Abstract
An unusual self-transferable virulence-resistance plasmid (pUO-StVR2) was found in nine multidrug-resistant (ACSSuT phenotype) Salmonella enterica serotype Typhimurium clinical isolates that were assigned to four different phage types and a single and distinctive XbaI pulsed-field gel electrophoresis profile. pUO-StVR2 is an IncFII plasmid of about 140 kb in length carrying the spvA, spvB, and spvC (Salmonella plasmid virulence) and rck (resistance to complement killing) genes. It also carries the oxa1/aadA1a (ampicillin resistance and streptomycin-spectinomycin resistance) gene cassette configuration located within a class 1 integron with qacEΔ1/sul1 (ammonium antiseptics resistance and sulfadiazine resistance); the transposon genes merA, tnpA, and tnpR (mercury resistance, transposase, and resolvase of Tn21, respectively); and the catA1 (chloramphenicol resistance) and tet(B) (tetracycline resistance) genes. The insertion of resistance genes into a Salmonella virulence plasmid constitutes a new and interesting example of plasmid evolution and presents a serious public health problem.
Several Salmonella enterica serotypes harbor plasmids which are essential for virulence. Although virulence plasmids from different serotypes are not identical, the sequences required for virulence are highly conserved (5, 16, 20, 21, 23, 33). Most of the Salmonella enterica serotype Typhimurium isolates carry a serovar-specific 90-kb virulence plasmid which belongs to the incompatibility group IncFII. However, irrespective of their plasmid content, serotype Typhimurium isolates have been assigned to different clones, which are usually defined by phage typing (2) and DNA fingerprinting (14, 20, 22, 31, 39). A particular serotype Typhimurium clonal group which includes phage type 104 (DT104) strains that are, at the least, resistant to ampicillin, chloramphenicol-florfenicol, streptomycin-spectinomycin, sulfonamides, and tetracycline (ACSSuT phenotype) has emerged worldwide and presents a global health problem (12, 22, 26, 31, 34). Recently, the emergence of other multidrug-resistant serotype Typhimurium clones in which the resistance is plasmid mediated has been reported (7, 16, 21, 38, 39). In these clones, the resistance genes are often part of transposons and/or integrons, elements that facilitate the intracellular movement of resistance genes. Among the different classes of integrons described, class 1 integrons are the most frequently found in Salmonella as well as in other gram-negative bacteria and they can be found within transposons and be chromosome and/or plasmid located (3-7, 11, 13-17, 19, 28-30, 38).
In the present work, we report an unusual self-transferable virulence plasmid responsible for multidrug resistance (ACSSuT) in serotype Typhimurium clinical isolates of various phage types sharing a common XbaI pulsed-field gel electrophoresis (PFGE) profile.
MATERIALS AND METHODS
Bacterial strains.
In previous works (reference 15 and unpublished data), 83 Salmonella enterica serotype Typhimurium isolates carrying class 1 integrons were identified. Nine of these isolates were selected for the study presented here. They originated from human feces and were collected at regional hospitals. All were received throughout 1993 to 1999 at the Laboratorio de Salud Pública (LSP) of the Principality of Asturias. The isolates were phage typed in the Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid, Spain, by using Anderson's scheme (2). As controls in different experiments, the following serotype Typhimurium strains carrying the 90-kb virulence plasmid were tested: LT2 (pSLT90) (Bayer AG Collection), ATCC 14028 (pSLT90) (American Type Culture Collection), and LSP14/92 (pUO-StV1), which is a DT104 (subtype l)-ACSSuT clinical isolate (14-16). For mating experiments, serotype Typhimurium LSP31/93 was used as plasmid donor and the rifampin-resistant Escherichia coli K-12 strain J53 and nalidixic acid-resistant Salmonella enterica serotype Panama LSP291/98 were used as recipients. These strains were tested for susceptibility to 15 antimicrobial agents (15) by disk diffusion (24) and to florfenicol by broth microdilution (25) by using a Sensititre semiautomatic system (Trek Diagnostics, East Grinstead, United Kingdom). Results were analyzed according to standards set by NCCLS (24, 25).
Plasmid and PFGE analysis.
Plasmid profiling, plasmid curing, and transfer of antibiotic resistance by conjugation were performed as described in references 14 and 16. Plasmid relationships were tested by restriction analysis using 5 U of HindIII, ClaI, and EcoRI (Amersham Pharmacia Biotech, Barcelona, Spain) separately and by hybridization using gene-specific probes (16). PFGE analysis was performed with XbaI as described in reference 14.
PCR amplification, purification, and sequencing of DNA.
The detection of Salmonella virulence plasmid genes, resistance genes, and transposon- and/or integron-related genes was performed with the PCR conditions and primers described previously (10, 14-16, 18) or designed for this work: repFIIA-F/B, CTGTCGTAAGCTGATGGC/CTCTGCCACAAACTTCAGC; traT-F/B, GATGGTTACACTGGTCAG/TCTGAGATCTGTACGTCG; rck-F/B, TCGTTCTGTCCTCACTGC/TCATAGCCCAGATCGATG; and pefA-F/B, GCACACGCTGCCAATGAA/CACAGACTTGAAGTCACC (accession number AE006471); and [tnpR-F/B], [GGCGACACCGTGGTGGTGCATAGC/CGGTAAGCCCCGCGTTGCTTGGC] (accession number AL513383). Restriction and sequencing of the PCR products were carried out as described in references 15 and 16, and the sequences obtained were compared to those registered in GenBank.
RESULTS AND DISCUSSION
Characterization of a self-transferable virulence-resistance plasmid of Salmonella enterica serotype Typhimurium.
In previous works (reference 15 and unpublished data), nine serotype Typhimurium isolates were identified which carried a class 1 qacEΔ1/sul1 (encoding ammonium antiseptics resistance and sulfadiazine resistance) integron, generating PCR products of 2,000 bp with 5′ CS and 3′ CS primers. By using PvuI, TaqI, and BglII endonucleases, the 2,000-bp PCR products yielded the same restriction pattern as the one described previously (15) for an integron-borne oxa1/aadA1a (encoding ampicillin resistance and streptomycin-spectinomycin resistance) cassette array. PCR amplification confirmed the presence of both oxa1 and aadA1a within the 2,000-bp amplicons. The oxa1/aadA1a cassette configuration is characteristic for the transposon Tn2603 (27, 40). The presence of transposon genes tnpA, tnpR, and merA (encoding the transposase, resolvase, and mercury reductase, respectively) was confirmed by PCR amplification (amplicons of about 327, 240, and 1,232 bp, respectively, were generated) in the nine isolates, but long PCR amplification (data not shown) failed to confirm a link between transposon and integron genes. On the other hand, amplification and sequencing also revealed the presence of the catA1 (encoding chloramphenicol resistance) and tet(B) (encoding tetracycline resistance) genes (accession numbers AP000342 and Y19113, respectively) in the nine isolates.
Plasmid analysis showed that the nine serotype Typhimurium isolates carried a large plasmid of about 140 kb together with other small plasmids but not the serotype Typhimurium 90-kb virulence plasmid (Fig. 1). However, when these isolates were tested by PCR for virulence plasmid genes (repFIIA [plasmid incompatibility group FII replicons]; traT [conjugative transfer]; spvA, spvB, and spvC [Salmonella plasmid virulence]; rck [resistance to complement killing]; and pefA [plasmid-encoded fimbria]), the expected amplicons (about 288, 483, 600, 1,060, 424, 474, and 442 bp, respectively) were generated with all except the pefA primers.
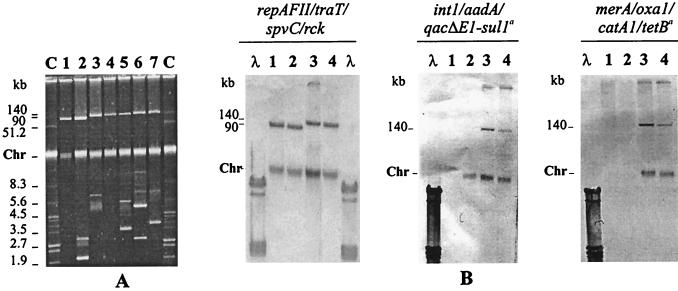
FIG. 1.
Plasmid analysis of representative serotype Typhimurium clinical isolates and control strains. (A) Plasmid profiles. Lanes: C, molecular size standard plasmids; 1 through 7, profiles generated by LT2 (lane 1), LSP14/92-DT104 (subtype l) (lane 2), LSP31/93- DT120 (lane 3), E. coli CT31 (lane 4), LSP28/98-DT104 (subtype b-low) (lane 5), LSP106/94-NT, LSP233/98-NT, LSP349/98-NT; 362/98-NT, LSP436/99-NT, and LSP559/99-NT (lane 6), and LSP21/99-RDNC (lane 7). (B) Lanes: 1 through 4, successive hybridizations of lanes 1 through 4 from panel A, with the probes cited at the top of the figure; λ, phage lambda DNA digested with PstI. Chr, chromosomal DNA.
To ascertain if the virulence and resistance genes were linked and whether they were located in the plasmid or chromosome, the isolate LSP31/93 was tested by curing and conjugation experiments. By curing, no segregants were found among the approximately 300 CFU tested. Moreover, the 140-kb plasmid was self-transferable to E. coli K-12 J53 and more effectively transferable to Panama LSP291/98. The transconjugants expressed the same resistance phenotype (ACSSuT) as the parental strain, and their plasmid DNAs generated the expected PCR products and hybridized with probes for the virulence plasmid, transposon and/or integron, and resistance genes (Fig. 1 and 2). This plasmid was labeled pUO-StVR2 (plasmid University of Oviedo-Salmonella enterica serotype Typhimurium virulence and resistance).
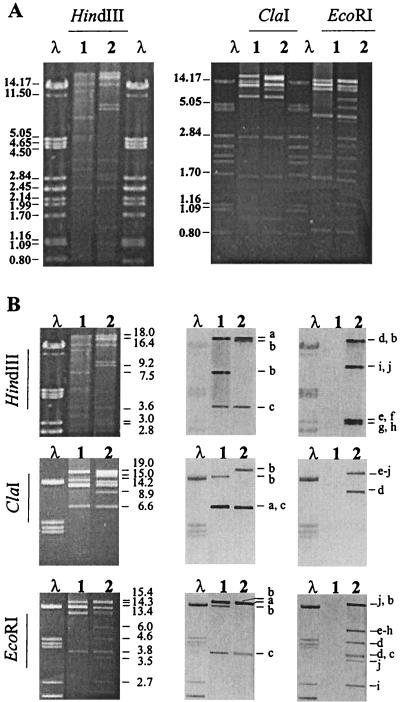
FIG. 2.
Restriction and hybridization analysis of serotype Typhimurium virulence plasmids. (A) Plasmid restriction profiles generated by HindIII, ClaI, and EcoRI (fragment sizes in kilobases). (B) Successive hybridizations of digested DNA with virulence plasmid gene probes (a, traT; b, rck; c, spvC) and resistance gene probes [d, merA; e, int1; f, oxa1; g, aadA1; h, qacΕΔ1-sul1; i, catA1; j, tet(B)]. Lanes: λ, phage lambda DNA digested with PstI (fragment sizes in kilobases); 1, pSLT90 from LT2; 2, pUO-StVR2 from E. coli CT31 transconjugant. Chr, chromosomal DNA.
To demonstrate the genetic relationship between pUO-StVR2 from the E. coli CT31 transconjugant and pSLT90 from LT2, both plasmids were tested by restriction analysis (Fig. 2A) with three endonucleases (HindIII, ClaI, and EcoRI) and by hybridization with probes derived from pSLT90 virulence genes (Fig. 2B). Plasmid restriction analysis showed that each plasmid revealed a distinct restriction profile with some fragments banding at identical positions. The hybridizations made in order to locate the virulence plasmid genes in the HindIII, ClaI, and EcoRI restriction patterns showed that the traT probe always hybridized to matching fragments (18, 6.6, and 14.3 kb, with each endonuclease), the spvC probe also always hybridized to matching fragments (3.6, 6.6, and 3.8 kb), and the rck probe in contrast hybridized to mismatching fragments in all restriction patterns (Fig. 2B).
To locate the transposon and/or integron and resistance genes in the restriction patterns of pUO-StVR2, hybridizations with specific probes were carried out (Fig. 2B). The most important findings were as follows. (i) The merA probe always hybridized to fragments (16.4, 8.9, and 4.6 and 3.8 kb with HindIII, ClaI, and EcoRI, respectively) different from those to which the other resistance gene probes hybridized. (ii) HindIII analysis showed three gene groups (probes hybridizing to the same fragment): catA1-tet(B) (an approximately 9.2-kb fragment), int1-oxa1 (an approximately 3-kb fragment), and aadA1-qacΕΔ1-sul1 (an approximately 2.8-kb fragment). The two last groups were generated from the integron and were expected on the basis of its DNA sequence (accession number AJ009819.1). (iii) ClaI analysis revealed a group including all the resistance genes tested: catA1-tet(B)-int1-oxa1-aadA1-qacΕΔ1-sul1 (an approximately 15-kb fragment). (iv) EcoRI analysis showed the int1-oxa1-aadA1-qacΕΔ1-sul1 group (an approximately 6-kb fragment), corresponding to the integron genes, which was separated from catA1 (an approximately 2.7-kb fragment) and tet(B) (about 15.4 and 3.5 kb). By this approach, linkage between the merA and integron genes could not be established.
Characterization of the isolates containing the virulence-resistance plasmid pUO-StVR2.
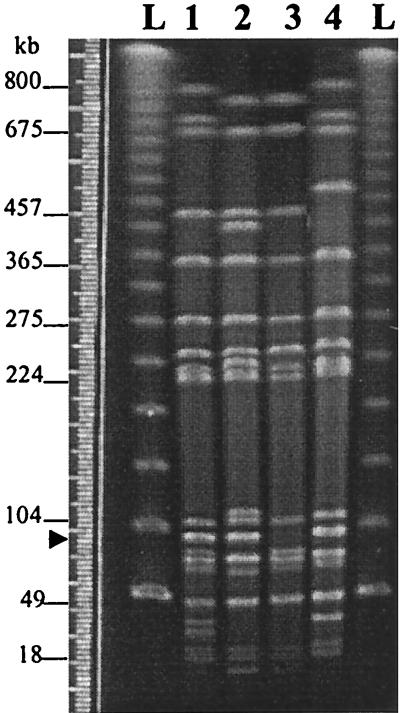
The nine isolates carrying pUO-StVR2 showed the ACSSuT phenotype, but they were florfenicol susceptible. Only one of them was phage type DT104 (subtype b-low), six were non-phage typeable (NT), and one was DT120, and another presented a nonrecognized phage lysis pattern (RDNC). To establish the genetic relationship among the nine isolates as well as their relationship with the serotype Typhimurium control strains, the isolate DNAs were analyzed by XbaI PFGE (Fig. 3). PFGE profiles were defined considering only the well-visualized fragments (size, ≥18 kb) The nine isolates generated identical profiles (PFGE3) which presented between 11 and 14 matching fragments and between 6 and 11 mismatching fragments regarding the ones from the control strains. These results support the fact that the nine isolates should be classified as members of a single clonal group, well differentiated from the control strains, one of which represented the prevalent DT104-ACSSuT clone (with PFGE2) (14, 22). Within this new clonal group, there is some heterogeneity with respect to phage type and plasmid content and a relationship between both markers seems to exist (Fig. 1).
FIG. 3.
PFGE analysis of serotype Typhimurium strains representing different clones. Lanes: L, PFGE ladder (New England Biolabs, Schwalbach, Germany); 1 through 4, XbaI PFGE profiles generated by serotype Typhimurium LT2 (PFGE1), LSP14/92 (PFGE3), LSP31/93 (PFGE4), and ATCC 14028 (PFGE2), respectively. Molecular sizes indicated in the figure correspond to the LT2 XbaI fragment sizes given in reference 12. The nine clinical isolates under study generated PFGE4. The arrow indicates the fragment corresponding to the 90-kb virulence plasmid.
Previous work has shown that changes in phage types can be the result of the loss or acquisition of plasmids (37). Since not all DT104 strains present the markers described for the so-called DT104-ACSSuT clone or clonal line (in our experience, NT strains could also be ascribed to this clone), it must be taken into account that phage typing alone is not a suitable method for the identification of a clone.
Hypothesis concerning the origin of the virulence-resistance plasmid pUO-StVR2.
Some interesting facts regarding the dispersion of virulence and resistance plasmids, as well as of the genes harbored by pUO-StVR2, have previously been published. (i) Serovar-specific virulence plasmids of different sizes have been found in Salmonella subspecies I (23, 33), but it has also been reported that some serotypes belonging to subspecies II, IIIa, and IV carry spv genes on the chromosome (5). (ii) At least three other types of virulence-resistance plasmids, each of them encoding different resistance genes, have been detected in Salmonella collected in different countries (9, 14, 16, 21). (iii) The integron reported here is found forming part of transposon Tn2603 (27, 40), and a similar integron has been found in a 140-kb IncFI plasmid of serotype Typhimurium non-DT104-resistant isolates collected in Italy (38). (iv) Both the catA1 and tet(B) genes are widely dispersed among Enterobacteriaceae and can be part of transposons (Tn9 and Tn10, respectively) and be located in plasmids (1, 8, 19, 32). These facts encourage us to suggest the following possible events leading to the generation of pUO-StVR2. First, in the animal reservoir, a serotype Typhimurium bacterium showing a distinctive PFGE profile and carrying a virulence plasmid (in association with other small plasmids) acquired one (or more) resistance plasmid(s). In a second step, recombination-transposition processes between the virulence and the resistance plasmid(s) could have taken place, generating the hybrid plasmid (pUO-StVR2). This hybrid plasmid belongs to the serotype Typhimurium virulence plasmid family (because it carries the repFIIA, traT, spv, and rck genes) but is of a larger size since it has gained several resistance genes [oxa1, aadA1a, qacEΔ1, sul1, merA, catA1, and tet(B)], some of them integron and/or probably transposon related. Finally, pUO-StVR2-carrying Salmonella enterica serotype Typhimurium strains spread and established in their reservoir and sporadically reach humans through the food chain. The latter is supported by the isolation of such strains from human feces during the period from 1993 to 1999. In addition to the strains described above, we found in the year 2000 three new serotype Typhimurium clinical isolates carrying the same resistance genes, including the integron-borne oxa1/aadA1a cassette configuration. To date, we have found no other Salmonella isolate presenting this cassette array among about 2,000 isolates (isolated between 1989 and 2000) tested for integrons and/or resistance plasmids (15, 23, 35, 36; unpublished data).
The findings presented above support the fact that different serotype Typhimurium clones or clonal groups, with distinctive PFGE profiles, display similar resistance phenotypes based on different resistance genes which can be located on the chromosome and/or on plasmids. In each clone, some resistance genes and their gene arrays seem to be characteristic, frequently including one or more transposon and/or integron structures. Serotype Typhimurium has a wide animal reservoir, composed of mainly cattle and swine, which is linked to humans via the food chain. Consequently, the selection and maintenance of different resistance genes in different serotypes and clones like the one reported here constitute a serious public health problem.
Acknowledgments
We thank M. A. González-Hevia (LSP, Principado de Asturias, Spain) and H.-P. Kroll (Bayer AG Pharma-Research Center, Wuppertal, Germany) for Salmonella strains, M. A. Usera and A. Aladueña (Salmonella Reference Center, Centro Nacional de Microbiología, CNM, Madrid, Spain) for serotyping and phage typing serotype Typhimurium isolates, the personnel of the National Salmonella Reference Laboratory (NRL—Salmonella, BgVV, Fg. 501, Berlin, Germany), especially E. Junker for his helpful assistance and A. Schroeter for his expert advice. We thank H.-P. Kroll also for his help in carrying out preliminary work.
This work was supported by grants from the Fondo de Investigación Sanitaria (FIS 00/1084), Ministry of Health and Consumption, Madrid, Spain, and the BgVV (F501-28), Ministry of Consumer Protection and Agriculture (BMVEL). B. Guerra was the recipient of a short-term FEMS fellowship. S. Soto is the recipient of grant Formación de Personal Investigador (AP98) from the Ministry of Education and Culture, Madrid, Spain.
REFERENCES
- 1.Alton, N. K., and D. Vapnek. 1979. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature 282:864-869. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, E. S., L. R. Ward, M. J. de Saxe, and J. D. H. de Sa. 1977. Bacteriophage typing designations of Salmonella typhimurium. J. Hyg. 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass, L., C. A. Liebert, M. D. Lee, A. O. Summers, D. G. White, S. G. Thayer, and J. Maurer. 1999. Incidence and characterization of integrons, genetic elements mediating multidrug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, E. F., and D. L. Hartl. 1998. Salmonella virulence plasmid: modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIIa, IV and VII isolates. Genetics 149:1183-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs, C. E., and P. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli, A. 2001. Importance of integrons in the diffusion of resistance. Vet. Res. 32:243-259. [DOI] [PubMed] [Google Scholar]
- 8.Chalmers, S., R. Sewitz, K. Lipkow, and P. Crellin. 2000. Complete nucleotide sequence of Tn10. J. Bacteriol. 182:2970-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, C., C. H. Chiu, W. Y. Wu, C. H. Chu, T. P. Liu, and J. T. Ou. 2001. Large resistance virulence plasmids in clinical isolates of Salmonella enterica serovar Choleraesuis. Antimicrob. Agents Chemother. 45:2299-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahlberg, C., and M. Hermansson. 1995. Abundance of Tn3, Tn21, and Tn501 transposase (tnpA) sequences in bacterial community DNA from marine environments. Appl. Environ. Microbiol. 61:3051-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fluit, A. C., and F. J. Smith. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 12.Glynn, M. K., C. Boop, W. Dewitt, P. Dabney, M. Moktar, and F. Angulo. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333-1338. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein, C., M. D. Lee, S. Sanchez, C. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White, and J. J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerra, B., I. Laconcha, S. M. Soto, M. A. González-Hevia, and M. C. Mendoza. 2000. Molecular characterization of emergent multiresistant Salmonella enterica serotype [4,5,12:i:-] organisms causing human salmonellosis. FEMS Microbiol. Lett. 190:341-347. [DOI] [PubMed] [Google Scholar]
- 15.Guerra, B., S. M. Soto, S. Cal, and M. C. Mendoza. 2000. Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob. Agents Chemother. 44:2166-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerra, B., S. M. Soto, J. M. Argüelles, and M. C. Mendoza. 2001. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:-]. Antimicrob. Agents Chemother. 45:1305-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liebert, C. A., J. Wireman, T. Smith, and A. O. Summers. 1997. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 63:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liebert, C. A., R. M. Hall, and A. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, S. L., and K. E. Sanderson. 1995. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J. Bacteriol. 177:3355-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llanes, C., V. Kirchgesner, and P. Plesiat. 1999. Propagation of TEM- and PSE-type β-lactamases among amoxicillin-resistant Salmonella spp. isolated in France. Antimicrob. Agents Chemother. 43:2430-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malorny, B., A. Schroeter, C. Bunge, B. Hoog, A. Steinbeck, and R. Helmuth. 2001. Evaluation of molecular typing methods for Salmonella enterica serovar Typhimurium DT104 isolated in Germany from healthy pigs. Vet. Res. 32:119-129. [DOI] [PubMed] [Google Scholar]
- 23.Martín, M. C., M. A. González-Hevia, J. A. Alvarez-Riesgo, and M. C. Mendoza. 2001. Salmonella serotype Virchow causing salmonellosis in a Spanish region. Characterization and survey of clones by DNA fingerprinting, phage typing and antimicrobial resistance. Eur. J. Epidemiol. 17:31-40. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Ng, L.-K., M. R. Mulvey, I. Martin, G. A. Petters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouellette, M., L. Bissonnette, and P. H. Roy. 1987. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc. Natl. Acad. Sci. USA 84:7378-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters, E. D. J., M. A. Leverstein-van Hall, T. A. Box, J. Verhoef, and A. C. Fluit. 2001. Novel gene cassettes and integrons. Antimicrob. Agents Chemother. 45:2961-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile elements. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 31.Ridley, A., and J. Threlfall. 1998. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT104. Microb. Drug Resist. 4:113-118. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, M. C. 1996. Tetracycline determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 33.Rotger, R., and J. Casadesús. 1999. The virulence plasmids of Salmonella. Int. Microbiol. 2:177-184. [PubMed] [Google Scholar]
- 34.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1997. Characterization of integrons and antibiotic resistance genes in Danish multiresistant Salmonella typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 35.Soto, S. M., N. Martínez, B. Guerra, M. A. González-Hevia, and M. C. Mendoza. 2000. Usefulness of genetic typing methods to trace epidemiologically Salmonella serotype Ohio. Epidemiol. Infect. 125:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soto, S. M., B. Guerra, A. del Cerro, M. A. González-Hevia, and M. C. Mendoza. 2001. Outbreaks and sporadic cases of Salmonella serovar Panama studied by DNA fingerprinting and antimicrobial resistance. Int. J. Food Microbiol. 71:35-43. [DOI] [PubMed] [Google Scholar]
- 37.Threlfall, E. J., L. R. Ward, and B. Rowe. 1978. The spread of multiresistant strains of Salmonella phage types 204 and 193 in Britain. Br. Med. J. 2:997-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tosini, F., P. Visca, A. M. Dionisi, C. Pezzella, A. Petruca, and A. Carattoli. 1998. Class 1 integron-borne multiple antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 42:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker, R. A., E. Lindsay, M. J. Woodward, L. R. Ward, and E. J. Threlfall. 2001. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype Typhimurium phage type U302 (MR U302) from humans, animals and foods. Microb. Drug Resist. 7:13-21. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto, T., M. Tanaka, R. Baba, and S. Yamagishi. 1981. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Mol. Gen. Genet. 181:464-469. [DOI] [PubMed] [Google Scholar]