Abstract
Kluyvera ascorbata produces a β-lactamase that results in an atypical susceptibility pattern, including low-level resistance to penicillins, cephalothin, and cefuroxime, but this resistance is reversed by clavulanate. Ten nucleotide sequences of the corresponding gene, blaKLUA, were obtained and were found to have minor variations (96 to 100%). Otherwise, blaKLUA was found to be similar (95 to 100%) to some plasmid-encoded CTX-M-type β-lactamases. Finally, mobilization of blaKLUA on a plasmid was found to be mediated probably by a genetic mobile element like ISEcp1.
Resistance to oxyimino-β-lactams such as cefotaxime, ceftazidime, or ceftriaxone in gram-negative rods has commonly been associated with the expression of acquired β-lactamases. Most of them are class A extended-spectrum β-lactamase (ESBL) derivatives of the TEM-1, TEM-2, or SHV-1 enzyme (4). Since the 1990s, a new step in resistance to β-lactams was discovered with the demonstration of the mobilization of a chromosomal β-lactamase on a plasmid, allowing the wide dissemination of β-lactamases and conferring the potential for epidemic problems. Many plasmid-mediated AmpC β-lactamases have been described, such as MIR-1 (Enterobacter cloacae), CMY-2 (Citrobacter freundii), DHA-1 (Morganella morganii), and ACC-1 (Hafnia alvei) (20). More recently, the discovery of SFO-1 suggested a probable mobilization from the chromosome of Serratia fonticola on a plasmid harbored in an E. cloacae isolate in Japan (16). To date, that was the only report of the mobilization of a chromosomally mediated class A β-lactamase.
The CTX-M-type β-lactamases (CTX-M-1 to CTX-M-15, Toho-1 and Toho-2, and UOE-1 and UOE-2), encoded by transferable plasmids, constitute a novel group of class A ESBLs whose origins are still unknown (2, 10, 11, 13, 15, 18, 21, 23). These CTX-M-type enzymes are not closely related to TEM or SHV ESBLs but share extensive similarities (70 to 75%) with the chromosomal Klebsiella oxytoca β-lactamases (23). Nevertheless, Kluyvera ascorbata, a species of the family Enterobacteriaceae that is rarely detected in medical practice (7, 24), was suggested as another probable progenitor because of the particular susceptibility patterns of some clinical isolates including resistance to cefotaxime and aztreonam but susceptibility to ceftazidime and the high potentiation effect of the reversal of resistance by clavulanate (D. Bertei et al., Abstr. 14th Réunion Interdisciplin. Chimiothér. Anti-Infect., abstr. 199, p. 10, 1994).
Bacterial strains.
Ten nonduplicate strains of K. ascorbata including the type strain were supplied by the Institut Pasteur Collection (Paris, France), and two strains were isolated at Tenon and Saint-Louis Hospitals (Paris, France), respectively. Each strain was identified on the basis of its behavior in culture and its biochemical characteristics by using the API 20E system (bioMérieux, Marcy-l'Etoile, France) and by using Biotype-100 carbon source utilization strips (bioMérieux) and Recognizer software (Taxolab, Institut Pasteur, Paris, France).
Antimicrobial agents and susceptibility testing.
The patterns of susceptibility to antibiotics usually active against members of the family Enterobacteriaceae were determined by the disk diffusion method, as described previously (8).
The MICs of 12 β-lactams (Table 1) including penicillins and cephalosporins in the presence or absence of β-lactamase inhibitors (clavulanate at 2 μg/ml or tazobactam at 4 μg/ml) were determined by a dilution method on Mueller-Hinton agar (Sanofi Diagnostics Pasteur, Marnes La Coquette, France) with an inoculum of 105 CFU/spot applied with a multi-inoculation device (Multipoint Inoculator A400; Denleytech, Woking, United Kingdom) (8).
TABLE 1.
MICs of 12 β-lactams for the 12 clinical K. ascorbata isolates, recipient strain E. coli XL-1, and the E. coli XL-1 transformant producing KLUA-1
| β-Lactam(s)a | No. of isolates for which MIC (μg/ml) was:
|
MIC (μg/ml) for E. coli XL-1 recipient
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | R+ | R− | |
| Amoxicillin | 1 | 1 | 4 | 4 | 2 | >128 | 4 | ||||||||
| Amoxicillin + CA | 3b | 3 | 4 | 2 | 1 | 1 | |||||||||
| Ticarcillin | 2 | 3 | 4 | 3 | >128 | 0.5 | |||||||||
| Ticarcillin + CA | 5b | 2 | 4 | 1 | 4 | 0.125 | |||||||||
| Piperacillin | 1 | 4 | 4 | 3 | 128 | 1 | |||||||||
| Piperacillin + TZ | 4b | 3 | 3 | 1 | 1 | 2 | 0.12 | ||||||||
| Cephalothin | 2 | 7 | 3 | >128 | 0.5 | ||||||||||
| Cefuroxime | 1 | 6 | 5 | 64 | 1 | ||||||||||
| Cefoxitin | 1 | 4 | 4 | 3 | 2 | 1 | |||||||||
| Cefotaxime | 3 | 7 | 2 | 128 | <0.03 | ||||||||||
| Ceftriaxone | 2 | 6 | 4 | 14 | <0.03 | ||||||||||
| Ceftazidime | 6 | 5 | 1 | 4 | 0.06 | ||||||||||
| Cefepime | 9 | 2 | 1 | 4 | <0.03 | ||||||||||
| Aztreonam | 4 | 6 | 2 | 8 | 0.12 | ||||||||||
| Imipenem | 1 | 6 | 5 | 0.25 | <0.03 | ||||||||||
CA, 2 μg of clavulanic acid per ml; TZ, 4 μg of tazobactam per ml.
MIC, <0.125 μg/ml.
R−, E. coli recipient; R+, E. coli with recombinant plasmid producing KLUA-1.
All strains tested a low level of resistance to penicillins (amoxicillin, ticarcillin, piperacillin) but also displayed a low level of resistance to cephalothin and cefuroxime (Table 1). Significant synergy with clavulanate was observed for amoxicillin and ticarcillin (32- to 512-fold). A low level of synergy was also observed between piperacillin and tazobactam, which correlated with the high level of activity of the ureidopenicillin piperacillin. Finally, all strains tested were highly susceptible to extended-spectrum cephalosporins, moxalactam, cefoxitin, aztreonam, and imipenem.
Kinetic and IEF analyses.
Subsequently, the resistance pattern was correlated with β-lactamase production. The β-lactamase contents were obtained from sonicated extracts of bacteria cultured overnight at 37°C in Trypticase soy broth (bioMérieux), prepared as described previously (8). Analytical isoelectric focusing (IEF) was performed in a polyacrylamide gel as described previously by the iodiometric procedure with benzylpenicillin for detection (8). We detected β-lactamases with different pIs, as follows: pI 6.9, two strains; pI 8.0, six strains; pI 8.4, four strains (see Table 3).
TABLE 3.
Genotypic characterization of β-lactamase contents of K. ascorbata strains and β-lactamase pIs
| Strain | pI | Result of PCR with primer(s):
|
Gene | Genbank accession no. | ||
|---|---|---|---|---|---|---|
| MA1-MA2 (208-751 bp; 543 bp) | MA3 (1-430 bp; 429 bp) | MA4 (461-883 bp; 422 bp) | ||||
| CIP 82.95T | 8.0 | + | + | + | blaKLUA-1 | AJ272538 |
| IP 15.79 | 8.4 | + | + | + | blaKLUA-2 | AJ251722 |
| IP 5.79 | 8.0 | + | + | + | blaKLUA-3 | AJ427461 |
| IP 8.79 | 8.0 | + | + | + | blaKLUA-4 | AJ427462 |
| IP 12.79 | 8.0 | + | + | + | blaKLUA-5 | AJ427463 |
| SL Ka99 | 6.9 | − | − | − | ||
| IP 2.89 | 6.9 | − | − | − | ||
| IP 3.89 | 8.0 | + | + | + | blaKLUA-8 | AJ427465 |
| TN Ka01 | 8.4 | + | + | + | blaKLUA-9 | AJ427466 |
| IP 43.50 | 8.0 | + | + | + | blaKLUA-10 | AJ427467 |
| IP 4450.94 | 8.4 | + | + | + | blaKLUA-11 | AJ427468 |
| IP 684.74 | 8.4 | + | + | + | blaKLUA-12 | AJ427469 |
Finally, the substrate and inhibition profiles of a single crude extract (type strain CIP 82.95T) were determined. This extract was produced from 4 liters of culture grown in brain heart infusion (Difco, Detroit, Mich.) and incubated at 37°C for 5 h after initial inoculation of 300 ml of a starter culture. Cells were harvested by centrifugation at 5,800 × g for 30 min at 4°C. The pellets (about 20 g) were washed by resuspension in 40 ml of ice-cold 0.1 M NaCl plus 0.05 M sodium azide and centrifuged as described above. The washed pellet was resuspended in 40 ml of the same solution and lysed by ultrasound treatment (three times for 10 s each time per gram of pellets; power = 50 W and frequency = 20 kHz with type 20-200 equipment [Alcatel, Paris, France]). The crude extract was cleared by centrifugation at 48,000 × g for 30 min at 4°C. Nucleic acids were precipitated by adding 0.2 M spermine (Sigma, Saint-Quentin Fallavier, France) and centrifugation (48,000 × g for 30 min at 4°C). The crude extract was then subjected to chromatography on a Sephadex G100 column. The active fractions were pooled, subjected to chromatography on Bio-Rex 70 resin, and then concentrated by ultrafiltration.
The kinetic constants Vmax, which was expressed relative to the Vmax for benzylpenicillin (which was set at 100%), and Km were determined by computerized microacidimetric assay in 0.1 M NaCl at pH 7.0 and 37°C (14). One unit of β-lactamase was defined as the amount of enzyme that hydrolyzed 1 μmol of benzylpenicillin per min at pH 7.0 and 37°C. Kinetic analysis showed that the activity of the enzyme was highest with cephalothin, but that cefuroxime and cefotaxime were also good substrates (Table 2). Conversely, the enzyme displayed a low level of activity with ceftazidime, aztreonam, and cephamycin. Tazobactam was the most efficient inhibitor (50% inhibitory concentration [IC50], 10 nM), closely followed by clavulanic acid (IC50, 20 nM). Sulbactam was the least potent inhibitor (IC50, 100 nM). Finally, chloride ions at a concentration of 0.5 M had no inhibitory effect.
TABLE 2.
Kinetic constants (Vmax, Km) of β-lactamase produced by K. ascorbata strain CIP 82. 95Ta
| Substrate | Vmax (relative) | Km (μM) | Vmax/Km (relative) |
|---|---|---|---|
| Benzylpenicillin | 100 | 17.5 | 100 |
| Ticarcillin | 7.5 | 10 | 13 |
| Cephalothin | 1,100 | 141 | 136 |
| Cefuroxime | 80 | 35 | 40 |
| Cefotaxime | 110 | 375 | 5.1 |
| Ceftazidime | <0.1 | 50 | <0.04 |
| Aztreonam | 0.3 | 105 | 0.06 |
| Cefoxitin | <0.1 | 110 | <0.02 |
Vmax was expressed relative to the Vmax for benzylpenicillin (which was set at 100%).
Genetic characterization of blaKLUA.
The identification of only one susceptibility pattern among the 12 strains of K. ascorbata tested suggested that this species possesses a naturally occurring, chromosomally mediated β-lactamase. Furthermore, this assumption was confirmed by the failure of both conjugation experiments and plasmid extraction with the High Pure plasmid isolation kit (Roche Biochemicals, Neuilly-sur-Seine, France).
On the basis of the observed pattern of susceptibility and enzymatic properties, we designed degenerate oligonucleotide primers from the sequences of genes encoding CTX-M-type enzymes (primer MA1-forward [5′-SCSATGTGCAGYACCAGTAA-3′] and primer MA2-reverse [5′-CCGCRATATGRTTGGTGGTG-3′]) and used them to amplify an internal fragment of the K. ascorbata bla genes. In each PCR, the cycling conditions were as follows: 35 cycles of 60 s at 94°C for denaturation, 30 s at 60°C for annealing, and 30 s at 72°C for elongation with 2 U of Taq DNA polymerase (Perkin-Elmer, Foster City, Calif.). Amplifications were obtained for all K. ascorbata strains producing a β-lactamase of pI 8.0 or 8.4. Nevertheless, the two bla genes encoding β-lactamase of pI 6.9 were not amplified with these CTX-M-specific primers (Table 3).
The PCR products were then purified with Qiagen (Courtaboeuf, France) columns, and the DNA sequences were determined twice on both strands by the procedure of Sanger et al. (22) by using fluorescent dye-labeled dideoxynucleotides, thermal cycling with Taq polymerase, and an ABI 373A DNA sequencer (Applied Biosystems, Foster City, Calif.). All PCR products consisted of a 540-bp fragment, which was found to be highly similar (>97% identity) to the cluster that included the blaCTX-M-2, blaCTX-M-4, blaCTX-M-5, blaCTX-M-6, blaCTX-M-7, and blaToho-1 genes.
Finally, other primers more specific for the blaCTX-M gene of this cluster were used to amplify the entire Kluyvera bla gene: primers M3-forward (5′-ATGATGACTCAGAGCATTCGC-3′) and M3-reverse (5′-GGGCAATCAGCTTATTCATGG-3′) and primers M4-forward (5′-TTGCTCGCTCGTTGGGTGAT-3′) and M4-reverse (5′-TATTGCATCAGAAACCGTGGG-3′) (Table 3). Amplifications with primer M3 or M4 were performed by using the same cycling conditions described above. The nucleotide sequences of K. ascorbata genes encoding a β-lactamase with a pI of 8.0 or 8.4 (blaKLUA-1 to blaKLUA-5 and blaKLUA-8 to blaKLUA-12) were determined (Table 3). High degrees of homology (>95%) were observed among all these blaKLUA genes coding for class A β-lactamases. Moreover, blaKLUA genes were also found to share extensive similarities (95 to 100%) with the genes producing the CTX-M-2, CTX-M-4, CTX-M-5, CTX-M-6, CTX-M-7, and Toho-1 β-lactamases; but they displayed lower levels of identity with the genes for the other CTX-M types (68 to 84%) (Table 4). However, this high level of similarity observed suggested that the chromosome-encoded β-lactamase of K. ascorbata is a more probable progenitor of at least some plasmid-encoded CTX-M enzymes than K. oxytoca (23). Finally, the highest degrees of homology between blaKLUA and chromosome-encoded bla genes were observed for class A enzymes produced by Kluyvera cryocrescens (≈77%), S. fonticola (≈74%), K. oxytoca (≈73%), and Citrobacter sedlakii (≈73%) (6, 9, 19; C. Humeniuk, unpublished data).
TABLE 4.
Identities of amino acid sequences of K. ascorbata β-lactamases and CTX-M, Toho, UOE β-lactamases
| Enzyme(s) | Range % identity (reference) with clusters including:
|
|||||
|---|---|---|---|---|---|---|
| KLUA-1, -2, -3, -4, -5, -8, -9, -10, -11, and -12 (this work) | CTX-M-1, -3, -10, -11, and -12 and UOE-1 (13, 18, 23a) | CTX-M-2, -4, -5, -6, and -7 and Toho-1 (3, 10, 11, 23) | CTX-M-9, -13, -14, and -15 and UOE-2 (21b) | Toho-2 (15) | CTX-M-8 (2) | |
| KLUA-1, -2, -3, -4, -5, -8, -9, -10, -11, and -12 | 96-100 | 77-82 | 95-100 | 78-81 | 68-71 | 82-84 |
| CTX-M-1, -3, -10, -11, and -12 and UOE-1 | 98-100 | 77-81 | 79-80 | 70-71 | 80-82 | |
| CTX-M-2, -4, -5, -6, and -7 and Toho-1 | 97-100 | 78-81 | 68-71 | 81-84 | ||
| CTX-M-9, -13, -14, and -15 and UOE-2 | 99-100 | 87-88 | 85-86 | |||
| Toho-2 | 100 | 75 | ||||
| CTX-M-8 | 100 | |||||
Only two bla genes encoding a β-lactamase of pI 6.9 were not amplified with the CTX-M-specific primers (Table 3). Characterization of a β-lactamase(s) produced by both of these strains is in progress. In fact, it appears that K. ascorbata could produce at least two different kinds of β-lactamase genes, as has been observed for K. oxytoca, in which two main groups of β-lactamase genes have been distinguished (blaOXY-1 and blaOXY-2; identity, 90%) (9). This heterogeneity of the K. ascorbata β-lactamases could be correlated with the diversity observed among CTX-M-type enzymes, which are divided into five clusters (Table 4).
Genetic environment of blaKLUA-1.
As the chromosomal β-lactamase genes of K. ascorbata are the probable progenitors of some of the plasmid-encoded extended-spectrum CTX-M-type enzymes, we investigated the genetic organization of blaKLUA-1 by cloning and sequencing the surrounding regions of this gene. Chromosomal DNA was prepared from K. ascorbata CIP 82.95T, partially digested with Sau3A, and ligated into the BamHI site of pBK-CMV (pBK Phagemid Vectors; Stratagene, La Jolla, Calif.), as described previously (17). The recombinant plasmid was introduced into Escherichia coli XL-1 by the standard CaCl2 procedure. Several transformants were selected on Mueller-Hinton agar (Sanofi Diagnostics Pasteur) supplemented with amoxicillin (40 μg · ml−1) and kanamycin (25 μg · ml−1) and were further characterized by determination of their antibiotic susceptibility profiles and pIs. The molecular sizes of the inserts were estimated from the results of restriction digestion and electrophoresis in 1 to 3% agarose gels, as described previously (17). Finally, both strands of the DNA sequence of a 7-kb insert were determined. The BLASTN program on the National Center for Biotechnology Information website (revision date, 20 July 2001; http://www.ncbi.nlm.nih.gov) was used for database searches (1).
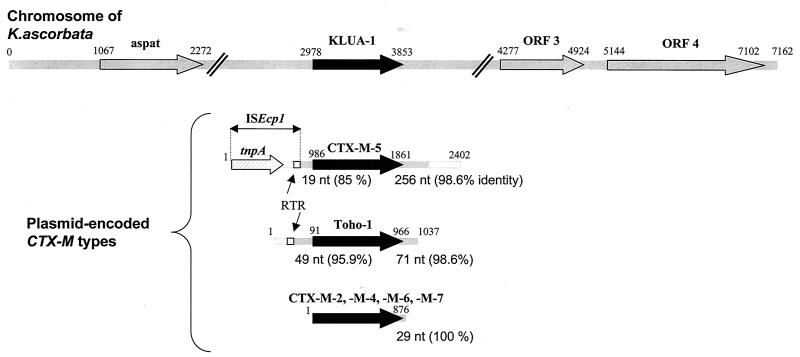
Three open reading frames (ORFs) were found immediately upstream (2,980 bp) and downstream (3,300 bp) of the blaKLUA gene (ORF 2) (Fig. 1). The protein corresponding to ORF 1 was 86% identical to an aspartate aminotransferase from E. coli. The deduced amino acid sequence of the protein encoded by ORF 3 was 43% identical to that of a putative protein from E. coli. The protein encoded by the fourth ORF showed various degrees of similarity (28 to 56%) to adhesive proteins (MisL, AidaI, VirG) known to contribute to the pathogenicities of several enterobacteria. Recently, the nucleotide sequence of blaKLUC, which encodes the chromosomal β-lactamase of K. cryocrescens, and its surrounding regions has been reported (6). Genes corresponding to an aspartate aminotransferase and a putative protein showing 94 and 66% identities with the proteins encoding by K. ascorbata ORF 1 and ORF 3, respectively, were found upstream and downstream of the blaKLUC gene. In fact, this particular genetic organization could be a characteristic of the genus Kluyvera.
FIG. 1.
Genetic environment of blaKLUA-1 gene encoding the β-lactamase produced by K. ascorbata strain CIP 82.95T. The regions surrounding the chromosomal blaKLUA-1 gene and the sequences adjacent to the plasmidic genes encoding β-lactamases CTX-M-2, CTX-M-4, CTX-M-5, CTX-M-6, CTX-M-7, and Toho-1 (2, 10, 11, 23) were compared. The RTR sequence and tnpA correspond to the right terminal repeat and transposase A, respectively, which are typical of ISEcp1.
Finally, no ampR gene was detected upstream of the blaKLUA-1 gene (Fig. 1). Indeed, some class A β-lactamases closely related to K. ascorbata enzymes are regulated by an ampR gene: cumR (Proteus vulgaris) (5), sedR (C. sedlakii), (19) and fonR (S. fonticola) (C. Humeniuk, unpublished data).
Arguments for mobilization of chromosomal blaKLUA gene.
The analysis of the sequences surrounding the blaCTX-M genes revealed the presence of sequences similar to those for CTX-M-5 on the K. ascorbata chromosome (5 nucleotides [nt] upstream and 179 nt downstream with 80 and 100% identities, respectively, with the chromosome of K. ascorbata), Toho-1 (49 nt upstream and 71 nt downstream with 95.9 and 98.6% identities, respectively), and CTX-M-2, -M-4, -M-6, and -M-7 (29 nt upstream with 100% identity) (Fig. 1). However, major differences between the adjacent sequences of the other blaCTX-M genes (CTX-M-1, -M-3, -M-8, -M-12, -M-13, -M-14, and -M-15; Toho-2; UOE-1; and UOE-2) and the chromosome of K. ascorbata were observed. This fact confirms that the β-lactamase of K. ascorbata is probably the progenitor of only some plasmid-encoded CTX-M-type enzymes. To date, this is the second report of the mobilization of a chromosomally mediated class A β-lactamase.
So far we have investigated the potential mechanism of mobilization of the blaKLUA gene by sequencing surrounding regions of the blaCTX-M-5 gene located on plasmid pCLL3417 (3). The GenBank accession number for this nucleotide sequence is AJ286192. Sequencing confirmed the presence of sequences similar to that of the K. ascorbata chromosome (19 nt upstream and 256 nt downstream with 85 and 98.6% identities, respectively) and revealed the presence of the novel insertion sequence, ISEcp1, described by Stapleton (P. D. Stapleton, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1457, 1999) (Fig. 1). Subsequently, this element was reported for several plasmid-encoded AmpC enzymes including CMY-5, LAT-1, and ACC-1, which were probably mobilized from chromosomal β-lactamases of C. freundii or H. alvei (20). It was also detected on plasmids carrying other blaCTX-M genes (CTX-M-3-like, CTX-M-14, CTX-M-15, Toho-1, Toho-2, and UOE-2) (11, 12, 15; GenBank accession numbers AF252622, AF252623, and AF311345).
Given the low level of clinical importance of K. ascorbata (7, 24), it is important to consider the possible transfer of the bla gene in veterinary practice or in the environment. Several preliminary reports have indeed described Salmonella enterica serovar Typhimurium as producing β-lactamases of the CTX-M2, CTX-M-4, CTX-M-5, and CTX-M-7 types in various countries (Argentina, Latvia, Russia, Greece) (3, 10, 23). If K. ascorbata acts as a reservoir for these plasmid-encoded CTX-M enzymes, the sequences of more chromosomal genes must be determined to confirm the existence of a single progenitor due to the heterogeneity observed in the CTX-M cluster (Table 4).
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of the insert reported in this paper is AJ272538.
Acknowledgments
This work was financed by a grant for “Réseau β-lactamases: de la clinique à la recherche” from the Ministère de l'Education Nationale, de la Recherche et de la Technologie, Paris, France.
REFERENCES
- 1.Altschul, S. F., L. M. Thomas, A. A. Shaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R., J. L. Sampaio, R. Labia, C. de Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datz, M., B. Joris, E. A. Azab, M. Galleni, J. Van Beeumen, J. M. Frère, and H. H. Martin. 1994. A common system controls the induction of very different genes. The class-A β-lactamase of Proteus vulgaris and the enterobacterial class C β-lactamase. Eur. J. Biochem. 226:149-157. [DOI] [PubMed] [Google Scholar]
- 6.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farmer, J. J., III, G. R. Fanning, G. P. Huntley-Carter, B. Holmes, F. W. Hickman, C. Richard, and D. J. Brenner. 1981. Kluyvera, a new (redefined) genus in the family Enterobacteriaceae: identification of Kluyvera ascorbata sp. nov. and Kluyvera cryocrescens sp. nov. in clinical specimens. J. Clin. Microbiol. 13:919-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitoussi, F., G. Arlet, P. A. G. Grimont, P. Lagrange, and A. Philippon. 1995. Escherichia hermannii: susceptibility pattern to β-lactams and production of β-lactamase. J. Antimicrob. Chemother. 36:537-543. [DOI] [PubMed] [Google Scholar]
- 9.Fournier, B., P. H. Roy, P. H. Lagrange, and A. Philippon. 1996. Chromosomal β-lactamase genes of Klebsiella oxytoca are divided into two main groups: blaOXY-1 and blaOXY-2. Antimicrob. Agents Chemother. 40:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gazouli, M., E. Tzelepi, A. Markogiannakis, N. J. Legakis, and L. S. Tzouvelekis. 1998. Two novel plasmid-mediated cefotaxime-hydrolysing β-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. FEMS Microbiol. Lett. 165:289-293. [DOI] [PubMed] [Google Scholar]
- 11.Ishii, Y., A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa. 1995. Cloning and sequence of the gene encoding a cefotaxime-hydolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISECp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 13.Kariuki, S., J. E. Corkill, G. Revathi, R. Musoke, and C. A. Hart. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazmierczak, A., A. Philippon, R. Chardon, R. Labia, and F. Le Goffic. 1973. Constantes enzymatiques (Km et Vmax) des β-lactamases mesurées par une méthode microacidimétrique couplée à l'ordinateur. Ann. Microbiol. Inst. Pasteur. 124B:259-268. [PubMed] [Google Scholar]
- 15.Ma, L., Y. Yshii, M. Ishiguro, H. Matsuzawa, and K. Yamaguchi. 1998. Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob. Agents Chemother. 42:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto, Y., and M. Inoue. 1999. Characterization of SFO-1, a plasmid-mediated inducible class A β-lactamase from Enterobacter cloacae. Antimicrob. Agents Chemother. 43:307-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadjar, D., M. Rouveau, C. Verdet, J. L. Donayb, J. L. Herrmann, P. H. Lagrange, A. Philippon, and G. Arlet. 2000. Outbreak of Klebsiella pneumoniae producing transferable AmpC-type beta-lactamase (ACC-1) originating from Hafnia alvei. FEMS Microbiol. Lett. 187:35-40. [DOI] [PubMed] [Google Scholar]
- 18.Oliver, A., J. C. Perez-Diaz, T. M. Coque, F. Baquero, and R. Canton. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrella, S., D. Clermont, I. Casin, V. Jarlier, and W. Sougakoff. 2001. Novel class A β-lactamase Sed-1 from Citrobacter sedlakii: genetic diversity of β-lactamases within the Citrobacter genus. Antimicrob. Agents Chemother. 45:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philippon, A. M., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabate, M., R. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequencing of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger, T., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 24.Yogev, R., and S. Kozlowski. 1990. Peritonitis due to Kluyvera ascorbata: case report and review. Rev. Infect. Dis. 12:399-402. [DOI] [PubMed] [Google Scholar]