Abstract
PenB is the third resistance determinant in the stepwise acquisition of multiple resistance genes in chromosomally mediated resistant Neisseria gonorrhoeae (CMRNG). Alterations in porIB, one of two alleles at the por locus that encodes the outer membrane protein porin IB (PIB), were recently reported to be responsible for the increased resistance to penicillin and tetracycline conferred by penB, but the specific mutations conferring antibiotic resistance were not identified experimentally. To determine which amino acids in PIB confer increased resistance, we transformed a recipient strain with chimeras of the porIB genes from strains FA1090 and FA140 (penB2). These studies revealed that two amino acid changes, G120D and A121D, were both necessary and sufficient to confer increased resistance to penicillin and tetracycline. Site-saturation and site-directed mutagenesis of Gly-120 and Ala-121 revealed that both a single mutation, G120K, and the double mutations G120R A121H and G120P A121P also conferred antibiotic resistance to the recipient strain. The identical mutations in PIA increased penicillin and tetracycline resistance either moderately or not at all. Analysis of porIB genes present in the GenBank database from 51 clinical isolates demonstrated that lysine and aspartate mutations at positions 120 and/or 121 also occur in nature. These studies demonstrate that charged amino acids at positions 120 and 121 in PIB are highly preferential for conferring resistance to penicillin and tetracycline in N. gonorrhoeae.
From 1945 to 1988, penicillin was the antibiotic of choice for treatment of gonococcal infections. During this time, however, the resistance of isolates gradually increased until treatment failure with penicillin became widespread and penicillin was discontinued as a first-line antibiotic. Resistance to other antibiotics, including erythromycin and tetracycline, also increased during this time. The antibiotics currently recommended for treatment of gonococcal infections are expanded-spectrum cephalosporins (i.e., ceftriaxone and cefixime) or fluoroquinolones (7), but resistance to fluoroquinolones threatens to make these antibiotics obsolete as well (8).
Resistance to penicillin and tetracycline in gonococci can be either plasmid mediated or chromosomally mediated (5). Plasmid-mediated resistance to penicillin is due to the production of a TEM-1-like β-lactamase, whereas plasmid-mediated resistance to tetracycline is due to expression of the TetM determinant acquired from Streptococcus pneumoniae (34). In contrast, chromosomally mediated resistance to penicillin and tetracycline in Neisseria gonorrhoeae results from the acquisition of multiple resistance genes, each of which confers an incremental increase in resistance until the cell becomes refractory to clinically achieved levels of the antibiotic. As demonstrated by the work of Sparling and others (15, 18, 39), these resistance genes can be transferred in the laboratory in a stepwise manner from a resistant strain to a susceptible strain by DNA uptake and homologous recombination.
The first determinant in the stepwise acquisition of penicillin resistance is the penA gene, which encodes altered forms of penicillin-binding protein 2 (PBP 2) that have a lower rate of acylation by penicillin (15, 16, 40). Insertion of a single aspartic acid residue preceding Asp-345 (Asp-345a) in PBP 2 is the major cause of the decreased rate of acylation by penicillin, although additional mutations in PBP 2 also contribute (2). The second resistance determinant is the mtr (multiple transferable resistance) locus, which increases resistance to diverse hydrophobic agents, including erythromycin and detergent-like fatty-acids (24, 35, 39). Resistance to these agents is most commonly due to a single base pair deletion in the regulatory region of the mtrCDE locus that results in increased expression of the energy-dependent Mtr efflux pump.
The third resistance determinant in chromosomally mediated resistant N. gonorrhoeae (CMRNG) is penB, which increases resistance to both penicillin and tetracycline. Previous reports showed that the penB genetic locus was closely linked to the por locus, which encodes an outer membrane porin protein through which small molecules and solutes diffuse into the periplasmic space (23). N. gonorrhoeae has two porins, PIA and PIB, encoded by alleles of a single por locus (6). In 1980, Cannon and colleagues observed a 98% cotransformation frequency of porIB and the penB gene, suggesting that por was separate from but closely linked to penB in the genome (4). However, Gill et al. (20) recently reported that resistance conferred by the penB gene in the intermediate-level penicillin-resistant strain FA140 is due to mutations within the porIB gene. The authors speculated that alterations in putative loop 3, which in the crystal structures of several porins is located within the β-barrel and forms the constriction of the pore, were responsible for increased resistance to antibiotics, but no experimental evidence was provided to support this hypothesis. Studies with other porins have shown that mutations in other regions can have significant effects on pore properties (31, 36). In order to delineate more clearly the molecular mechanisms of chromosomally mediated antibiotic resistance in N. gonorrhoeae, we set out to determine the specific amino acid alteration(s) in PIB that confers resistance to penicillin and tetracycline and to define the types of amino acids at these positions that are capable of mediating resistance.
MATERIALS AND METHODS
Strains.
N. gonorrhoeae strain FA1090 (5) and the third-level transformant FA140 (FA19 × FA48) (37) have been described previously. Strain FA19 penA4 mtr was derived from FA19 (38) by stepwise transformation with genomic DNA from FA6140 (18). FA19 penA4 mtr (PIB) was constructed as described below. Escherichia coli MC1061 cells were transformed with porIB-containing plasmids, and colonies were grown on Luria-Bertani (LB) agar containing 50 μg of kanamycin/ml, 350 μg of erythromycin/ml, or 34 μg of chloramphenicol/ml.
Construction of strain FA19 penA4 mtr (PIB).
Attempts to transform the coding sequence of porIB into a strain containing the porIA gene, i.e., FA19 penA4 mtr, were unsuccessful. Therefore, we generated a recipient strain of N. gonorrhoeae that contains the first two resistance determinants, penA4 and mtr, and the FA1090 porIB gene to facilitate homologous recombination between donor and recipient DNAs. The porIB gene was amplified from FA1090 genomic DNA with Taq polymerase and transformed into strain FA19 penA4 mtr. The 5′ primer (por-S1, 5′-CGAGCTCGCCGTCTGAACCATCTACCGCGCCGACCTTAC-3′) was complementary to a region ∼1,000 bp upstream of the porIB coding sequence and contained at its 5′ end a SacI restriction enzyme site (boldfaced) and the specific 10-bp uptake sequence (US) (underlined) necessary for uptake of DNA into gonococci (17, 21). The 3′ primer (por-U1, 5′-CGTCTGAGGCCGTCTGAATATGGATAGATTCGTCATTCCCGC-3′) was complementary to a region ∼300 bp downstream of the porIB gene and contained both an US and an XbaI restriction site at its 5′ end. The 2.3-kb PCR amplification product was subcloned into the SacI and XbaI sites of a modified pBluescript vector, pBSC-KS, with the porIB gene in the direction opposite from the lac promoter. pBSC-KS has the chloramphenicol resistance gene from pACYC184 (9) in place of the β-lactamase resistance gene and a deletion of the BglI site. The kanamycin resistance gene was subsequently cloned into the unique BglI site located 40 bp downstream of the porIB stop codon. FA19 penA4 mtr was transformed with this plasmid, and transformants were selected on GCB agar plates containing 50 μg of kanamycin/ml. Replacement of the endogenous porIA gene with the FA1090 porIB gene was confirmed via sequencing of the por gene amplified by PCR from genomic DNA (University of North Carolina—Chapel Hill [UNC-CH] Sequencing Facility).
Generation of porIB mutant constructs.
The mature coding sequences (i.e., lacking the region encoding the 19-amino-acid leader sequence) of the porIB gene from strains FA1090 and FA140 plus 300 bp of downstream sequence were amplified from genomic DNA with por-U1 and a 5′ primer (por-S2, CGAGCTCGCCGTCTGAAGATGTCACCCTGTACGGTGCCATCAAA) complementary to bases 58 to 84 of the porIB gene. The 5′ primer also contained an US and a SacI restriction site at its 5′ end. The PCR products were digested with SacI and XbaI and subcloned into similarly digested pBSC-KS. Silent unique restriction sites (HindIII, EcoRI, and ClaI) were incorporated into the FA1090 and FA140 porIB genes at codons 110, 187, and 274, which surround the codon differences between the two genes. porIB chimeras were constructed by digestion and DNA fragment exchange. Constructs containing single or double mutations were created via four-primer PCR (26) with the FA1090 porIB gene as a template. To aid in selection, the erythromycin resistance gene (ermC) was inserted into the unique BglI site of the plasmids as described above for the kanamycin resistance gene. All constructs were verified by sequencing.
A library of porIB genes containing randomized codons at positions 120 and 121 was also generated by four-primer PCR. The porIB gene from strain FA1090 containing the silent restriction sites was used as the template in the first set of reactions. The external primers were por-S2 and por-U1. The internal down primer (5′-384GGATTCCCAAGCATTGACGTTNNNNNNGGTGTTTTTCAGGGGGCTGTT337-3′) contained an equal mixture of all four nucleotides at the six base positions encoding amino acids 120 and 121 in PIB and 21 bases of porIB sequence on either side. The internal up primer (5′-364AACGTCAATGCTTGGGAATCC384-3′) was complementary to 21 bases of por DNA on the 5′ end of the randomized sequence. Sequencing of an aliquot of the final PCR product verified that all four nucleotides were represented equally at each of the six randomized positions. The PCR products were either used directly to transform gonoccocal cells or ligated into pBSC-KS. For the latter, E. coli MC1061 cells were transformed with the ligation mixture, and a plasmid library was selected in LB broth with chloramphenicol. Sequencing of an aliquot of the library again confirmed the presence of all four bases at roughly equal intensities at each of the six randomized positions.
PCR amplification of the porIB gene from transformants.
Colonies were resuspended in 50 μl of distilled H2O and boiled for 10 min, and 5-μl aliquots were used as the DNA template for PCR with Taq DNA polymerase. PCR primers were por-S1 and por-U1. Amplification conditions were 94°C for 5 min; 30 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 1 min, 15 s; and a final extension of 72°C for 7 min. For all recombinants, the entire amplified gene was sequenced to verify homologous recombination of the mutation(s).
Genetic transformation.
N. gonorrhoeae was transformed as described previously (38). Briefly, cells were passaged on GCB agar, and a single piliated colony was streaked onto a fresh GCB plate and allowed to grow overnight. Cells were scraped from the plate and gently resuspended to a cell density of 108/ml (optical density at 560 nm, 0.18) in prewarmed GC broth containing 10 mM MgCl2 and supplements I and II (29). Following addition of NaHCO3 to a final concentration of 10 mM, aliquots (900 μl) of diluted cells were mixed with 100 μl of 5- to 50-μg/ml donor DNA in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) and incubated for 5 h at 37°C under a humidified 5% CO2 atmosphere. Cells were plated on GCB agar with the appropriate antibiotic and grown for 24 to 48 h at 37°C under 5% CO2. Alternatively, cells at 104, 103, or 102 CFU/ml were incubated with 5 to 50 μg of DNA on GCB agar for 16 to 20 h and then transferred to selection plates (22).
MIC assays.
N. gonorrhoeae colonies were suspended in GCB broth to a density of 104/μl, and 5 μl was spotted onto GCB agar plates (15) containing increasing concentrations of penicillin (0.06, 0.125, 0.3, 0.35, 0.4, 0.5, 0.75, and 1.0 μg/ml) or tetracycline (0.156, 0.313, 0.45, 0.625, 0.75, 1.0, and 1.5 μg/ml). The plates were then incubated at 37°C under 5% CO2 for 24 h. Alternatively, cells were resuspended in GCB broth, and 1,000 colonies were spread to GCB agar plates containing increasing concentrations of either penicillin or tetracycline. The MIC was defined as the minimal concentration of antibiotic at which no more than 5 colonies were observed after 24 h. The two methods gave very similar results. MICs are presented as average values from at least four separate experiments.
Growth assay.
N. gonorrhoeae cells were plated onto GCB agar plates from frozen stocks and grown overnight at 37°C under 5% CO2. Nonpiliated colonies were passaged on GCB plates and grown for 16 to 20 h. Cells were gently scraped from the plates and suspended at a final density of ∼14 Klett units per 10 ml of GCB broth plus supplements I and II. Side-arm flasks containing the bacterial suspensions were shaken at 37°C under 5% CO2, and cell densities were measured every hour with a Klett reader.
SDS-PAGE and Western blotting.
N. gonorrhoeae cells were scraped from GCB agar plates and diluted to 5 × 107 CFU/ml in GC broth. The cells were pelleted, suspended in 45 μl of 1× Laemmli buffer, and heated at 80°C for 5 min. Four microliters of the cell lysates was loaded onto two 10% polyacrylamide gels, and total proteins were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). One of the gels was stained with Coomassie brilliant blue R-250, while the other gel was electroblotted onto nitrocellulose membranes in transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol, 0.01% SDS [pH 8.3]). The membrane was probed sequentially with H5 monoclonal antisera, which recognize FA1090 porin protein (a gift from Janne Cannon, UNC-CH), and an anti-mouse secondary antibody conjugated to horseradish peroxidase. Porin proteins were visualized by incubation with Supersignal chemiluminescent substrate (Pierce/Endogen, Rockford, Ill.).
Outer membranes.
N. gonorrhoeae outer membranes were purified as described by Lamden and Heckels (30). Briefly, cells were scraped from GCB agar plates and suspended in 5 ml of lithium acetate buffer (0.2 M lithium chloride, 0.1 M sodium acetate, 0.01 M EDTA [pH 5.8]). The cells were broken by 15 passages through a 22-gauge needle. Cell debris was pelleted at low speed (11,000 × g), and outer membranes were pelleted at 100,000 × g for 2 h. Protein levels were determined via a modified Bradford assay. Four micrograms of outer membrane proteins in 1× Laemmli sample buffer was incubated either at room temperature or at 80°C for 5 min and subjected to SDS-PAGE. Western blotting was performed to detect monomeric and trimeric forms of the porin proteins by use of the H5 monoclonal antisera to the FA1090 porin protein as described for whole-cell lysates.
RESULTS
Identification of specific mutations in FA140 PIB that confer intermediate-level resistance to penicillin and tetracycline.
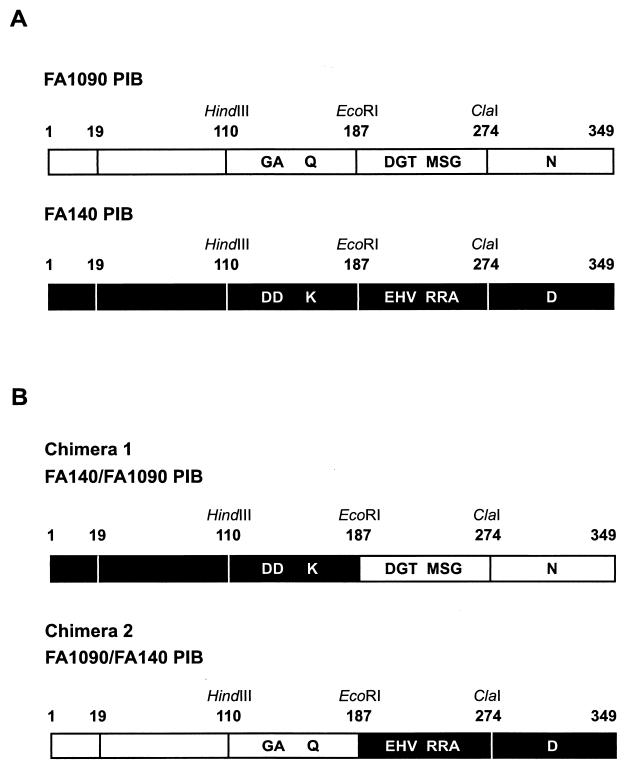
The PIB proteins of strains FA1090 and FA140 (penA2 mtr penB2) differ by 10 amino acids (Fig. 1A), but only the porin from FA140 confers intermediate levels of penicillin and tetracycline resistance. To determine which of these 10 amino acid differences is (are) responsible for conferring resistance to penicillin and tetracycline, we created silent unique restriction sites surrounding the sequences encoding these amino acids to allow construction of porin chimeras and to aid in identifying strains transformed with the recombinant genes (Fig. 1A). Chimera 1 was created by fusing codons 1 to 187 of FA140 porin (containing three alterations) to codons 188 to 349 (containing seven alterations) of FA1090 porin, while chimera 2 was the reverse, beginning with the PIB sequence from FA1090 and switching to the FA140 sequence at codon 188 (Fig. 1B). A novel strain of N. gonorrhoeae, FA19 penA4 mtr (PIB), which contains the first two resistance determinants from FA6140 and the porIB gene from FA1090, was constructed to serve as the recipient strain for transformation experiments with the chimeras (as described in Materials and Methods).
FIG. 1.
FA1090 and FA140 PIB proteins. (A) HindIII, EcoRI, and ClaI silent restriction sites were inserted at codons 110, 187, and 274, respectively, flanking the amino acid differences between the PIB proteins of strains FA1090 and FA140. Residues 1 to 19 constitute the cleavable signal sequence. (B) porIB chimeras were generated via DNA sequence exchange at the EcoRI site.
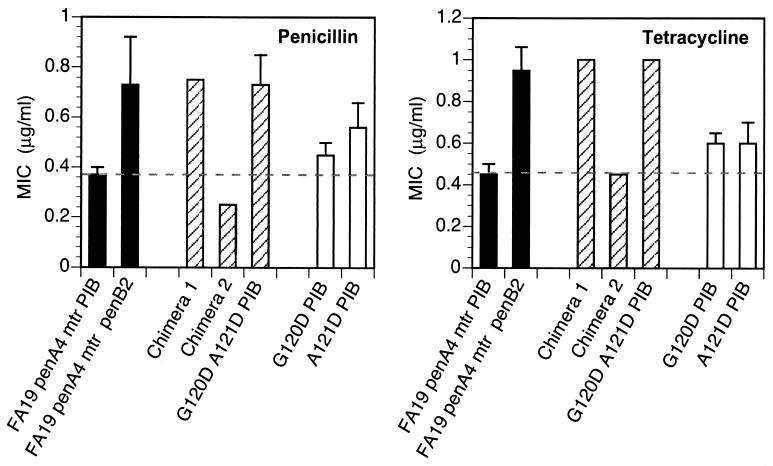
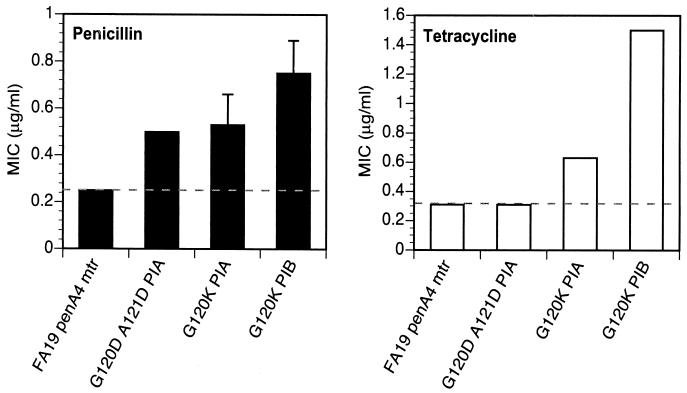
FA19 penA4 mtr (PIB) was transformed with the porIB chimeras, and transformants were selected on GCB agar plates containing 0.55 μg of tetracycline/ml. Transformation with genomic DNA isolated from strain FA140 (positive control) gave rise to resistant colonies at a frequency of 8 × 10−5. Transformation with chimera 1 (FA14019-187/FA1090188-349 porIB) resulted in the growth of tetracycline-resistant colonies at a frequency of 4 × 10−5, whereas transformation with chimera 2 (FA109019-187/FA140188-349 porIB) failed to generate resistant colonies. Thus, only transformation with the chimera encoding the FA140 porIB N-terminal sequence resulted in tetracycline-resistant colonies on selection plates. Nine of 10 porIB genes amplified by PCR from separate transformants showed the expected digestion pattern with HindIII and EcoRI, confirming that genomic recombination had occurred. Sequencing of the PIB gene from the one clone that did not show the expected digestion pattern revealed that it differed from the PIB of the recipient strain by only 2 amino acids, G120D and A121D. MICs of penicillin and tetracycline for each of the 10 transformants were the same as for strains obtained by transformation with the complete FA140 sequence (Fig. 2). Thus, alteration of the amino acids at positions 120 and 121 (Gly and Ala, respectively) to aspartic acid is sufficient to confer the same level of resistance to penicillin and tetracycline as the PIB from FA140.
FIG. 2.
MICs of penicillin and tetracycline for N. gonorrhoeae strains harboring porIB chimeras. Solid bars represent MICs for the recipient strain, FA19 penA4 mtr (PIB), and the isogenic strain, FA19 penA4 mtr penB, which contains the PIB from strain FA140. Strains with PIB chimeras (striped bars) and strains with a single aspartate mutation at position 120 or 121 (open bars) were created by transformation of strain FA19 penA4 mtr (PIB) with mutant porIB DNA. MICs shown are averages from five experiments. Error bars, standard deviations. Dashed lines denote the respective MICs for the recipient strain.
To verify that other residues in FA140 PIB do not contribute to the penB phenotype, FA19 penA4 mtr (PIB) was transformed with a plasmid containing the FA1090/FA140 porIB chimera (chimera 2) and a linked erythromycin resistance gene (ermC) to aid in the selection of transformed colonies. The ermC gene was incorporated into the plasmid downstream of the porIB stop codon and was flanked on its 3′ end by 300 bp of N. gonorrhoeae sequence. Recombination within the por gene in the erythromycin-resistant transformants was confirmed by PCR amplification and restriction digestion. The resistance to penicillin and tetracycline of the resultant strain was essentially identical to that of the recipient strain (Fig. 2). Thus, these data further confirm that mutations at positions 120 and 121 are the only changes capable of conferring increased resistance.
To determine whether aspartic acid mutations at both residues were necessary to confer full resistance to penicillin and tetracycline, FA1090 porIB constructs containing either a single G120D or a single A121D amino acid alteration and the flanking ermC gene were generated and used to transform FA19 penA4 mtr (PIB) to erythromycin resistance. Following confirmation by sequencing that the mutations had recombined into the genome, MIC analysis revealed that mutation of either Gly-120 or Ala-121 in PIB to aspartic acid conferred a moderate increase in resistance to penicillin and tetracycline over that of the recipient strain (Fig. 2). However, resistance to either antibiotic was 1.5-fold lower than that of strains harboring aspartic acid mutations at both positions. Therefore, both the G120D and the A121D mutation are necessary to confer full resistance to penicillin and tetracycline.
Isolation of penicillin- and tetracycline-resistant strains transformed with porIB DNA containing randomized codons at positions 120 and 121.
To determine the importance of specific amino acid mutations in PIB proteins at residues 120 and 121, we randomized the codons at positions 120 and 121 by multiplex PCR and used these constructs to transform the recipient strain to increased penicillin or tetracycline resistance. Transformants were selected on GCB plates containing either 0.57 μg of tetracycline/ml or 0.37 μg of penicillin/ml, and 20 isolates were chosen for further analysis. The MIC of penicillin for each transformant was 0.75 μg/ml, identical to that for the strain with G120D and A121D double PIB mutations. However, the MIC of tetracycline for all 20 transformants was 1.5 μg/ml, compared with 1.0 μg/ml for the strain with the double aspartic acid PIB mutations. The porIB genes from each of the 20 resistant transformants were amplified by PCR and sequenced. Eighteen porIB genes harbored a lysine codon at residue 120, while one other had an arginine at that position (Table 1). Eight different charged and uncharged amino acids were found at residue 121. We also isolated one transformant with prolines at both residues 120 and 121. Overall, these data suggest that mutations at positions 120 and/or 121, particularly to charged amino acids, confer intermediate-level resistance to penicillin and tetracycline.
TABLE 1.
Amino acid residues at positions 120 and 121 in PIB proteins from strains isolated from transformation of FA19 penA4 mtr (PIB) with porIB DNA harboring randomized codon mutations at these positions
| No. of clones | Amino acid 120 | Amino acid 121 |
|---|---|---|
| 2 | K | F |
| 4 | K | H |
| 1 | K | M |
| 3 | K | N |
| 4 | K | R |
| 2 | K | T |
| 2 | K | Y |
| 1 | R | H |
| 1 | P | P |
Analysis of charged and uncharged amino acid mutations at positions 120 and 121 in PIB.
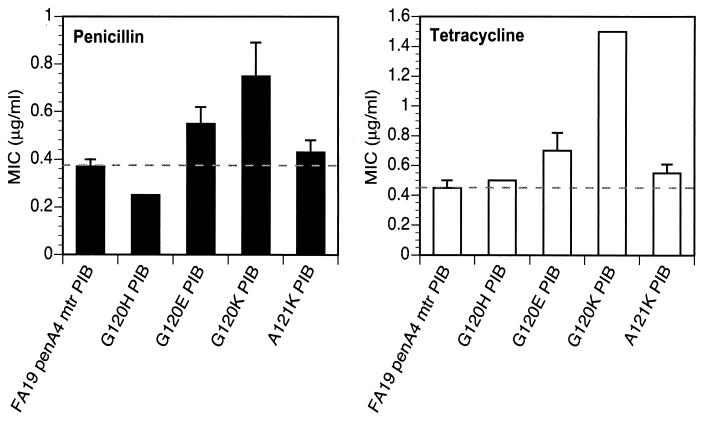
We also mutated Gly-120 and Ala-121 to several charged amino acids to investigate further the role of charged side chains in conferring resistance to penicillin and tetracycline. FA19 penA4 mtr (PIB) was transformed with porIB constructs containing either a G120H, G120E, G120K, or A121K mutation and the linked erythromycin resistance gene, and cells were selected on plates containing 6.0 μg of erythromycin/ml. The MICs of penicillin and tetracycline for the strain containing the G120H PIB mutation were essentially identical to those for the recipient strain, whereas the MICs for the strain with the G120E PIB mutation were similar to those of strains harboring either the G120D or the A121D single PIB mutation (Fig. 3). Mutation of Gly-120 to Lys conferred the same level of resistance to the recipient strain as each of the random mutants containing a G120K mutation, whereas mutation of Ala-121 to Lys conferred only moderate resistance to both antibiotics (Fig. 3). These data demonstrate that a single mutation of G120 to lysine in PIB is the only change needed to confer full intermediate-level resistance to penicillin and tetracycline, which is consistent with the results from our randomized screen.
FIG. 3.
MICs of penicillin and tetracycline for N. gonorrhoeae strains harboring single-amino-acid mutations in PIB. Strains with the single-amino-acid mutation G120H, G120E, G120K, or A121K in PIB were created by transformation of strain FA19 penA4 mtr (PIB) with altered porIB constructs. MICs are averages from five experiments. Error bars, standard deviations. Dashed lines denote the respective MICs for the recipient strain, FA19 penA4 mtr (PIB).
To investigate the consequences of side chain charge versus size in mediating resistance to penicillin and tetracycline, PIB constructs with uncharged amino acids at positions 120 and 121 (G120L A121L or G120N A121N) were transformed into the recipient strain. Leucine or asparagine residues were chosen because their sizes closely resemble those of their charged counterparts lysine and aspartic acid, respectively. The MICs of penicillin and tetracycline for the strains with these double PIB mutations were essentially identical to those of the recipient strain (data not shown), demonstrating that the charge of the side chain, and not its size, is the most important attribute for conferring resistance to the two antibiotics.
Rate of growth of N. gonorrhoeae strains.
Because mutations in residues 120 and 121 apparently alter the flux of antibiotics across the outer membrane, we determined whether these alterations might also alter the passage of metabolites. Therefore, we compared the growth rate of the recipient strain, FA19 penA4 mtr (PIB), with those of isogenic strains containing either the G120K or the G120D A121D PIB mutations. All strains exhibited similar growth curves and growth rates in GCB broth over three separate experiments (data not shown). Doubling times were 1.27 ± 0.06 h for the strain with the FA1090 PIB, 1.13 ± 0.06 h for the G120D A121D PIB mutant, and 1.18 ± 0.03 h for the G120K PIB mutant. Therefore, mutations at residues 120 and/or 121 in PIB do not appear to compromise the growth rate of the cell, at least in GCB medium.
Determination of porin expression levels in penB strains.
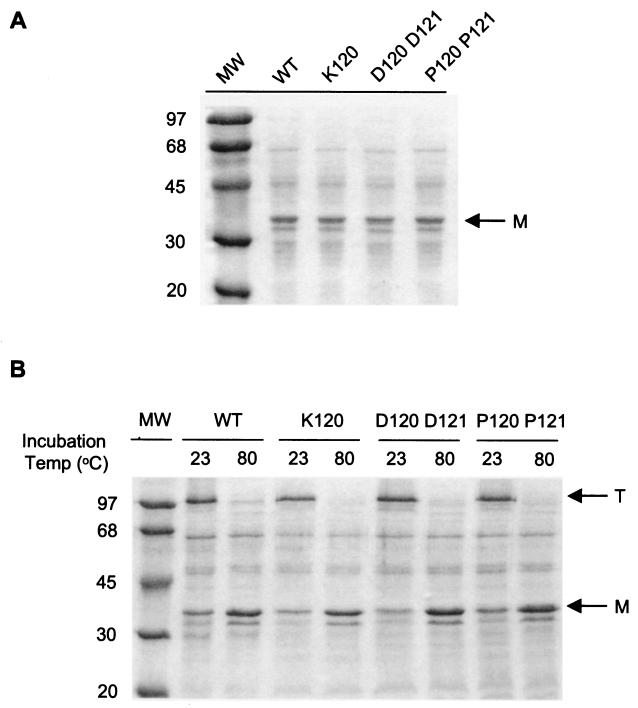
To determine whether the mutations in PIB altered either the level of porin expression or the propensity of the porin monomers to form trimers at the cell surface, we compared the levels of PIB protein expressed in strain FA19 penA4 mtr (PIB) to those in strains expressing PIB with G120K, G120D A121D, G120K A121R, or G120P A121P mutations. Coomassie blue staining (Fig. 4A) and Western blotting (data not shown) of whole-cell lysates showed no detectable differences in PIB expression among the tested strains over several experiments. To test the effects of the mutations on the integrity of porin trimers, outer membranes from the recipient and penB strains were incubated in SDS-PAGE loading buffer either at room temperature or at 80°C and subjected to SDS-PAGE. Protein staining with Coomassie blue indicated that porin trimers were present in equal amounts in all of the strains examined when they were incubated at room temperature and completely dissociated into monomers at 80°C (Fig. 4B). These results were further confirmed by immunoblotting (data not shown). Taken together, these data suggest that resistance to penicillin and tetracycline in penB strains is not a result of altered porin levels or the propensity of the porin to form trimers at the cell surface but is presumably due to alterations in the porin channel itself.
FIG. 4.
SDS-PAGE analysis of PIB expression. (A) N. gonorrhoeae strains (5 × 107 CFU/ml) containing either the FA1090 (WT) or a mutant (K120, D120 D121, or P120 P121) PIB protein were incubated at 80°C for 5 min in Laemmli loading buffer and subjected to SDS-PAGE. (B) Approximately 4 μg of outer membranes prepared from the strains described in the legend to panel A was incubated in Laemmli loading buffer at either 23 or 80°C for 5 min prior to electrophoresis. Proteins were stained with Coomassie brilliant blue. Arrows indicate porin monomers (M) and trimers (T).
The G120K mutation in PIA confers increased penicillin and tetracycline resistance.
N. gonorrhoeae has two porins, PIA and PIB, whose genes are alleles of a single locus (6). To date, increased resistance to penicillin and tetracycline in penB isolates has been correlated only with alterations in PIB and not with alterations in PIA. Since these proteins are 67% identical, we investigated whether mutation of Gly-120 to lysine in loop 3 of PIA, which is the residue equivalent to Gly-120 in PIB (Fig. 5), could also confer resistance to penicillin and tetracycline. FA19 penA4 mtr was transformed with porIA mutant DNA, and transformants were selected at a frequency of 1.6 × 10−5 on GCB plates containing 0.24 μg of penicillin/ml. MIC analysis of the transformed strain revealed that while the G120K mutation in PIA increases both tetracycline and penicillin resistance over that of the recipient strain, it does not increase resistance to the same levels as an isogenic strain harboring the G120K mutation in PIB (Fig. 6). This was especially evident in the levels of tetracycline resistance. FA19 penA4 mtr was also transformed with porIA G120D G121D DNA. The resultant strain showed only a small increase in resistance to penicillin and no increase in resistance to tetracycline (Fig. 6). These data suggest that structural differences in loop 3 or potentially in the barrel regions between PIB and PIA are critical for these mutations to increase resistance.
FIG. 5.
Alignment of PIA and PIB protein sequences. PIA and PIB proteins from strains FA19 and FA1090, respectively, were aligned with the Clustal-X program (41). The figure was generated from the aligned sequences by the Boxshade program (available on the World Wide Web).
FIG. 6.
MICs of penicillin and tetracycline for N. gonorrhoeae strains with PIA mutations. Strains harboring PIA with either a G120D A121D double mutation or a G120K single mutation were created via transformation of strain FA19 penA4 mtr with the altered porIA DNA. MICs are averages from four experiments. Error bars, standard deviations. MICs for the strain with the G120K PIB mutation are shown for comparison. Dashed lines indicate the respective MICs for the recipient strain, FA19 penA4 mtr.
Identification of PIB amino acid mutations at residues 120 and 121 in clinical strains.
To determine the prevalence of naturally occurring charged amino acids at PIB positions 120 and 121, we examined the loop 3 sequences of 51 clinical isolates whose por sequences are present in the GenBank database (11, 19, 27). These strains represented 14 distinct PIB serovars and separate geographical locations, including the United States, Singapore, Great Britain, and Kenya. Of these, 15 strains (29.4%) had lysine and aspartate codons at positions 120 and 121, respectively, 6 strains (11.8%) had an aspartate residue at position 120 or 121, and 3 strains (5.9%) harbored aspartate codons at both positions. We also analyzed the loop 3 sequences of 45 PIA proteins. At residue 120, 21 strains (46.6%) had an aspartate residue whereas the rest of the isolates had a glycine. Only a glycine was seen at position 121. These data indicate that mutations of residues 120 and/or 121 are common in clinical isolates, especially mutations to charged amino acids.
DISCUSSION
The focus of this study was to investigate the structure-function aspects of porin-mediated resistance in N. gonorrhoeae. We showed that alteration of residues 120 and 121 of PIB to aspartic acids confers intermediate-level resistance to penicillin and tetracycline. We also showed that a single-amino-acid mutation in PIB, G120K, is sufficient to confer full intermediate-level antibiotic resistance when expressed in a penA mtr recipient strain. Moreover, lysine and aspartic acid residues were found at positions 120 and/or 121 in the PIB proteins of approximately 47% of clinical isolates in the GenBank database, indicating that these changes occur in nature.
Our data demonstrated that mutations in PIB at positions 120 and/or 121, particularly to charged amino acids, are capable of conferring antibiotic resistance in N. gonorrhoeae. A single Asp mutation at either position 120 or 121 conferred only partial resistance to the antibiotics, showing that Asp mutations at both these positions were necessary for conferring the same level of resistance as the FA140 PIB. Moreover, mutation of Gly-120 to Glu conferred similar levels of resistance to the recipient strain as PIB containing a single Gly-120-to-Asp mutation. Although we did not examine the effects of double Glu mutations at positions 120 and 121 on resistance, we suspect that this mutant would show levels of resistance similar to those of the double Asp mutant. In contrast, only a single G120K mutation was necessary to confer the same level of penicillin resistance and even higher levels of tetracycline resistance to the recipient strain. The latter observation explains both the abundance of lysine and the lack of aspartates in the randomized library screen. The expected frequency of selecting a single lysine codon at position 120 is 1 of 32 (since 2 of 64 codons encode lysine) compared to 1 of 1,024 for transformants harboring aspartic acid codons at both positions [the product of the frequency of aspartate codons at each position, (1/32)2].
Gram-negative bacteria have developed two main porin-mediated mechanisms to increase resistance to antibiotics. One of these mechanisms, in which the bacteria become resistant to antibiotics following a loss of porin expression, is observed in many gram-negative bacteria, including Pseudomonas aeruginosa and Klebsiella pneumoniae (25, 32, 42). Because certain antibiotics diffuse selectively through a particular porin, loss of that porin species increases resistance to these antibiotics. The second porin-mediated mechanism, as described in this study, utilizes structural alterations in porin channels that presumably decrease the flux of antibiotics across the outer membrane. The latter mechanism is utilized by N. gonorrhoeae because it expresses only a single porin allele, and gonococci lacking porin are not viable (6). Interestingly, Enterobacter aerogenes has been reported to increase resistance to antibiotics via both mechanisms (10, 13).
Previous protein sequence alignments of N. gonorrhoeae PIB with other porins placed residues 120 and 121 in loop 3, which folds into the lumen of the barrel and constricts the porin channel (14). Recently, the structure of the Comomonas acidovorans Omp32 porin, which has higher homology to gonococcal PIB than other porins whose structures are known, was solved at a resolution of 2.1 Å (45). The amino acids of the C. acidovorans Omp32 monomer that correspond to N. gonorrhoeae PIB residues 120 and 121, as determined from an alignment of these two porin protein sequences, are Thr-102 and Ser-103. In the crystal structure of Omp32, these two residues are located on a small α-helix in loop 3 with their side chains pointing into the channel, in an ideal position to affect the permeation of antibiotics. Gill et al. (20) predicted that residues in loop 3 of PIB were important for mediating resistance to antibiotics, although no supporting evidence was presented. We have confirmed and extended their work, and we show that residues 120 and 121 in loop 3 are responsible for conferring porin-mediated resistance to both penicillin and tetracycline.
Crystal structures of Rhodobacter capsulatus and E. coli porins were the first to show that loop 3 folds into the barrel and constricts pore size (12, 43). The importance of loop 3 in the permeation, ion conductance, and ion selectivity of several porins has been investigated in detail. These studies have related alterations in the constriction loop to changes in pore properties. Specifically, studies with the phosphate starvation-induced E. coli porin PhoE revealed that mutation of a single lysine residue (Lys-125) to glutamic acid in loop 3 reverses ion selectivity from anionic to cationic and inhibits interaction with polyphosphate (1). Deletions and site-directed mutagenesis of loop 3 in E. coli OmpF and OmpC resulted not only in changes in ion selectivity but also in changes in sugar permeation and antibiotic susceptibility (28, 31, 33, 36). As with the PIB of N. gonorrhoeae, mutations in loop 3 of E. aerogenes porin conferred reduced susceptibility to cephalosporins (10). In vitro studies with E. coli porins have shown that mutations in the barrel also confer altered antibiotic susceptibilities and pore properties (31, 33, 36). It is not clear whether residues other than 120 and 121 in PIB contribute to antibiotic resistance in clinical strains of N. gonorrhoeae; however, our data suggest that mutations in loop 3 are the predominant means by which intermediate-level resistance to penicillin and tetracycline is conferred in penB strains.
Antibiotic resistance in both laboratory and clinical strains of N. gonorrhoeae has been linked to PIB serovars and not to PIA (44). This may be because an aspartate residue at position 120 in PIA confers only modest resistance to penicillin and no resistance to tetracycline over that of wild-type PIA. However, the mechanism underlying the differential susceptibility to tetracycline and penicillin observed in this mutant is currently unknown. The PIA sequences from clinical isolates in the GenBank database show only a glycine or an aspartate at this position. However, since a G120K mutation in PIA confers modest levels of resistance to penicillin and tetracycline over those of wild-type PIA, it is intriguing that a lysine mutation has not developed naturally in PIA proteins. Reasons for this may include biological differences between PIA and PIB proteins, which may contribute to differing selection pressures, including infection localization and serum resistance properties (3, 6).
The mechanism(s) by which the mutations in loop 3 of N. gonorrhoeae PIB confer antibiotic resistance is presently unknown. Gill et al. (20) speculated that aspartic acid residues in loop 3 of the FA140 PIB decrease antibiotic flux via anionic repulsion. However, we believe this mechanism is unlikely, since we show in this study that a basic lysine residue at position 120 also mediates intermediate-level resistance to both penicillin and tetracycline. Our data suggest that mutations at residues 120 and/or 121 in PIB alter the conformation of loop 3 to decrease the flux of antibiotics into the periplasmic space. This model is further supported by the recovery of a penB transformant with proline residues in this region, which are known to perturb protein structure due to the incorporation of a secondary amide linkage into the peptide backbone. Previous studies also have shown that expression of the penB phenotype in the gonococcus depends on prior acquisition of the mtr resistance gene (39). Electrophysiological and biochemical studies of purified PIB proteins to address the mechanism of resistance to penicillin and tetracycline conferred by mutations in PIB and the apparent synergism between mtr and penB are currently in progress.
Acknowledgments
This work was supported by grant AI-36901 from the National Institutes of Health.
We gratefully acknowledge the assistance and invaluable advice of Janne Cannon. We also thank Janne Cannon for providing the H5 monoclonal antisera, Joanne Dempsey and Nan Guyer for help and constructive discussions, and Mei Hu for technical support and suggestions.
REFERENCES
- 1.Bauer, K., M. Struyve, D. Bosch, R. Benz, and J. Tommassen. 1989. One single lysine residue is responsible for the special interaction between polyphosphate and the outer membrane porin PhoE of Escherichia coli. J. Biol. Chem. 264:16393-16398. [PubMed] [Google Scholar]
- 2.Brannigan, J. A., I. A. Tirodimos, Q. Y. Zhang, C. G. Dowson, and B. G. Spratt. 1990. Insertion of an extra amino acid is the main cause of the low affinity of penicillin-binding protein 2 in penicillin-resistant strains of Neisseria gonorrhoeae. Mol. Microbiol. 4:913-919. [DOI] [PubMed] [Google Scholar]
- 3.Cannon, J. G., T. M. Buchanan, and P. F. Sparling. 1983. Confirmation of association of protein I serotype of Neisseria gonorrhoeae with ability to cause disseminated infection. Infect. Immun. 40:816-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon, J. G., D. G. Klapper, E. Y. Blackman, and P. F. Sparling. 1980. Genetic locus (nmp-1) affecting the principal outer membrane protein of Neisseria gonorrhoeae. J. Bacteriol. 143:847-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon, J. G., and P. F. Sparling. 1984. The genetics of the gonococcus. Annu. Rev. Genet. 38:111-133. [DOI] [PubMed] [Google Scholar]
- 6.Carbonetti, N. H., V. I. Simnad, H. S. Seifert, M. So, and P. F. Sparling. 1988. Genetics of protein I of Neisseria gonorrhoeae: construction of hybrid porins. Proc. Natl. Acad. Sci. USA 85:6841-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1998. 1998 Guidelines for treatment of sexually transmitted diseases. Morb. Mortal. Wkly. Rep. 47:1-118. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2000. Fluoroquinolone resistance in Neisseria gonorrhoeae, Hawaii, 1999, and decreased susceptibility to azithromycin in N. gonorrhoeae, Missouri, 1999. Morb. Mortal. Wkly. Rep. 49:833-837. [PubMed] [Google Scholar]
- 9.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevalier, J., J. M. Pages, and M. Mallea. 1999. In vivo modification of porin activity conferring antibiotic resistance to Enterobacter aerogenes. Biochem. Biophys. Res. Commun. 266:248-251. [DOI] [PubMed] [Google Scholar]
- 11.Cooke, S. J., H. de la Paz, C. La Poh, C. A. Ison, and J. E. Heckels. 1997. Variation within serovars of Neisseria gonorrhoeae detected by structural analysis of outer-membrane protein PIB and by pulsed-field gel electrophoresis. Microbiology 143:1415-1422. [DOI] [PubMed] [Google Scholar]
- 12.Cowan, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Ghosh, R. A. Pauptit, J. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 13.De, E., A. Basle, M. Jaquinod, N. Saint, M. Mallea, G. Molle, and J. M. Pages. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41:189-198. [DOI] [PubMed] [Google Scholar]
- 14.Derrick, J. P., R. Urwin, J. Suker, I. M. Feavers, and M. C. Maiden. 1999. Structural and evolutionary inference from molecular variation in Neisseria porins. Infect. Immun. 67:2406-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dougherty, T. J., A. E. Koller, and A. Tomasz. 1980. Penicillin-binding proteins of penicillin-susceptible and intrinsically resistant Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 18:730-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowson, C. G., A. E. Jephcott, K. R. Gough, and B. G. Spratt. 1989. Penicillin-binding protein 2 genes of non-β-lactamase-producing, penicillin-resistant strains of Neisseria gonorrhoeae. Mol. Microbiol. 3:35-41. [DOI] [PubMed] [Google Scholar]
- 17.Elkins, C., C. E. Thomas, H. S. Seifert, and P. F. Sparling. 1991. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J. Bacteriol. 173:3911-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruki, H., and P. F. Sparling. 1986. Genetics of resistance in a non-β-lactamase-producing gonococcus with relatively high-level penicillin resistance. Antimicrob. Agents Chemother. 30:856-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fudyk, T. C., I. W. Maclean, J. N. Simonsen, E. N. Njagi, J. Kimani, R. C. Brunham, and F. A. Plummer. 1999. Genetic diversity and mosaicism at the por locus of Neisseria gonorrhoeae. J. Bacteriol. 181:5591-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill, M. J., S. Simjee, K. Al-Hattawi, B. D. Robertson, C. S. Easmon, and C. A. Ison. 1998. Gonococcal resistance to β-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob. Agents Chemother. 42:2799-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman, S. D., and J. J. Scocca. 1991. Factors influencing the specific interaction of Neisseria gonorrhoeae with transforming DNA. J. Bacteriol. 173:5921-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunn, J. S., and D. C. Stein. 1996. Use of a non-selective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol. Gen. Genet. 251:509-517. [DOI] [PubMed] [Google Scholar]
- 23.Guymon, L. F., D. L. Walstad, and P. F. Sparling. 1978. Cell envelope alterations in antibiotic-sensitive and-resistant strains of Neisseria gonorrhoeae. J. Bacteriol. 136:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez-Alles, S., M. Conejo, A. Pascual, J. M. Tomas, V. J. Benedi, and L. Martinez-Martinez. 2000. Relationship between outer membrane alterations and susceptibility to antimicrobial agents in isogenic strains of Klebsiella pneumoniae. J. Antimicrob. Chemother. 46:273-277. [DOI] [PubMed] [Google Scholar]
- 26.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 27.Hobbs, M. M., T. M. Alcorn, R. H. Davis, W. Fischer, J. C. Thomas, I. Martin, C. Ison, P. F. Sparling, and M. S. Cohen. 1999. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J. Infect. Dis. 179:371-381. [DOI] [PubMed] [Google Scholar]
- 28.Jeanteur, D., T. Schirmer, D. Fourel, V. Simonet, G. Rummel, C. Widmer, J. P. Rosenbusch, F. Pattus, and J. M. Pages. 1994. Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc. Natl. Acad. Sci. USA 91:10675-10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellogg, D. S., W. L. Peacock, W. E. Deacon, L. Browh, and C. I. Perkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to colonial variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamden, P. R., and J. E. Heckels. 1979. Outer membrane protein composition and colonial morphology of Neisseria gonorrhoeae strain P9. FEMS Microbiol. Lett. 5:263-265. [Google Scholar]
- 31.Lou, K. L., N. Saint, A. Prilipov, G. Rummel, S. A. Benson, J. P. Rosenbusch, and T. Schirmer. 1996. Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis. J. Biol. Chem. 271:20669-20675. [PubMed] [Google Scholar]
- 32.Martinez-Martinez, L., S. Hernandez-Alles, S. Alberti, J. M. Tomas, V. J. Benedi, and G. A. Jacoby. 1996. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 40:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misra, R., and S. A. Benson. 1988. Isolation and characterization of OmpC porin mutants with altered pore properties. J. Bacteriol. 170:528-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morse, S. A., S. R. Johnson, J. W. Biddle, and M. C. Roberts. 1986. High-level tetracycline resistance in Neisseria gonorrhoeae is result of acquisition of streptococcal tetM determinant. Antimicrob. Agents Chemother. 30:664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769-775. [DOI] [PubMed] [Google Scholar]
- 36.Saint, N., K. L. Lou, C. Widmer, M. Luckey, T. Schirmer, and J. P. Rosenbusch. 1996. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterization. J. Biol. Chem. 271:20676-20680. [PubMed] [Google Scholar]
- 37.Sarubbi, F. A., Jr., P. F. Sparling, E. Blackman, and E. Lewis. 1975. Loss of low-level antibiotic resistance in Neisseria gonorrhoeae due to env mutations. J. Bacteriol. 124:750-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarubbi, F. A., Jr., E. Blackman, and P. F. Sparling. 1974. Genetic mapping of linked antibiotic resistance loci in Neisseria gonorrhoeae. J. Bacteriol. 120:1284-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sparling, P. F., F. A. Sarubbi, Jr., and E. Blackman. 1975. Inheritance of low-level resistance to penicillin, tetracycline, and chloramphenicol in Neisseria gonorrhoeae. J. Bacteriol. 124:740-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spratt, B. G. 1988. Hybrid penicillin-binding proteins in penicillin-resistant strains of Neisseria gonorrhoeae. Nature 332:173-176. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., F. Plewniak, and O. Poch. 1999. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 27:2682-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trias, J., and H. Nikaido. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, M. S., A. Kreusch, E. Schiltz, U. Nestel, W. Welte, J. Weckesser, and G. E. Schulz. 1991. The structure of porin from Rhodobacter capsulatus at 1.8 Å resolution. FEBS Lett. 280:379-382. [DOI] [PubMed] [Google Scholar]
- 44.Woodford, N., K. M. Bindayna, C. S. Easmon, and C. A. Ison. 1989. Associations between serotype and susceptibility to antibiotics of Neisseria gonorrhoeae. Genitourin. Med. 65:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeth, K., K. Diederichs, W. Welte, and H. Engelhardt. 2000. Crystal structure of omp32, the anion-selective porin from Comamonas acidovorans, in complex with a periplasmic peptide at 2.1 Å resolution. Structure Fold Des. 8:981-992. [DOI] [PubMed] [Google Scholar]