phenotype by overexpression of lisR.
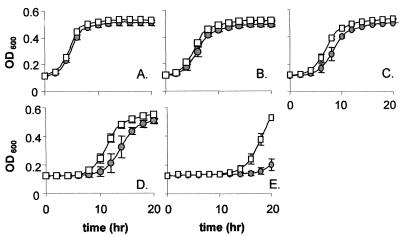
To determine whether overexpressing lisR reversed the enhanced nisin resistance displayed by LO28ΔlisK, we compared the growth of LO28ΔlisK-NICE containing pNZ8048-lisR to that when it contained pNZ8048. The level of nisin required to affect the growth of the strains (greater than 100 μg/ml) was in excess of that required to induce the NICE system maximally (45 μg/ml), thus allowing the induction of lisR and the examination of the growth kinetics under nisin inhibition. The results confirmed that induction of lisR results in a reversion to nisin sensitivity, overcoming the ΔlisK mutation (Fig. 3). As in the parental strain, this sensitivity manifests itself as an increased lag period relative to that of LO28ΔlisK-NICE(pNZ8048), which is 6 h in the presence of 300 μg of nisin/ml (Fig. 3E). Overexpression of LisR by nisin was also found to result in a reversion to wild-type levels of ethanol sensitivity (data not shown) though a reversion to cephalosporin resistance was not apparent. For fear that the use of nisin was interfering with the ability of the NICE system to complement cephalosporin sensitivity, an alternative overexpression vector, pNZ44, was used. This vector is a derivative of pNZ8048 in which a constitutive P44 promoter replaces the inducible Pnis (29). It was found that LO28ΔlisK containing pNZ44-lisR exhibited a reversion to cephalosporin resistance, though the degree of reversion varied depending on the antibiotic used (Table 2). This system was also used to determine if the levels of the three genes associated with nisin resistance increased in response to LisR overexpression (Fig. 2B). It was found that, while increases in the levels of transcript corresponding to lmo2487 (gene C) and, to a much lesser extent, lmo2229 (PBP) were observed, no change in the level of lmo1021 (histidine kinase) was detected. The variation in the extent to which complementation of the cephalosporin sensitivity phenotype occurred may reflect either the involvement of an alternate response regulator that interacts with LisK or, more likely, variations in the affinity of LisR for the three promoters, analogous to the differential expression of genes within the regulon of the closely related CsrR response regulator in Streptococcus pyogenes (10, 31).
FIG. 3.
Complementation of the nisin resistance phenotype of LO28ΔlisK by using the NICE system. Growth of LO28ΔlisKNICE(pNZ8048) (squares) and LO28ΔlisKNICE(pNZ8048-lisR) (circles) in TSB-YE with 0 (A), 50 (B), 100 (C), 200 (D), and 300 (E) μg of nisin powder/ml. Growth was determined by using a Spectra Max 340 spectrophotometer (Molecular Devices) over a 20-h period. Error bars, standard deviations from the means of quadruplicate experiments.
As well as examining the regulation and function of the PBPs, histidine kinase, and gene C we are currently attempting to identify both additional genes under the control of LisR (using the inducible overexpression strategy alluded to previously) and the stimulus which triggers LisK, so that we can gain a more precise understanding of the molecular mechanisms underlying this phenomenon.
In conclusion, we have shown that the LisRK two-component signal transduction system, in addition to playing a role in the response of bacteria to acid, ethanol, hydrogen peroxide, and in vivo stresses, has a major role in the growth of this potentially lethal food pathogen in the presence of lantibiotic bacteriocin and in determining the sensitivity to antibiotics within the cephalosporin family. A link between nisin resistance and cephalosporin sensitivity, previously observed by Gravesen et al. (14), in spontaneous mutants has been confirmed, three genes associated with this phenotype have been shown to controlled through LisK, and it has been shown that complete or partial complementation of the phenotypes displayed by LO28ΔlisK can be reversed by overexpressing lisR.
Finally, in the process of determining the role of LisRK in nisin resistance, we have also confirmed that the NICE system functions in L. monocytogenes. The importance of this finding stems from the lack of inducible gene expression systems for Listeria and the adaptability of the NICE system, which permits precise regulation of expression levels and which can facilitate the production of very high levels of recombinant proteins. The relatively high nisin resistance of strain LO28 makes it especially suitable for use with this system to facilitate the identification of the LisRK regulon and potentially genes controlled by other transcriptional regulators in Listeria. It should also prove a useful tool for others interested in expressing foreign proteins in L. monocytogenes.
Acknowledgments
We thank Kathryn J. Boor for providing plasmid pKSV7, Stephen McGrath for pNZ44, and Michiel Kleerebezem for plasmids pNZ9530 and pNZ8048.
This project was funded by the Irish Government under the National Development Plan 2000-2006.
REFERENCES
- 1.Boucher, P. E., K. Murakami, A. Ishihama, and S. Stibitz. 1997. Nature of DNA binding and RNA polymersae interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J. Bacteriol. 179:1755-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breukink, E., C. van Kraaij, R. A. Demel, R. J. Siezen, O. P. Kuipers, and B. de Kruijff. 1997. The C-terminal region of nisin is responsible for the initial interaction of nisin with the target membrane. Biochemistry 36:6968-6976. [DOI] [PubMed] [Google Scholar]
- 3.Breukink, E., I. Wiedmann, C. van Kraaij, O. P. Kuipers, H. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Cotter, P. D., N. Emerson, C. G. M. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotter, P. D., C. G. M. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 7.Dahl, J. L., B. Y. Wei, and R. J. Kadner. 1997. Protein phosphorylation affects binding of the Escherichia coli transcriptional activator UhpA to the uhpT promoter. J. Biol. Chem. 272:1910-1919. [DOI] [PubMed] [Google Scholar]
- 8.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichenbaum, Z., M. J. Federele, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Federle, M. J., and J. R. Scott. 2002. Identification of binding sites for the group A streptococcal global regulator CovR. Mol. Microbiol. 43:1161-1172. [DOI] [PubMed] [Google Scholar]
- 11.Garcera, M. J., M. G. Elferink, A. J. Driessen, and W. N. Konings. 1993. In vitro pore-forming activity of the lantibiotic nisin. Role of protonmotive force and lipid composition. Eur. J. Biochem. 212:417-422. [DOI] [PubMed] [Google Scholar]
- 12.Giammarinaro, P., M. Sicard, and A. M. Gasc. 1999. Genetic and physiological studies of the CiaH-CiaR two-component signal-transducing system involved in cefotaxime resistance and competence of Streptococcus pneumoniae. Microbiology 145:1859-1869. [DOI] [PubMed] [Google Scholar]
- 13.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 14.Gravesen, A., K. Sorensen, F. M. Aarestrup, and S. Knochel. 2001. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb. Drug Resist. 7:127-135. [DOI] [PubMed] [Google Scholar]
- 15.Guenzi, E., A. M. Gasc, M. A. Sicard, and R. Hakenbeck. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12:505-515. [DOI] [PubMed] [Google Scholar]
- 16.Hakenbeck, R., A. Konig, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakenbeck, R., T. Grebe, D. Zahner, and J. B. Stock. 1999. Beta-lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol. Microbiol. 33:673-678. [DOI] [PubMed] [Google Scholar]
- 18.Hof, H., T. Nichterlein, and M. Kretschmar. 1997. Management of listeriosis. Clin. Microbiol. Rev. 10:345-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-534. [PubMed] [Google Scholar]
- 20.Kallipolitis, B. H., and H. Ingmer. 2001. Listeria monocytogenes response regulators important for stress tolerance and pathogenesis. FEMS Microbiol. Lett. 204:111-115. [DOI] [PubMed] [Google Scholar]
- 21.Kleerebezem, M., M. M. Beerthuyzen, E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klijn, A. W. P. E. 2001. Spontaneous resistance against lacticin 3147 in Lactococcus lactis and Listeria monocytogenes. M.Sc. thesis. National University of Ireland, Cork, Ireland.
- 23.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 24.Liu, W., and F. M. Hulett. 1998. Comparison of the PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology 144:1443-1450. [DOI] [PubMed] [Google Scholar]
- 25.Mantovani, H. C., and J. B. Russell. 2001. Nisin resistance of Streptococcus bovis. Appl. Environ. Microbiol. 67:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, I., J. M. Ruysschaert, D. Sanders, and C. J. Giggard. 1996. Interaction of the lantibiotic nisin with membranes revealed by fluorescence quenching of an introduced tryptophan. Eur. J. Biochem. 239:156-164. [DOI] [PubMed] [Google Scholar]
- 27.Masnier-Patin, S., and J. Richard. 1996. Cell wall changes in nisin-resistant variants of Listeria innocua grown in the presence of high nisin concentrations. FEMS Microbiol. Lett. 140:29-35. [DOI] [PubMed] [Google Scholar]
- 28.Mazotta, A. S., and T. J. Montville. 1999. Characterization of fatty acid composition, spore germination and thermal resistance in a nisin-resistant mutant of Clostridium botulinum 169B and in the wild-type strain. Appl. Environ. Microbiol. 65:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67:608-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mead, P., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Chapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, A. A., N. C. Engleberg, and V. J. DiRita. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40:976-990. [DOI] [PubMed] [Google Scholar]
- 32.Ming, X., and M. A. Daeschel. 1995. Correlation of cellular phospholipid content of nisin resistance of Listeria monocytogenes ScottA. J. Food Prot. 58:416-420. [DOI] [PubMed] [Google Scholar]
- 33.Nishino, K., and A. Yamaguchi. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell-Motherway, M., G. F. Fitzgerald, D. van Sinderen. 1997. Cloning and sequence analysis of putative histidine protein kinases isolated from Lactococcus lactis MG1363. Appl. Environ. Microbiol. 63:2454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connell-Motherway, M., D. van Sinderen, F. Morel-Deville, G. F. Fitzgerald, S. D. Ehrlich, and P. Morel. 2000. Six putative two-component regulatory systems isolated from Lactococcus lactis subsp. cremoris MG1363. Microbiology 146:935-947. [DOI] [PubMed] [Google Scholar]
- 36.O'Driscoll, B. 1997. Characterisation of the acid tolerance response in Listeria monocytogenes. Ph.D. thesis. National University of Ireland, Cork, Ireland.
- 37.O'Sullivan, M. C. 1998. Inhibition of Listeria innocua by bacteriocins of lactic acid bacteria; characterization of bacteriocin resistant isolates of Listeria innocua 4202. M.Sc. thesis, National University of Ireland, Cork, Ireland.
- 38.Pavan, S., P. Hols, J. Delcour, M.-C. Geoffroy, C. Grangette, M. Kleerebezem, and A. Mercenier. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl. Environ. Microbiol. 66:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popham, D. L., and P. Setlow. 1995. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J. Bacteriol. 177:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ripio, M. T., G. Dominguez-Bernal, M. Lara, M. Suarez, and J. A. Vazquez-Boland. 1997. A Gly-145-Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 42.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Temple, M. E., and M. C. Nahata. 2000. Treatment of listeriosis. Ann. Pharmacother. 34:656-661. [DOI] [PubMed] [Google Scholar]
- 44.Vicente, M. F., J. C. Perez-Daz, F. Baquero, M. Angel de Pedro, and J. Berenguer. 1990. Penicillin-binding protein 3 of Listeria monocytogenes as the primary lethal target for beta-lactams. Antimicrob. Agents Chemother. 34:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiedemann, I., E. Breukink, C. van Kraaij, O. P. Kuipers, G. Bierbaum, B. de Kruijff, and H. G. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]
- 46.Wong, S., D. Street, S. I. Delgado, and K. C. Klontz. 2000. Recalls of foods and cosmetics due to microbial contamination reported to the U.S. Food and Drug Administration. J. Food Prot. 63:1113-1116. [DOI] [PubMed] [Google Scholar]
- 47.Zahner, D., T. Grebe, E. Guenzi, J. Krauss, M. van der Linden, K. Terhune, J. B. Stock, and R. Hackenbeck. 1996. Resistance determinants for beta-lactam antibiotics in laboratory mutants of Streptococcus pneumoniae that are involved in genetic competence. Microb. Drug Resist. 2:187-191. [DOI] [PubMed] [Google Scholar]
- 48.Zahner, D., K. Kaminski, L. M. van, L. M., T. Mascher, M. Meral, and R. Hakenbeck. 2002. The ciaR/ciaH regulatory network of Streptococcus pneumoniae. Mol. Microbiol Biotechnol 4:211-216. [PubMed] [Google Scholar]