Resistance to erythromycin in Streptococcus pneumoniae was first detected in 1967 in the United States and subsequently worldwide (11, 20). The corresponding mechanism was rapidly identified as ribosomal methylation, which had been primarily reported as being responsible for erythromycin resistance in staphylococci (44). Further spread of resistance was then noted in a few countries, such as France, where hospitals observed a sharp increase in the proportion of resistant pneumococci, which reached approximately 20% in 1984 (16). This trend was observed several years before the emergence and spread of penicillin resistance in pneumococci in France. Recently, an increasing number of countries have noted changes in the evolution of macrolide resistance. In some of them, such as the United States, increased incidence has been correlated with the emergence of a new mechanism of erythromycin resistance—efflux (39). This review is devoted to the mechanisms responsible for resistance to macrolides and related antibiotics in pneumococci.
THE MACROLIDES
Macrolides have a common structure formed by a large lactone ring. Erythromycin is a mixture of antibiotics that includes erythromycin A, which is the active compound and which has a 14-membered lactone ring with two sugars, l-cladinose and an amino sugar. Other commercially available macrolides derived from erythromycin A include clarithromycin, dirithromycin, roxithromycin, and azithromycin, which has an enlarged, 15-membered ring resulting from a nitrogen insertion. The structural modifications of erythromycin A resulted in improved pharmacokinetic profiles and better tolerance, but cross-resistance between members of this class of antimicrobial agents was still observed. Certain 16-membered macrolides are also available in a few countries (spiramycin, josamycin, midecamycin, and miocamycin) or for veterinary use (tylosin). The recently developed ketolides telithromycin and ABT773 are derived from clarithromycin and have two major modifications, replacement of l-cladinose by a keto function and an 11- to 12-carbamate extension with an arylalkyl modification in telithromycin, the latter of which may partially explain its increased intrinsic activity and activity against erythromycin-resistant strains, as discussed below (13, 33). In telithromycin and ABT773, modification at the C-6 position prevents inactivation of the molecule in acid medium.
RIBOSOME BINDING SITE AND MODE OF ACTION OF ERYTHROMYCIN
The ribosome structure and contact points between the ribosome and erythromycin A were recently identified by crystallography studies (35). The bacterial ribosome is formed by a small, 30S subunit and a large, 50S subunit. The latter is composed of 23S rRNA and of a minimum of 30 proteins. The secondary structure of 23S rRNA is folded due to base pairing and forms six domains numbered I to VI, while the tertiary structure of the molecule is maintained by its interactions with proteins. Stoichiometric binding of erythromycin A to the 50S subunit causes inhibition of protein synthesis.
The binding site of erythromycin is composed of domain V sequences near the peptidyltransferase center, where the polypeptide chain is synthesized. Hairpin 35 in domain II is in the vicinity of this binding site (1, 17). High-resolution X-ray structures of the 50S ribosomal subunit of Deinococcus radiodurans complexed with erythromycin A showed that the 2′-OH group of the desosamine sugar of the antibiotic appears to form three hydrogen bonds with adenines at positions 2058 and 2059 (Escherichia coli numbering) (35). The dimethylamino group of the desosamine sugar also appears to interact with A2505. The 6-OH of the lactone ring may form a hydrogen bond with A2062, the 11-OH and 12-OH may form one hydrogen bond with U2609, but the cladinose sugar does not seem to be involved in interactions with 23S rRNA. Although footprinting experiments have implicated adenine at position 752 (domain II) in the binding of erythromycin, no direct interaction has been shown between the two structures, at least in the ribosome of D. radiodurans (17, 35). The binding site of erythromycin A is located within the tunnel that serves as a channel for the growing peptide. The surface of this tunnel is formed by domains I to V of 23S rRNA, by several ribosomal proteins including the globular structures of ribosomal proteins L22 and L4, and by a β hairpin of L22 (27). Erythromycin does not inhibit the peptidyltransferase activity but prevents the extension of the peptide chain by blocking the polypeptide exit tunnel and provokes the premature release of peptidyl-tRNA (24). Moreover, erythromycin also prevents ribosomal assembly at an early stage of protein synthesis (6).
MECHANISMS OF RESISTANCE TO MACROLIDES
A common mechanism shared by bacteria for becoming resistant to antimicrobial agents is the diminution of the affinity of the antibiotic for its target. This effect may result from enzymatic detoxification of the drug or, conversely, from target modification. A third possibility is diminished access to the target secondary to active efflux or decreased uptake of the molecules. The resistance of Streptococcus pneumoniae to erythromycin is due to modification of the ribosomal target by methylation or mutation and active efflux of the drug; drug modification has not been reported in this species.
RIBOSOMAL METHYLATION: THE MLSB RESISTANCE PHENOTYPE
As already mentioned, ribosomal modification by methylation was the first mechanism of resistance to erythromycin elucidated and remained unique for decades. It is secondary to the acquisition of an erm gene (erythromycin ribosome methylase) usually carried by transposable elements in pneumococci. This gene encodes a ribosomal methylase which dimethylates pneumococcal 23S rRNA at a single site, adenine at position 2058 (44). As previously alluded to, the A2058 nucleotide is a key nucleotide for the binding of erythromycin. The modification markedly reduces the affinity of erythromycin for its target, probably by preventing direct access to the target or by modifying the conformation of the binding site. Cross-resistance to macrolides, lincosamides, and streptogramin B antibiotics (Table 1), which gave its name to the MLSB resistance phenotype, is due to the overlapping binding sites of the drugs (44).
TABLE 1.
Macrolide-lincosamide-streptogramin B resistance in S. pneumoniae due to gene acquisition
| Gene(s) | Resistance phenotype(s)a | Phenotype forb:
|
Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 14-M and 15-M | K | 16-M | L | SB | SA | SA+B | |||
| erm(B) | MLSB (i) | R | S | R | r or R | r or R | S | S | 33 |
| MLSB (c) | R | R | R | R | R | S | S | ||
| erm(A) | MLSB (c) | R | R | R | R | R | S | S | 38 |
| mef(A) | M | R | S | S | S | S | S | S | 39 |
| erm(B) + mef(A) | MLSB + M | R | ND | R | R | R | S | S | 23 |
c, constitutive; i, inducible.
14-M, 15-M, and 16-M, 14-, 15-, and 16-membered macrolides, respectively; K, ketolides; L, lincosamides; SB, streptogramin B; SA, streptogramin A; SA+B, streptogramin A and B; r, low-level resistance; R, high-level resistance; S, susceptibility; ND, not determined.
erm DETERMINANTS
The erm(B) determinant, initially called erm(AM), was first characterized on plasmid pAM77 in Streptococcus sanguis A1 isolated from dental plaque in 1978 (18). The gene is widely distributed, not only in S. pneumoniae but also in a variety of other streptococcal and enterococcal species, in enterobacteria, and in staphylococci, indicating easy exchange of genetic information even between phylogenetically remote species. In pneumococci, the gene is borne by conjugative transposons related to Tn1545, Tn1545-like elements, or a Tn917-like element that is part of a larger composite transposon, Tn3872 (8, 22). Transposition occurs from chromosome to chromosome of strains of S. pneumoniae. Both clonal spread of resistant strains and horizontal transfer of the element account for the high prevalence of the erm(B) gene in erythromycin-resistant pneumococci in certain countries. In one study, sequences homologous to the structural gene for the integrase of Tn1545, an enzyme required for the movements of the element, were found in all 36 S. pneumoniae strains resistant to erythromycin studied (30). Strains belonging to the 23F or 6B lineage appear to have erm(B) as part of Tn3872 or a modified form of Tn916 and Tn1545. Tn1545-like elements may also be exchanged between pneumococci by transformation. However, this mode of transfer, which is considered essential for the spread of beta-lactam resistance by alteration of genes for penicillin binding proteins in pneumococci, has not been shown for erythromycin resistance.
Although widely predominant, erm(B) is not the only representative of the erm gene class in pneumococci. The presence of an erm(A) gene has been reported for a single strain, isolated in Greece, to which it conferred cross-resistance to erythromycin and clindamycin (38) and for one strain with a resident erm(B) gene (2). This determinant, first detected in Streptococcus pyogenes, was initially designated ermTR and was subsequently included in the erm(A) gene class because of its close relatedness to erm(A) in Staphylococcus aureus (31).
REGULATION OF erm(B) EXPRESSION AND THE MLSB RESISTANCE PHENOTYPE
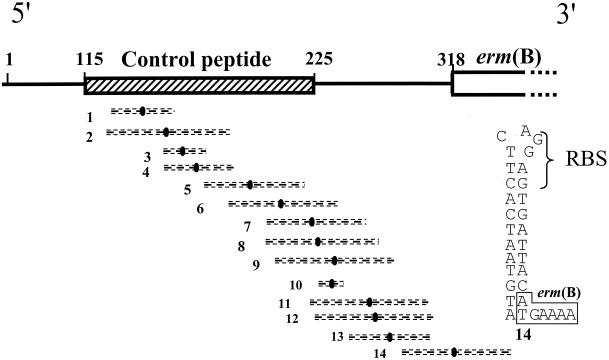
The methylase encoded by erm(B) may be constitutively or inducibly synthesized. When expression is constitutive, the erm(B) mRNA is active, and its translation by the ribosomes allows constitutive methylation of the ribosomes, probably while they are synthesized (45). When resistance is inducible, erm(B) mRNA is synthesized, but in an inactive conformation, and becomes active only in the presence of inducing macrolides. Although for erm(B) the mechanism of induction has not been thoroughly studied, a model which can be inferred from the translational regulation model of erm(C) in S. aureus (44) has been proposed and can be summarized as follows. The 5′ end of erm(B) presents a series of inverted repeats which are responsible for the lack of methylase synthesis in the absence of erythromycin (Fig. 1). Fourteen pairs of repeats have been identified which could form alternative stem-loop structures by base pairing (18). As shown in Fig. 1, one of these stem-loops sequesters the ribosome binding site and initiation codon for the methylase. Thus, the methylase cannot be produced, since the initiation motifs for translation of the enzyme are not accessible to the ribosomes. Induction is related to the presence of sequences coding for a small leader peptide of 36 amino acids upstream from the methylase gene. In the presence of low concentrations of erythromycin, binding of the antibiotic to a ribosome translating the leader peptide causes the ribosome to stall, in turn destabilizing the pairing of the inverted repeats and inducing conformational rearrangements in the mRNA. In particular, displacement of the stem-loop shown in Fig. 1 unmasks the initiation sequences for the methylase, allowing synthesis to proceed by the ribosomes that are not complexed with erythromycin or by those that are methylated. Methylation of some ribosomes might occur through transient rearrangements of the stem-loop structures, which would lead to the synthesis of a basal level of the methylase. For a given erm gene, the inducing capacity of the macrolides depends on the antibiotic structure. The global structure of the drug, rather than the number of atoms in the lactone ring, determines the inducing capacity of a macrolide. As an example, erythromycin is an inducer for the production of most Erm methylases, whereas ketolides, which have a similar lactone ring, are not. A lack of inducing ability of ketolides has been related to the replacement of one of the erythromycin sugars, l-cladinose, by a keto function (4, 32). It is likely that the intimate mode of action of a macrolide determines its capacity to act as an inducer, since proper ribosome stalling is required for the induction of methylase production. For erm(B), the commercially available macrolides (including the 14-, 15-, and 16-membered macrolides), lincosamides, and streptogramin B antibiotics are inducers of methylase synthesis to various degrees, leading to cross-resistance to these antimicrobial agents.
FIG. 1.
Schematic representation of the structure of the mRNA from the inducible erm(B) gene from pAM77. The sequences of the control peptide (hatched box) and of the methylase [(erm(B)] are shown. Numbers 1 to 14 indicate inverted repeats with their symmetry axes (solid ovals flanked by broken lines). The secondary structure which is putatively formed by inverted repeat 14 and which would sequester the initiation sequence for the methylase in the absence of erythromycin (18) is shown at the right. RBS, ribosome binding site.
It has been shown for erm(A) and erm(C), both in laboratory mutants and in clinical isolates, that constitutive expression is due to deletions, duplications, or point mutations in the attenuator sequence leading to derepressed production of the methylase (45). In pneumococci, the constitutive expression of MLSB resistance is infrequently found (33). However, despite the fact that the vast majority of pneumococci express erythromycin resistance inducibly, it has been shown by primer extension analysis of five strains that various proportions of ribosomes are methylated even in the absence of erythromycin (46). This paradox has been explained for certain strains by the presence of mutations in the stem-loop structure that sequester the initiation sequences for the methylase. Fusion of the mutated erm(B) attenuator with a lacZ reporter gene has confirmed that the expression of the methylase can be partly derepressed in certain strains (32). Other additional features, such as differences in the promoter strength or in the copy number of the erm(B) gene, may also account for the various levels of ribosomal methylation.
MACROLIDE EFFLUX
Physiological pumps conferring erythromycin resistance by efflux have been described for several gram-positive organisms, such as Cmr from Corynebacterium glutamicum, which belongs to the major facilitator superfamily class of pumps (19), but not for S. pneumoniae. However, acquired resistance to macrolides conferred by active efflux has been detected recently in this species (39). The gene responsible for efflux was initially called mefE and was subsequently assigned to the mef(A) gene class because of its close relatedness to the mefA gene in S. pyogenes (31). The Mef(A) pump belongs to the major facilitator superfamily class. It contains 12 transmembrane domains spanning the cytoplasmic membrane, and efflux is driven by the proton motive force (7). Few substrates have been identified, and the pump seems to be specific to erythromycin and its derivatives, including azithromycin. Resistance appears to be induced with erythromycin and is expressed at moderate levels, with erythromycin MICs of between 1 and 64 μg/ml (generally between 8 and 32 μg/ml). Because the 16-membered macrolides, the lincosamides, and the streptogramin B antibiotics are not substrates of the pump, these antimicrobial agents remain active, even after induction with erythromycin. Resistance to erythromycin combined with susceptibility to clindamycin, whether the cells are induced or not induced with erythromycin, defines the M resistance phenotype.
The mef(A) gene is transferable among pneumococci (9) and is a member of a group of closely related, large transposable elements (15, 34). Although the 7,244-bp transposon Tn1207.1 is apparently intact, it is defective for transfer (34), as is the 5.4- to 5.5-kb MEGA element, which is devoid of the transposase gene (15). Downstream from mef(A) lies a gene that putatively encodes an ATP binding cassette transporter and whose role in the expression of resistance remains questionable. The cloned mef(A) gene alone is sufficient to confer resistance, although it is not possible to exclude the possibility that the pump interacts with other proteins (7).
STREPTOGRAMINS AND TELITHROMYCIN
Both ribosomal methylation and drug efflux alter the activities of erythromycin A and its derivatives. Several strategies should allow MLSB resistance in pneumococci to be overcome: the use of methylase or efflux inhibitors, synergistic combinations with another antimicrobial agents, and the development of noninducing macrolides or of macrolides that have alternative ribosome binding sites or that are not substrates for the efflux pump. Two types of drugs have been developed with activities against MLSB-resistant pneumococci, the streptogramins and the ketolides. The streptogramins (pristinamycin and quinupristin-dalfopristin) are composed of two streptogramin factors, A and B, with synergistic activity resulting from a dual interaction with the ribosome (3). As mentioned above, Erm methylation of the ribosome affects the activity of the B component. However, synergy is maintained, most probably because of the mode of action of the streptogramins. Although the mechanism for synergy is not fully understood, the binding of factor A to its target may induce a conformational change in the ribosome leading to an increase in its affinity for factor B (3). The ribosomal alteration must be sufficiently marked to overcome the loss of affinity for the B molecule that results from rRNA methylation. The bactericidal activity of the streptogramin combination against pneumococci is also generally conserved in vitro (28).
The ketolides, like the macrolides, bind to the bacterial ribosome and exert their antibacterial effect by inhibition of protein synthesis. Despite the similarity between the macrolides and the ketolides, in terms of mechanism of action and therefore cross-resistance, recent data indicated that the ketolides have activity against MLSB-resistant pneumococci (21, 36). This finding appears to be due to two differences from the macrolides: the strength and nature of ribosome binding and the weak ability of the ketolides to act as inducers of macrolide resistance (12, 32). It has been shown that macrolides interact with two sites within the bacterial ribosome, domains II and V of 23S rRNA, with the interaction at domain II being relatively weak. The ketolides also interact with domains II and V but appear to have a 10-fold higher binding affinity (17). As discussed previously, MLSB resistance arises when the binding of the macrolides within domain V is compromised, principally through methylation. In contrast, the ketolides retain in part their ability to bind to MLSB-resistant ribosomes probably because of their stronger interaction with domain II. However, as previously mentioned, crystal studies with D. radiodurans 50S ribosomal subunits do not support the notion of direct contacts between the 14-membered macrolides and A752 or any other domain II residue (35). Alone, the increased ribosome binding property probably does not account for the activity of telithromycin against macrolide-resistant pneumococci. Another additional feature of the ketolides is their inability to induce MLSB resistance. Lack of induction of MLSB resistance with telithromycin is due to the replacement of the l-cladinose moiety at the C-3 position of the lactone ring by a ketone group (4). The basal production of methylase may affect weakly the activities of telithromycin and ABT773 because of their affinities for domain II. However, constitutive resistance or high-level basal production of methylase remains a stumbling block for the ketolides (32).
Compared to erythromycin, telithromycin is a weak inducer or substrate for the MefA pump. This fact is reflected by the difference in the increase in MICs due to this mechanism, 50-fold versus 500- to 2,000-fold, respectively (37).
RESISTANCE INDUCED BY RIBOSOMAL MUTATIONS
In vitro selection of E. coli mutants highly resistant to erythromycin has been of considerable value for characterization of the site of binding of this antibiotic to the ribosome. The clinical importance of this mechanism was recognized several years ago for microorganisms such as Helicobacter pylori and Mycobacterium avium but only recently for pneumococci (43).
Studies with pneumococcal mutants obtained in the laboratory have revealed that several structures participating in the binding of macrolides, domains V and II of 23S rRNA and proteins L22 and L4, can display mutations responsible for macrolide resistance (Table 2) (5, 40). Most mutations affect 23S rRNA and are similar to those reported for other bacterial species (43). S. pneumoniae has four copies of the rrl gene for 23S rRNA, and transformation experiments with mutated rrl have shown that susceptibility to erythromycin decreases as the number of the mutated gene copies increases (41). Since high-level erythromycin resistance can be achieved only when at least two copies are mutated, this finding may explain why resistance conferred by RNA mutation is rare in pneumococci compared to H. pylori or M. avium, which contain only one or two copies of the rrl gene.
TABLE 2.
MICs of macrolides and related antibiotics for ribosomal mutants of S. pneumoniae selected in vitro
| Resistance phenotype | Gene (product) | Alteration(s) | MIC (μg/ml) ofa:
|
Reference(s) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZM | CLR | ERY | SPI | JOS | TEL | SGB | PRI | CLI | ||||
| MLSB | rrl (23S rRNA domain V) | A2058G or A2058U | >32->200 | 16-100 | >32->100 | 6.25 | ND | 0.06-1 | 12.5 | 0.5 | 0.2-4 | 5, 40 |
| ML | rrl (23S rRNA domain V) | A2059G | >32 | 2 | 8 | ND | ND | 0.015 | ND | 0.25 | 2 | 5, 40 |
| ML (low level) | rrl (23S rRNA domain V) | C2610U | 0.125 | 0.03 | 0.06 | ND | ND | 0.008 | ND | 0.5 | 0.5 | 5 |
| MSB | rrl (23S rRNA domain V) | C2611A, C2611G, or C2611U | 0.5-12.5 | 0.06-100 | 0.06->100 | 0.78-1.56 | ND | 0.01-0.39 | 50->100 | 0.5 | 0.2-2 | 5, 40 |
| MKLSB | rrl (23S rRNA domain II) | A752 deletion | >32 | >32 | >32 | ND | ND | 4 | ND | 1 | 1 | 5 |
| MSB | rplD (L4 protein) | G69C and 67QSQKb | 0.39-1.56 | 0.05-0.2 | 0.05-0.2 | ND | 0.78-1.56 | 0.006-0.1 | 3.12 | ND | 0.05-0.2 | 40 |
| MS | rplV (L22 protein) | G95D, P99Q, A93E, P91S, and G83E | 0.06-1 | 0.125-1 | 0.25-1 | ND | ND | 0.06-0.25 | ND | 1-2 | 0.03-0.12 | 5 |
AZM, azithromycin; CLR, clarithromycin; ERY, erythromycin; SPI, spiramycin; JOS, josamycin, TEL, telithromycin; SGB, streptogramin B; PRI, pristinamycin IA; CLI, clindamycin. ND, not determined.
The alteration is underlined.
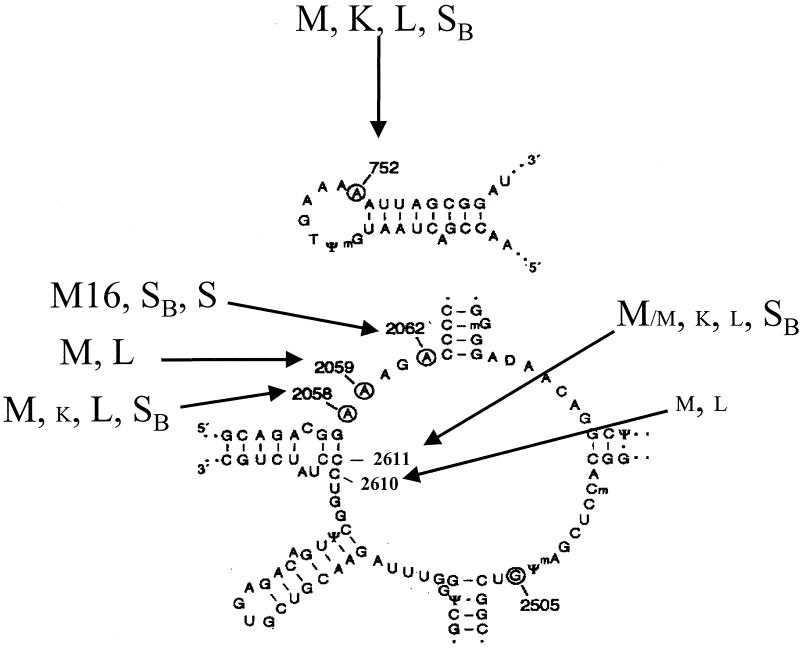
The resistance phenotype conferred by alterations in the 23S rRNA target varies not only according to the number of mutated copies but also according to the nature of the substituted base (Fig. 2) (43). Point mutations at position A2058 or A2059 are associated with phenotypes similar to those previously reported for other organisms. A2058G and A2058U substitutions confer the highest level of MLSB resistance, with MICs of erythromycin and related macrolides of between 32 and >200 μg/ml (5, 40). Telithromycin appears to be moderately affected (MICs of 0.06 to 1 μg/ml), probably because of the alternative interaction with domain II. Streptogramins retain activity, since synergy between the A and B factors is maintained.
FIG. 2.
Secondary structures of hairpin 35 in domain II (top) and in domain V (bottom) of 23S rRNA in E. coli. Nucleotides which are protected by erythromycin are circled (43). Arrows indicate mutations conferring macrolide resistance on S. pneumoniae. The corresponding phenotype is indicated (K, ketolides; L, lincosamides; M, macrolides; M16, 16-membered macrolides; SB, streptogramin B; S, streptogramin A and streptogramin B). Small capital letters denote low-level resistance.
The A2059G mutation confers a high level of resistance to erythromycin, azithromycin, and 16-membered macrolides, a moderate level of resistance to clarithromycin and clindamycin, but no resistance to streptogramins, defining the ML resistance phenotype (5, 40).
Mutations at position 2611 destabilize the base pairing G2057-C2611 in the single-strand structure of the central loop (Fig. 2). However, the C2611U substitution generally has a weak impact on the MICs of macrolides. Tait-Kamradt et al. (40) found higher levels of resistance to streptogramin B antibiotics conferred by C2611A and C2611G substitutions (Table 2).
The C2610U change has been reported only for pneumococci and yields a slight increase in the MICs of macrolides and clindamycin (5).
While telithromycin activity is only moderately altered by mutations in domain V, mutation of the adenine at position 752 in hairpin 35 (domain II) has a deleterious effect on the activity of the drug. A mutant combining a deletion of this base and a domain V mutation is resistant not only to 14- and 15-membered macrolides but also to telithromycin (MIC, 4 mg/liter), confirming the importance of domain II in the mechanism of action of this antibiotic (5).
Various mutations in the rplV (L22) and rplD (L4) genes have been shown to play a role in resistance in laboratory mutants and in transformants of a susceptible S. pneumoniae strain obtained with mutated genes (5, 40). The mutations in the L22 protein are located in a β-hairpin extension at the C terminus of the protein (5, 42). They confer resistance to streptogramins and low-level resistance to macrolides, whereas clindamycin does not seem to be affected (Table 2). The MICs of telithromycin are increased but remain below 0.25 μg/ml. The mutations in the L4 protein occur in a region of 32 amino acids highly conserved in various species and interfere with the binding of the protein to rRNA (40). These mutations generally confer an MSB resistance phenotype. The MICs of macrolides against the mutant strains are moderately increased. Studies by three-dimensional cryoelectron microscopy of erythromycin-resistant ribosomes of E. coli have shown that L4 and L22 mutants have substantial changes in the polypeptide tunnel (14). The L4 mutant which does not bind erythromycin has a narrowing of the tunnel entrance which probably decreases the capacity of erythromycin to come into contact with its target. In contrast, the L22 mutant has an enlargement of the entrance and could bind erythromycin but in an ineffective way.
Many of the mutations selected in vitro have been predictive of those found in clinical isolates (Table 3). The A2059G mutation confers an ML resistance phenotype (29, 41). A C2611G mutation was found in an isolate from Finland that was resistant to macrolides and highly resistant to streptogramin B antibiotics (29). Two types of rplD mutations in clinical isolates have been characterized (26, 29, 41). Sixteen isolates from Eastern Europe which were resistant to penicillin G and a Finnish isolate contained substitutions of three amino acids (69GTG71 → 69TPS71) and displayed an MSB resistance phenotype with a high level resistance to macrolides (26, 29). A Canadian isolate had a six-amino-acid insertion (underlined), 71GREKGTGR72, and displayed a similar phenotype but with a moderate level of resistance to all macrolides, including telithromycin (MIC, 3.12 μg/ml) (41). Recently, three strains isolated in Japan and for which the MICs of erythromycin were 64 or 128 μg/ml were reported to have an L22 mutation (D. J. Farrell, I. Morrissey, S. Bakker, D. Felmingham, J. Poehlsgaard, and S. Douthwaite, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1811, p. 100, 2001). The recent report of the emergence of an L22 mutant during treatment with azithromycin of fatal pneumococcal pneumonia emphasizes the clinical importance of mutations as a resistance mechanism (25). In summary, if, as expected, L4 and L22 mutants selected in vivo or in vitro have similar phenotypes, the MICs are surprisingly higher for the clinical isolates. The reasons for this difference are unknown but may be related, at least for L4 mutations, to differences in the types of mutations. Alternatively, other mechanisms of resistance to macrolide-lincosamide-streptogramin B antibiotics may also be present in wild strains.
TABLE 3.
MICs of macrolides and related antibiotics for ribosomal mutants of clinical isolates of S. pneumoniae
| Product | Mutation | No. of isolates | MIC (μg/ml) ofa:
|
Reference(s) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZM | CLR | ERY | SPI | JOS | TEL | QUI | PRI or Q-D | CLI | ||||
| 23S rRNA domain V (no. of mutated copies) | A2059G (2 or 3) | 5 | >100->512 | 12.5->512 | 50->100 | 512 | >100 | 0.01-0.25 | 3.12-32 | 2-4 | 0.78-2 | 29, 41 |
| A2059C (2 to 4) | 7 | 128-512 | 512->512 | 256->512 | 512->512 | ND | 0.06-0.12 | 16-32 | 1-2 | 1-2 | 29 | |
| A2062C (4) | 1 | 0.5 | ND | <0.25 | 512 | 64 | <0.0075 | 32 | 2 | <0.015 | 10 | |
| C2611G (4) | 1 | 128 | >512 | >512 | 16 | ND | 0.5 | >64 | 2 | 1 | 29 | |
| L4 protein | 69TPS71 | 17 | >100->512 | 12.5->512 | >100->512 | 64 | 100 | 0.03-0.2 | 12.5-25 | 2 | 0.05-0.2 | 29, 41 |
| Insertion (71GREKGTGR72)b | 1 | 12.5 | 12.5 | 6.25 | ND | 6.25 | 3.12 | 25 | ND | 0.05 | 41 | |
| L22 protein | Duplication (102KRTAHITRTAHITVA116)b | 1 | >1 | ND | >1 | ND | ND | ND | ND | >1 | ND | 25 |
QUI, quinupristin; Q-D, quinupristin-dalfopristin; see Table 2, footnote b, for other definitions.
The mutation is underlined.
A clinical isolate with an A2062C mutation not obtained so far in vitro had a particular phenotype of a high level of resistance to spiramycin and streptogramin B and a moderate level of resistance to streptogramins A and B and to the combination (10). It remained susceptible to 14- and 15-membered macrolides, to telithromycin, and to clindamycin. This new phenotype confirms the notion that the binding sites of 14- and 16-membered macrolides are distinct.
CONCLUSION
In recent years, both the incidence of macrolide resistance in pneumococci and the variety of resistance mechanism have increased sharply. The emergence of resistance mechanisms conferred by mutational alterations, in particular, is intriguing. This type of resistance may have remained undetected in the past because of a lack of adequate techniques or, alternatively, resistant mutants may have emerged and spread recently. It is conceivable that the use of new, long-acting macrolides with different pharmacokinetics may have contributed to modulation of the selective pressure exerted against pneumococci and to selection of new resistance genotypes. The variety of resulting phenotypes makes it particularly challenging to detect the nature of resistance in clinical isolates and may lead to difficulties in or make impossible the detection of resistance, depending on the individual drug(s) being tested.
Acknowledgments
We thank Milton Saier for laboratory hospitality.
REFERENCES
- 1.Ban, N., P. Nissen, P., J. Hansen, P. B. Moore, and, T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 2.Betriu, C., M. Redondo, M. L. Palau, A. Sanchez, M. Gomez, E. Culebras, A. Boloix, and J. J. Picazo. 2000. Comparative activities of linezolid, quinupristin-dalfopristin, moxifloxacin, and trovafloxacin against erythromycin-susceptible and -resistant streptococci. Antimicrob. Agents Chemother. 44:1838-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer, D., and K. Pepper. 1998. The streptogramin antibiotics: update on their mechanism of action. Exp. Opin. Investig. Drugs 7:591-599. [DOI] [PubMed] [Google Scholar]
- 4.Bonnefoy, A., A. M. Girard, C. Agouridas, and J. F. Chantot. 1997. Ketolides lack inducibility properties of MLS(B) resistance phenotype. J. Antimicrob. Chemother. 40:85-90. [DOI] [PubMed] [Google Scholar]
- 5.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champney, W. S., and C. L. Tober. 2000. Specific inhibition of 50S ribosomal subunit formation in Staphylococcus aureus cells by 16-membered macrolide, lincosamide, and streptogramin B antibiotics. Curr. Microbiol. 41:126-135. [DOI] [PubMed] [Google Scholar]
- 7.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 8.Courvalin, P., and C. Carlier. 1987. Tn1545: a conjugative shuttle transposon. Mol. Gen. Genet. 206:259-264. [DOI] [PubMed] [Google Scholar]
- 9.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon, J. M. S. 1967. Pneumococcus resistant to erythromycin and lincomycin. Lancet ii:474. [DOI] [PubMed]
- 12.Douthwaite, S., and W. S. Champney. 2001. Structures of ketolides and macrolides determine their mode of interaction with the ribosomal target site. J. Antimicrob. Chemother. 48(Suppl. T1):1-8. [DOI] [PubMed] [Google Scholar]
- 13.Felmingham, D., G. Zhanel, and D. Hoban. 2001. Activity of the ketolide antibacterial telithromycin against typical community-acquired respiratory pathogens. J. Antimicrob. Chemother. 48(Suppl. T1):33-42. [DOI] [PubMed] [Google Scholar]
- 14.Gabashvili, I. S., S. T. Gregory, M. Valle, R. Grassucci, M. Worbs, M. C. Wahl, A. E. Dahlberg, and J. Frank. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 8:181-188. [DOI] [PubMed] [Google Scholar]
- 15.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 16.Geslin, P., A. Buu-Hoi, A. Fremaux, and J. F. Acar. 1992. Antimicrobial resistance in Streptococcus pneumoniae: an epidemiological survey in France, 1970-1990. Clin. Infect. Dis. 15:95-98. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, L. H., P. Mauvais, and S. Douthwaite. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31:623-631. [DOI] [PubMed] [Google Scholar]
- 18.Horinouchi, S., W. H. Byeon, and B. Weisblum. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J. Bacteriol. 154:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jager, W., J. Kalinowski, and A. Puhler. 1997. A Corynebacterium glutamicum gene conferring multidrug resistance in the heterologous host Escherichia coli. J. Bacteriol. 179:2449-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klugman, K. P. 1990. Pneumococcal resistance to antibiotics. Clin. Microbiol. Rev. 3:171-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclercq, R. 2001. Overcoming antimicrobial resistance: profile of a new ketolide antibacterial, telithromycin. J. Antimicrob. Chemother. 48(Suppl. B):9-23. [DOI] [PubMed] [Google Scholar]
- 22.McDougal, L. K., F. C. Tenover, L. N. Lee, J. K. Rasheed, J. E. Patterson, J. H. Jorgensen, and D. J. LeBlanc. 1998. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2312-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGee, L., K. P. Klugman, A. Wasas, T. Capper, A. Brink, and The Antibiotics Surveillance Forum of South Africa. 2001. Serotype 19F multiresistant pneumococcal clone harboring two erythromycin resistance determinants [erm(B) and mef(A)] in South Africa. Antimicrob. Agents Chemother. 45:1595-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menninger, J. R. 1985. Functional consequences of binding macrolides to ribosomes. J. Antimicrob. Chemother. 16(Suppl. A):23-34. [DOI] [PubMed] [Google Scholar]
- 25.Musher, D. M., M. E. Dowell, V. D. Shortridge, R. K. Flamm, J. H. Jorgensen, P. Le Magueres, and, K. L. Krause. 2002. Emergence of macrolide resistance during treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:630-631. [DOI] [PubMed] [Google Scholar]
- 26.Nagai, K., P. C. Appelbaum, T. A. Davies, L. M. Kelly, D. B. Hoellman, A. T. Andrasevic, L. Drukalska, W. Hryniewicz, M. R. Jacobs, J. Kolman, J. Miciuleviciene, M. Pana, L. Setchanova, M. K. Thege, H. Hupkova, J. Trupl, and P. Urbaskova. 2002. Susceptibilities to telithromycin and six other agents and prevalence of macrolide resistance due to L4 ribosomal protein mutation among 992 pneumococci from 10 Central and Eastern European countries. Antimicrob. Agents Chemother. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 28.Pankuch, G. A., C. Lichtenberger, M. R. Jacobs, and P. C. Appelbaum. 1996. Antipneumococcal activities of RP 59500 (quinupristin-dalfopristin), penicillin G, erythromycin, and sparfloxacin determined by MIC and rapid time-kill methodologies. Antimicrob. Agents Chemother. 40:1653-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pihlajamaki, M., J. Kataja, H. Seppälä, J. Elliot, M. Leinonen, P. Huovinen, and J. Jalava. 2002. Ribosomal mutations in Streptococcus pneumoniae clinical isolates. Antimicrob. Agents Chemother. 46:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poyart-Salmeron, C., P. Trieu-Cuot, C. Carlier, and P. Courvalin. 1991. Nucleotide sequences specific for Tn1545-like conjugative transposons in pneumococci and staphylococci resistant to tetracycline. Antimicrob. Agents Chemother. 35:1657-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosato, A., H. Vicarini, A. Bonnefoy, J. F. Chantot, and R. Leclercq. 1998. A new ketolide, HMR 3004, active against streptococci inducibly resistant to erythromycin. Antimicrob. Agents Chemother. 42:1392-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosato, A., H. Vicarini, and R. Leclercq. 1999. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J. Antimicrob. Chemother. 43:559-562. [DOI] [PubMed] [Google Scholar]
- 34.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mefA in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlünzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath., and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 36.Shortridge, V. D., P. Zhong, Z. Cao, J. M. Beyer, L. S. Almer, N. C. Ramer, S. Z. Doktor, and R. K. Flamm. 2002. Comparison of in vitro activities of ABT-773 and telithromycin against macrolide-susceptible and -resistant streptococci and staphylococci. Antimicrob. Agents Chemother. 46:783-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutcliffe, J. 1999. Resistance to macrolides mediated by efflux mechanisms. Curr. Opin. Investig. Drugs 1:403-412. [Google Scholar]
- 38.Syrogiannopoulos, G. A., I. N. Grivea, A. Tait-Kamradt, G. D. Katopodis, N. G. Beratis, J. Sutcliffe, P. C. Appelbaum, and T. A. Davies. 2001. Identification of an erm(A) erythromycin resistance methylase gene in Streptococcus pneumoniae isolated in Greece. Antimicrob. Agents Chemother. 45:342-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Steptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unge, J., A. Aberg, S. Al-Kharadaghi, A. Nikulin, S. Nikonov, N. Davydova, N. Nevskaya, M. Garber, and A. Liljas. 1998. The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance. Structure 6:1577-1586. [DOI] [PubMed] [Google Scholar]
- 43.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong, P., Z. Cao, R. Hammond, Y. Chen, J. Beyer, V. D. Shortridge, L. Y. Phan, S. Pratt, J. Capobianco, K. A. Reich, R. K. Flamm, Y. S. Or, and L. Katz. 1999. Induction of ribosome methylation in MLS-resistant Streptococcus pneumoniae by macrolides and ketolides. Microb. Drug Resist. 5:183-188. [DOI] [PubMed] [Google Scholar]