Abstract
Six high-level evernimicin-resistant Enterococcus faecium isolates were identified among 304 avilamycin-resistant E. faecium isolates from animals and 404 stool samples from humans with diarrhea. All four animal isolates, and one of the human isolates, were able to transfer resistance to a susceptible E. faecium strain. The resulting transconjugants all tested positive for the presence of emtA, a gene encoding a methyltransferase previously linked with high-level evernimicin resistance. The four transconjugants derived from animal isolates all carried the same plasmid, while a differently sized plasmid was found in the isolate from humans. This study demonstrated a low incidence of high-level evernimicin resistance mediated by the emtA gene in different E. faecium isolates of animal and human origin.
There is a growing concern regarding the emergence of multiply antimicrobial-resistant enterococci as important nosocomial pathogens (7). This has increased the interest in either developing new antibiotics or modifying older antibiotics with activity against multiply resistant staphylococci and enterococci. One of these agents is evernimicin, an oligosaccharide antibiotic with activity against a broad range of gram-positive pathogenic bacteria including glycopeptide-resistant enterococci, methicillin-resistant staphylococci, and penicillin-resistant streptococci (9, 12, 14, 15, 17). Evernimicin inhibits protein synthesis in Staphylococcus aureus by binding with high affinity to a single site on the 50S subunit (13). The evernimicin binding site overlaps the binding site of another oligosaccharide antibiotic, avilamycin. Avilamycin has been used as a growth promoter for food animals in the European Union for several years, and as a consequence avilamycin resistance has been observed among Enterococcus faecium isolates from broilers in Denmark and other countries (4). High-level (>64 μg/ml) avilamycin resistance is mediated by mutations in the gene encoding ribosomal protein L16; these strains also exhibit low-level (MIC, 2 to 12 μg/ml) cross-resistance to evernimicin. (2). Recently, a new methyltransferase, which confers high-level (MIC, >64 μg/ml) evernimicin resistance through methylation of 23S rRNA, was cloned from an avilamycin-resistant E. faecium strain isolated from a broiler in Denmark (11). The present study was conducted to determine the occurrence of high-level evernimicin resistance among avilamycin-resistant enterococcal isolates from food animals and humans in Denmark.
A total of 304 avilamycin-resistant (MIC, ≥16 μg/ml) E. faecium and 14 Enterococcus faecalis isolates were obtained from the continuous monitoring for antimicrobial resistance among broilers and pigs in Denmark (4). These isolates were screened for high-level (MIC, >128 μg/ml) evernimicin resistance by the E-test according to the manufacturer's guidelines (AB Biodisk, Solna, Sweden); four positive isolates were identified. In March 1998 a total of 254 human stool samples, submitted for examination of diarrheal pathogens, were examined for the presence of avilamycin-resistant enterococci by plating a loopful of fecal material on Mueller-Hinton II agar plates containing 20 μg of avilamycin/ml. All patients sampled had a history of diarrhea but no history of either a recent hospital stay or antimicrobial treatment. One avilamycin-resistant E. faecium isolate was identified; this isolate also exhibited high-level evernimicin resistance. In June 2001 an additional 150 fecal samples from humans were, in relation to another study, screened for the presence of enterococcal isolates resistant to erythromycin and tetracycline with Slanetz and Bartley agar plates containing erythromycin (20 μg/ml) and tetracycline (10 μg/ml). Isolates obtained from these samples were also screened for avilamycin resistance; one isolate was recovered which also tested positive for high-level evernimicin resistance. All six isolates tested positive in PCR for the emtA gene with the following oligonucleotides as primers: 5′-GGTCAGCAGATCACTTGTTT-3′ and 5′-TGAACAATTCTAAGTCCTCG-3′.
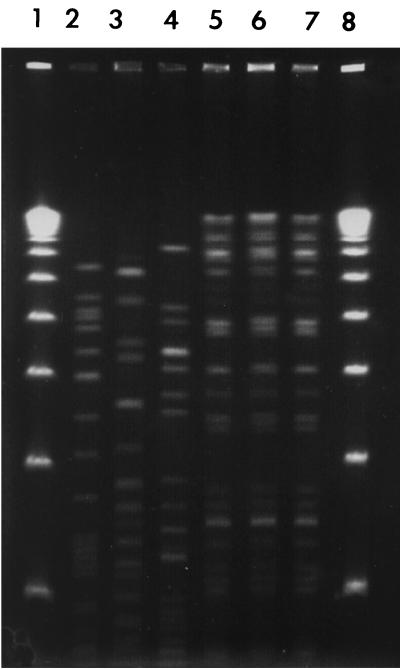
The six evernimicin-resistant isolates were subjected to pulsed-field gel electrophoresis (PFGE) typing with the restriction enzyme SmaI. DNA purification and enzyme digestion were performed as previously described (8). Four different PFGE types were identified: three of the four broiler isolates belonged to the same PFGE type, and the two isolates from humans gave different PFGE patterns (Fig. 1). We screened all six isolates for the ability to transfer evernimicin resistance to E. faecium BM4105 (resistant to rifampin and fusidic acid) by the filter mating procedure described previously (6). Transconjugants were isolated from within the inhibition zone of an E-test strip laid down on Mueller-Hinton II agar plates containing rifampin (50 μg/ml) and fusidic acid (10 μg/ml). All four animal isolates and one of the human isolates yielded transconjugants. Transconjugants were screened for susceptibility to avilamycin, bacitracin, chloramphenicol, erythromycin, gentamicin, kanamycin, penicillin, streptomycin, quinupristin-dalfopristin, tetracycline, and vancomycin as previously described (3). Resistance to avilamycin was cotransferred, but no other drug resistance markers appeared to be cotransferred. Plasmid DNA was extracted from the five transconjugants with the Qiagen Plasmid Midi kit (Qiagen, Valencia, Calif.) and restricted with either EcoRI or PvuII. EcoRI digestion of the plasmids from the four broiler isolates yielded three different bands, while PvuII digestion yielded five bands. The plasmids from the four broiler isolates were approximately 36 kb and indistinguishable even though the four isolates fell into two different PFGE patterns. In contrast, the human isolate harbored a plasmid of >100 kb. EcoRI digestion yielded 22 bands, and PvuII digestion yielded 18 bands. To determine if high-level evernimicin resistance was linked to the presence of the emtA gene (11), we performed a Southern blot analysis on the restricted plasmids (8). A digoxigenin-labeled emtA fragment was prepared by PCR with the oligonucleotides described above. The emtA probe hybridized to DNA fragments of approximately 6.9 and 12.5 kb in the EcoRI and PvuII restriction digests, respectively, of the transconjugants from the four broiler isolates (data not shown). In contrast, the emtA probe hybridized to fragments of 3.3 and 4.5 kb in the EcoRI and PvuII digests of the transconjugant of the human isolate.
FIG. 1.
PFGE profile of six high-level evernimicin-resistant E. faecium isolates from humans and broilers. Lanes 1 and 8, 50-kb ladder; lane 2, isolate 1 from humans; lane 3, isolate 2 from humans; lanes 4 to 7, isolates 1 to 4, respectively, from broilers.
The present study demonstrated that high-level evernimicin resistance has not been widely disseminated among E. faecium strains isolated from the animal or human population in Denmark. In the four isolates from broilers, three of which appeared clonal, the emtA gene was carried by a similarly sized plasmid in each case. In contrast, a plasmid of different size was observed in the single human isolate capable of transferring evernimicin resistance. Since the emtA gene is part of a transposon (11), it is possible that it was transposed from one plasmid to a different plasmid. Several studies have indicated that bacteria from animals and humans share the same resistance genes and that exchange probably occurs fairly frequently (10, 16, 19). This includes resistance genes in enterococci where the genes encoding resistance to important human antibiotics such as vancomycin (vanA) and quinupristin-dalfopristin [vat(E) and erm(B)] probably have spread from the animal reservoir to humans and thereby perhaps shortened the life span of these antibiotics (1, 5, 18).
Acknowledgments
We are grateful to Berith Kummerfelt, René Hendriksen, Betina Elemark, Dorte Nielsen, and Christina Aaby Svendsen for technical assistance.
The E-test was supplied by Schering-Plough Research Institute, Kenilworth, N.J.
REFERENCES
- 1.Aarestrup, F. M. 2000. Occurrence, selection and spread of resistance to antimicrobial agents used for growth promotion for food animals in Denmark. APMIS Suppl. 101:1-48. [PubMed] [Google Scholar]
- 2.Aarestrup, F. M., and L. B. Jensen. 2000. Presence of variations in ribosomal protein L16 corresponding to susceptibility of enterococci to oligosaccharides (avilamycin and evernimicin). Antimicrob. Agents Chemother. 44:3425-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aarestrup, F. M., Y. Agersø, P. G. Smith, M. Madsen, and L. B. Jensen. 2000. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37:127-137. [DOI] [PubMed] [Google Scholar]
- 4.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aarestrup, F. M., P. Butaye, and W. Witte. 2002. Nonhuman reservoirs of enterococci, p. 55-99. In M. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 6.Clewell, D. B., F. Y. An, B. A. White, and C. Gawron-Burke. 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J. Bacteriol. 162:1212-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen, L. B., P. Ahrens, L. Dons, R. N. Jones, A. M. Hammerum, and F. M. Aarestrup. 1998. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J. Clin. Microbiol. 36:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, R. N., D. E. Low, and M. A. Pfaller. 1999. Epidemiologic trends in nosocomial and community-acquired infections due to antibiotic-resistant gram-positive bacteria: the role of streptogramins and other newer compounds. Diagn. Microbiol. Infect. Dis. 33:101-112. [DOI] [PubMed] [Google Scholar]
- 10.Levy, S. B. 1997. Antibiotic resistance: an ecological imbalance. Ciba Found. Symp. 207:1-9. [DOI] [PubMed] [Google Scholar]
- 11.Mann, P. A., L. Xiong, A. S. Mankin, A. S. Chau, C. A. Mendrick, D. J. Najarian, C. A. Cramer, D. Loebenberg, E. Coates, N. J. Murgolo, F. M. Aarestrup, R. V. Goering, T. A. Black, R. S. Hare, and P. M. McNicholas. 2001. EmtA, a rRNA methyltransferase conferring high-level evernimicin resistance. Mol. Microbiol. 41:1349-1356. [DOI] [PubMed] [Google Scholar]
- 12.Marshall, S. A., R. N. Jones, M. E. Erwin, et al. 1999. Antimicrobial activity of SCH27899 (Ziracin), a novel everninomicin derivative, tested against Streptococcus spp.: disk diffusion/etest method evaluations and quality control guidelines. Diagn. Microbiol. Infect. Dis. 33:19-25. [DOI] [PubMed] [Google Scholar]
- 13.McNicholas, P. M., D. J. Najarian, P. A. Mann, D. Hesk, R. S. Hare, K. J. Shaw, and T. A. Black. 2000. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 44:1121-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakashio, S., H. Iwasawa, F. Y. Dun, K. Kanemitsu, and J. Shimada. 1995. Everninomicin, a new oligosaccharide antibiotic: its antimicrobial activity, post-antibiotic effect and synergistic bactericidal activity. Drugs Exp. Clin. Res. 21:7-16. [PubMed] [Google Scholar]
- 15.Schouten, M. A., A. Voss, and J. A. Hoogkamp-Korstanje. 1999. Antimicrobial susceptibility patterns of enterococci causing infections in Europe. Antimicrob. Agents Chemother. 43:2542-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teuber, M. 2001. Veterinary use and antibiotic resistance. Curr. Opin. Microbiol. 4:493-499. [DOI] [PubMed] [Google Scholar]
- 17.Urban, C., N. Mariano, K. Mosinka-Snipas, C. Wadee, T. Chahrour, and J. J. Rahal. 1996. Comparative in-vitro activity of SCH 27899, a novel everninomicin, and vancomycin. J. Antimicrob. Chemother. 37:361-364. [DOI] [PubMed] [Google Scholar]
- 18.Werner, G., I. Klare, H. Heier, K. H. Hinz, G. Bohme, M. Wendt, and W. Witte. 2000. Quinupristin/dalfopristin-resistant enterococci of the satA (vatD) and satG (vatE) genotypes from different ecological origins in Germany. Microb. Drug Resist. 6:37-47. [DOI] [PubMed] [Google Scholar]
- 19.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996-997. [DOI] [PubMed] [Google Scholar]