Abstract
CagA is a bacterial effector protein of Helicobacter pylori that is translocated via a type IV secretion system into gastric epithelial cells. We previously described that H. pylori require CagA to disrupt the organization and assembly of apical junctions in polarized epithelial cells. In this study, we provide evidence that CagA expression is not only sufficient to disrupt the apical junctions but also perturbs epithelial differentiation. CagA-expressing cells lose apicobasal polarity and cell–cell adhesion, extend migratory pseudopodia, and degrade basement membranes, acquiring an invasive phenotype. Expression of the CagA C-terminal domain, which contains the tyrosine phosphorylated EPIYA motifs, induces pseudopodial activity but is not sufficient to induce cell migration. Conversely, the N terminus targets CagA to the cell–cell junctions. Neither domain is sufficient to disrupt cell adhesion or cell polarity, but coexpressed in trans, the N terminus determines the localization of both polypeptides. We show that CagA induces a morphogenetic program in polarized Madin–Darby canine kidney cells resembling an epithelial-to-mesenchymal transition. We propose that altered cell–cell and cell matrix interactions may serve as an early event in H. pylori-induced carcinogenesis.
Keywords: differentiation, polarity, cell junctions, type IV secretion system, epithelial-to-mesenchymal transition
Several pathogenic bacteria have evolved the capacity to inject effector proteins into host cells as a way of manipulating specific cellular functions. Usually, these effector proteins are used to modify the bacterial attachment site, reorganize the actin cytoskeleton underneath adhered bacteria, trigger internalization, alter intracellular traffic, or induce cell death. CagA is an example of such a prokaryotic effector protein, translocated by adhered Helicobacter pylori into the host epithelial cell. CagA is unique, however, in that its chronic presence increases the risk of long-term complications for the host, such as the development of peptic ulcers and gastric adenocarcinoma.
We have previously shown that H. pylori use CagA to attach near the intercellular junctions and disrupt the organization and function of the apical junctional complex (AJC) of cultured epithelial cells (1). Within the host cell, CagA is phosphorylated by c-Src and Lyn kinases at tyrosine residues located in its C terminus (2–6), and this phosphorylation results in the activation of receptor tyrosine kinase (RTK)-like signaling pathways (7–11). Both the AJC and RTK signaling are important in regulating basic epithelial functions, such as establishing cell polarity and controlling cell division and migratory behavior during normal epithelial differentiation and wound healing. Mutations in genes involved in these pathways are frequently associated with oncogenic transformation.
To explore CagA's intrinsic biological properties in a well differentiated epithelial monolayer, we expressed the protein in polarized Madin–Darby canine kidney (MDCK) cells by means of mammalian expression vectors. We studied both the activity of the full-length protein, as well as of selected domains of CagA. We also tagged CagA with enhanced green fluorescent protein (GFP) to follow its localization in live cells and to compare the behavior of CagA-expressing cells with surrounding control cells by time-lapse microscopy. We found that CagA is not only sufficient to disrupt the epithelial junctions but also profoundly alters the differentiation and behavior of polarized epithelia.
Materials and Methods
Cell Culture and Transfection. MDCK II cells were obtained from W. J. Nelson (Stanford University, Stanford, CA). HEK 293 cells (Invitrogen) were used for immunoblotting and immunoprecipitation because of their higher transfection efficiency. AGS gastric adenocarcinoma cells were obtained from American Type Culture Collection. All cell types were grown in 5% CO2 atmosphere at 37°C in DMEM supplemented with 10% FBS (GIBCO/BRL). Transfection was carried out by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. In the polarized MDCK monolayers, transfection efficiency was on average 9% for GFP, 7% for GFP-NT-CagA (amino acids 1–877 of CagA), and 4% for both GFP-CT-CagA (amino acids 871–1216 of CagA) and GFP-CagA. To determine whether the number of CagA-expressing cells was lower because of cytotoxicity, we stained transfected monolayers with propidium iodide (Molecular Probes) at 24 h posttransfection. The average percentage of transfected cells that had propidium iodide incorporation was not statistically different between GFP (7.2%, n = 359) and the CagA constructs: GFP-NT-CagA (6.6%, n = 332, t test P = 0.8), GFP-CT-CagA (9.9%, n = 354, t test P = 0.2), and GFP-CagA-expressing cells (8.3%, n = 313, t test P = 0.6).
Plasmids and Constructs. CagA constructs were generated by cloning PCR products into vectors pIRES-puro and pEGFP-C3 (BD Biosciences Clontech) and verified by nucleotide sequencing. The following primers were designed to amplify the cagA sequence of strain G27 (12): Full-length gene (amino acids 1–1216) CagA-FW (ATGACTAACGAAACCATTAACC)/CagA-REV (GGTGGTTTCCAAAAATCTTAA), N terminus (amino acids 1–877) CagA-FW/NT-REV (GAGTTGAATGCAAAACTTGGA), and C terminus (amino acids 871-1216) CT-FW (GAGTTGAATGCAAAACTTGG)/CagA-REV. To generate the monomeric red fluorescent protein (RFP)-NT-CagA construct, the GFP sequence was replaced with the monomeric RFP cDNA (13) in the pEGFP-C3 vector. A list of additional vectors, CagA fragments, and controls can be found as Fig. 5 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site. This list includes several controls for the localization of CagA fragments to different cellular domains.
Quantitative Confocal Immunofluorescence. A list of antibodies used can be found in Tables 1 and 2, which are published as supporting information on the PNAS web site. Cells were processed as described in ref. 14. Apically exposed E-cadherin was visualized by using an antibody to the extracellular domain (15) added to the apical side of unpermeabilized monolayers polarized on Transwell polycarbonate filters (Corning Costar). Three-dimensional immunofluorescence images were reconstructed from 0.5-μm confocal optical sections by using volocity 3.5 (Improvision, Lexington, MA). To measure the perimeter of cellular junctions, we stained the monolayers with antibodies to ZO-1 and identified transfected cells through GFP fluorescence. The confocal optical sections from random fields were collapsed into single projections. imagej software (http://rsb.info.nih.gov/ij) was then used to select and measure the apical surface of individual cells. The data were then transferred to an excel worksheet and analyzed by using the spss 11.0 statistical software package (SPSS, Chicago).
Time-Lapse Imaging and Analysis. Cells plated on glass coverslips were placed in a sealed chamber kept at 37°C and imaged on a Zeiss Axiovert 200M microscope by using a 40× dry differential interference contrast microscopy (DIC) objective. Time-lapse images in the GFP and DIC channels were collected by a charge-coupled device camera (CoolSNAP-HQ, Photometrics, Tucson, AZ) and stored digitally every 10 min. After recording, the files were collated into digital movie sequences. Analysis of cell movement, speed, and translocation from the point of origin were done by using volocity 3.5 software. The outline of each cell at each time point was traced manually, and the centroid of individual cells was calculated at each time point. All coordinates were normalized to a starting point of 0,0, and plots of the trajectory of each cell were generated from the x–y coordinate data. The length of each plot defines the trajectory of each cell during the observation period. The shortest distance between the origin and the final position of each cell constitutes the translocation distance. Speed is calculated by dividing the trajectory length by the elapsed time, and the translocation rate was defined as the translocation divided by the elapsed time.
Invasion Assay. MDCK cells were plated at high density (2.5 × 105 cells per cm2) and polarized for 2 days on type IV collagen-rich Biocoat Matrigel invasion chambers (Becton Dickinson) before transfection with the CagA constructs. For inhibition of collagenases, GM6001 (Chemicon International, Temecula, CA) was added to the culture media at the time of plating at a final concentration of 12.5 μM. After immunostaining, randomly selected fields were imaged by confocal microscopy through the entire cell monolayer, including the filter and underlying cellular processes. Invading pseudopodia were defined as actin-rich structures that penetrated through the extracellular matrix into the space below the filters. We used the 3D reconstructed fluorescent phalloidin images to calculate the size of invading pseudopodia relative to the rest of the cell. The sum of the voxel intensities for the actin fluorescence of each pseudopodium was divided by that of the total cell actin signal to determine the percent of actin signal in the invading pseudopodia.
Results
CagA Alters the Epithelial Phenotype. Within 2 days of transfection, CagA-expressing cells acquire an elongated, spindle-shape morphology, lose their apicobasal orientation, extend long pseudopodia between adjacent cells, and sink below the monolayer, losing their connections with the apical junctions of neighboring cells (Fig. 1). To characterize the cellular events involved in this phenotypic transition, we monitored the morphology, apical junctions, and state of polarization of CagA-expressing cells at different times after transfection.
Fig. 1.
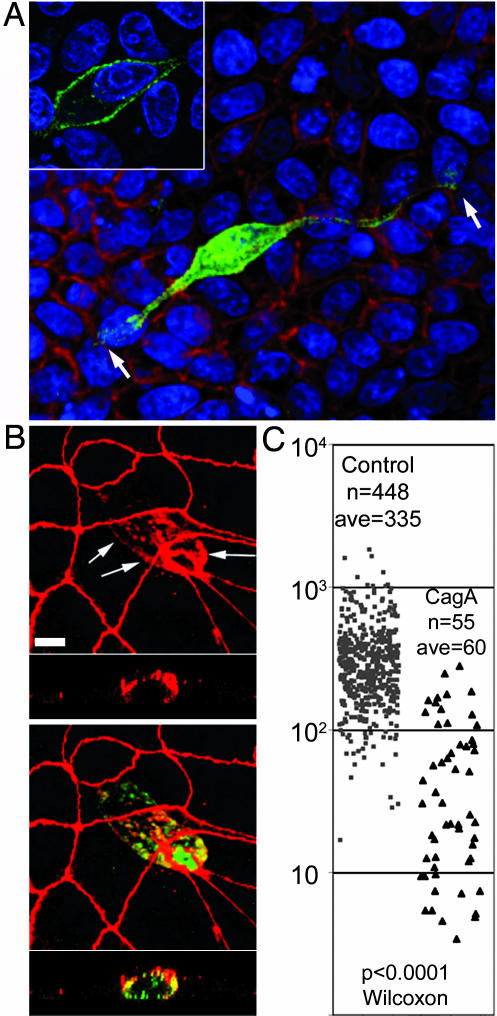
CagA perturbs the morphology and apical junctions of polarized epithelia. (A and B) Confocal immunofluorescence 3D reconstructions of confluent MDCK monolayers expressing GFP-CagA (green) for 24 h. (A) Cells were counterstained for F-actin (red) and the cell nuclei (blue). Shown is an x–y view seen from the bottom of the monolayer with one elongated CagA-expressing cell (green), spanning about nine cell diameters (arrows). (Inset) Optical section through the cell body of the CagA-expressing cell to show the subcellular localization of the protein. (B) CagA-expressing cells (green) stained for the tight junction protein ZO-1 (red) show mislocalization of ZO-1 to the basolateral membrane (arrows). Note also the reduced perimeter of the apical junctions in the CagA-expressing cell. The bottom strips are z sections. (C) The apical surface areas of control (gray squares) vs. CagA-expressing (black triangles) cells are plotted as a scatter plot. n, number of cells measured; ave, average cell surface area. The P values derived from a Wilcoxon nonparametric statistical test are noted. (Scale bars: 10 μm.)
To determine whether CagA is sufficient to disrupt cell–cell junctions, we analyzed the localization of the tight junction scaffolding protein ZO-1 in CagA-expressing cells. In nonconfluent monolayers, CagA preferentially localized to sites of junction formation that also contained ZO-1 (Fig. 4D; and see Fig. 6, which is published as supporting information on the PNAS web site). In confluent monolayers, however, CagA distributed throughout the cell periphery and induced the mislocalization of ZO-1 to the basolateral membrane (Fig. 1B). Furthermore, in CagA-expressing cells, the apical junction perimeter and the surface area of the apical membrane became markedly reduced as the cells penetrated the basolateral space. We measured the apical area of control and GFP-CagA-expressing cells within the same monolayer 20 h after transfection (Fig. 1C). The average apical area for the control cells was 335 μm2 (n = 448) versus 60 μm2 (n = 55) for CagA-expressing cells (Wilcoxon test P < 0.0001), indicating that CagA induces changes in epithelial morphology that are paralleled by disruption of the organization of the apical junctions and a reduction in the size of the apical junction perimeter and the area of the apical membrane.
Fig. 4.
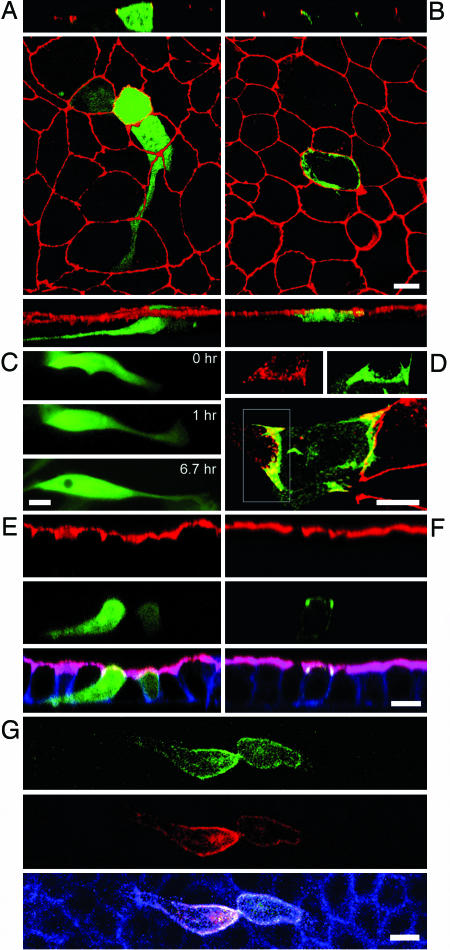
Differential localization and functional activity of CagA subdomains. Confocal immunofluorescence 3D reconstructions of GFP-CT-CagA (A) or GFP-NT-CagA-expressing (B) cells within confluent MDCK monolayers stained for ZO-1 (red). CagA-GFP fusions are green. (A and B Top) z sections. (Bottom) 3D reconstructed side views. (C) Three panels from a 6.7-h time-lapse movie show a cell expressing GFP-CT-CagA within a confluent monolayer. Extension of a long basal pseudopodium is observed, but the cell does not migrate. (D) A subconfluent MDCK monolayer expressing GFP-NT-CagA was stained with anti-ZO-1 antibodies to show the distribution of NT-CagA at areas of junction formation (yellow). (Inset) Area of a forming junction with separate ZO-1 (red) and GFP-NT-CagA (green) signals. (E and F) Neither the N- nor C-terminal portions of CagA are sufficient to disrupt apicobasal polarity. Shown are confocal z sections of MDCK monolayers expressing GFP-CT-CagA (E) and GFP-NT-CagA (F). Cells were stained for F-actin (blue) and an antibody to gp135 (red). (G) Coexpression of the two CagA fragments reveals a molecular interaction between the two domains. The NT-CagA fragment was tagged with red fluorescent protein (red) and coexpressed with GFP-CT-CagA (green) in a confluent monolayer. The cells were also stained for F-actin (blue). (Scale bars: 10 μm.)
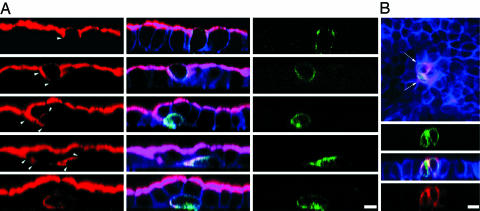
Loss of Cell Polarity After CagA Expression. Aberrant distribution of junctional proteins suggests that CagA perturbs junction function and affects cell polarity. The effect of CagA on cell polarity was analyzed by following the distribution of gp135, a membrane glycoprotein that is normally located exclusively in the apical membrane of polarized MDCK cells. As shown in Fig. 2A, CagA induced abnormal localization of this apical marker to the basolateral membrane. This loss of polarity occurred soon after CagA was expressed, because gp135 was seen in the basolateral membrane even before changes in cell morphology can be detected. At later stages, CagA-expressing cells elongated and migrated underneath neighboring cells, and gp135 was found throughout the basolateral membrane and concentrated at the trailing pole of elongated cells. Abnormal distribution of gp135 was found in 37% of CagA-expressing cells and 0% of cells transfected with the same vector driving GFP expression (unpaired t test P < 0.0001).
Fig. 2.
CagA causes loss of apicobasal polarity and opens the tight junctions. Confocal 3D reconstructions of MDCK monolayers polarized on Transwell filters, then transfected to express GFP-CagA (green), and stained for F-actin (blue). (A) Monolayers were stained for the apical membrane glycoprotein gp135 (red). Arrowheads show mislocalization of gp135 in the basolateral membrane as cells lose apicobasal polarity. Note also different stages in the loss of connection to the apical surface. (B) Polarized MDCK monolayer expressing GFP-CagA (green) was fixed without permeabilization and stained from the apical side with an antibody to the extracellular domain of E-cadherin (red). Arrows point to E-cadherin exposed on the apical surface of GFP-CagA-expressing cells. z sections show that anti-E-cadherin antibodies reach the paracellular space of GFP-CagA-expressing cells. (Scale bars: 10 μm.)
The localization of basolateral membrane proteins was also altered in CagA-expressing cells. E-cadherin is usually absent from the apical surface of epithelia and exposed if cell polarity is lost (16) or if the fence function of the junctions, which controls the diffusion of integral membrane proteins, fails. Antibodies to the extracellular domain of E-cadherin (17) added to the apical side of nonpermeabilized epithelial monolayers do not stain the apical surface and cannot reach basolateral epitopes. However, these antibodies stained the apical surface of ≈60% of CagA-expressing cells but not neighboring control cells (Fig. 2B). The paracellular barrier of the tight junctions was also disrupted in CagA-expressing cells, because these antibodies applied to the apex of nonpermeable cells penetrated the paracellular space to stain basolateral E-cadherin.
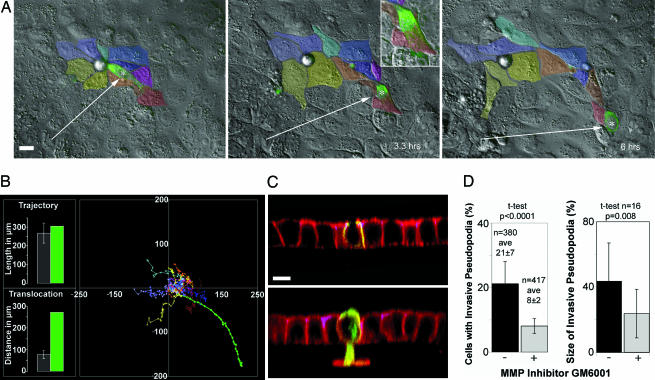
CagA-Expressing Cells Lose Cell Adhesion and Become Migratory. Loss of polarity, cell elongation, and loss of junction barrier function suggest that CagA may be causing the disruption of cell–cell adhesion and inducing cell migration. To test this hypothesis, we used time-lapse microscopy and examined the behavior of GFP-CagA-expressing cells within live-confluent monolayers. Cells were imaged by both differential interference contrast microscopy (DIC) and GFP-fluorescence at 10-min intervals for 6–16 h. The movement and spatial relationships of control and CagA-expressing cells were followed and compared with each other (Fig. 3A and Movies 1 and 2, which are published as supporting information on the PNAS web site). We observed that CagA-expressing cells extend pseudopodial processes between adjacent cells and also move between and under neighboring cells in a manner consistent with the loss of cell–cell adhesion (Fig. 3A). To describe this phenomenon quantitatively, we tracked the movement of individual CagA-expressing cells and surrounding control cells and plotted each cell trajectory (Fig. 3B). The average trajectory length of control cells was similar to that of the CagA-expressing cell, indicating that all cells moved at similar speeds. However, the translocation distance of the CagA-expressing cell was four times longer, because this cell moved without adhesive constraints from its neighbors. Analysis of several independent experiments showed that control cells moved with a speed of 43 ± 12 μm/hr (n = 18) and with a translocation rate of 13 ± 4 μm/hr (see Fig. 7, which is published as supporting information on the PNAS web site). This movement resulted in a modest translocation rate because cells are joined to each other. Cells expressing CagA moved at comparable speeds (62 ± 9 μm/hr, unpaired t test P = 0.24, n = 4). In contrast, they translocated almost four times further from their points of origin (44 ± 10 μm/hr, unpaired t test P = 0.007), indicating a loss of cell adhesion and movement independent of their neighbors.
Fig. 3.
CagA-expressing cells acquire a migratory and invasive phenotype. (A) Differential interference contrast microscopy (DIC) and fluorescence images of three frames from a 6-h time-lapse movie (see Movies 1 and 2) where the movement of a GFP-CagA-expressing cell (asterisk) and its contacting neighboring cells (color-coded) was followed. The CagA-expressing cell migrates from its original position and loses contact with most of the neighboring cells. (B) Graph of the trajectories of control cells color-coded in A and the CagA-expressing cell (green plot). The average trajectory length and translocation distances were compared between the control cells (gray bars) and the GFP-CagA-expressing cell (green bars) in the bar graphs. Error bars are one standard deviation from the mean. (C) Cells polarized on Matrigel-coated filters were transfected to express different CagA-GFP fusions (green), stained for F-actin (red), and imaged by confocal microscopy. (C Upper) z section of a monolayer expressing GFP-NT-CagA. These cells do not invade through the Matrigel. (C Lower) GFP-CagA-expressing cell with a large actin-rich pseudopodium that invades the basement membrane and crosses the filter. (D) GFP-CagA-expressing cells were imaged and invasive processes counted and measured volumetrically in the presence or absence of an MMP inhibitor. The percentage of cells with invasive pseudopodia and the percentage of actin signal below the basement membrane were calculated and plotted. Error bars represent one standard deviation from the mean. n, number of cells counted or measured; ave, averages. The P values derived from t tests are displayed. (Scale bars: 10 μm.)
The acquisition of migratory behavior in the context of loss of apicobasal polarity and the severing of intercellular junctions is reminiscent of epithelial-to-mesenchymal transitions (EMTs). This morphogenetic developmental program is observed at specialized sites during embryogenesis but is also thought to play an important part in the loss of epithelial characteristics that occurs during carcinogenic progression (reviewed in ref. 18).
CagA-Expressing Cells Are Able to Invade Through Extracellular Matrix. The EMT is not completely defined at a molecular level, but it is thought to involve a change in the developmental programming of the epithelial cells that results in invasive behavior. Epithelial cells that undergo this mesenchymal transition are able to invade the extracellular matrix by expressing matrix metalloproteinases (MMPs) and degrading their underlying basement membrane. To test whether CagA-expressing cells are able to penetrate basement membranes, we prepared MDCK monolayers on filters coated with Matrigel (a basement membrane-like substrate containing type IV collagen and laminin). As shown in Fig. 3C, ≈21% (n = 380) of CagA-expressing cells generated basal pseudopodia that invaded the Matrigel matrix and penetrated into the underlying filter pores. In contrast, only 0.6% of control cells (n = 1,890) penetrated the same matrix (t test P < 0.0001). We also investigated whether MMPs are involved in this invasive behavior by determining whether the number and size of CagA-induced invasive processes would respond to treatment with GM6001, a synthetic peptide inhibitor of MMPs (19). MDCK monolayers polarized on Matrigel-coated filters were pretreated with the MMP inhibitor before transfection. GFP-CagA-expressing cells were imaged 24 h after transfection and invasive processes counted. The percent of invasive CagA-expressing cells was reduced from 21 ± 7 (n = 380) without the inhibitor to 8 ± 2% (n = 417, t test P < 0.0001) in the presence of inhibitor (Fig. 3D). We also quantified volumetrically the actin signal below the basement membrane of CagA-expressing cells and compared it with the total actin signal to determine the relative size of the invasive pseudopodia (Fig. 3D). Noninhibited cells had an average of 43% of their actin in pseudopodia, versus 24% in the inhibited cells (n = 16, t test P = 0.008). Thus, our data are consistent with the notion that CagA induces invasive behavior in epithelial cells and possibly explain the results seen by others using general transcriptional analysis, namely that H. pylori infection is associated with induction of MMPs (20, 21).
CagA Functional Domains Are Important for Localized Signaling. To understand the contribution of different CagA domains to the loss of epithelial differentiation, we expressed different regions of the protein in polarized MDCK monolayers. H. pylori infection induces cellular elongation in unpolarized gastric adenocarcinoma cells (AGS) (2), and the tyrosine phosphorylation of CagA by src-family kinases is necessary for this process (9, 11). We therefore expressed the C-terminal 346 aa of CagA (CT-CagA), containing the phosphorylation sites within EPIYA motifs, and asked whether this region of the molecule is sufficient to activate the EMT-like behavior observed with the full-length molecule. We found that CT-CagA is tyrosine-phosphorylated in the host cell, and its phosphorylation could be prevented by the src inhibitor PP1 (Figs. 8 and 9, which are published as supporting information on the PNAS web site) (9). Furthermore, polarized epithelial cells within a confluent monolayer expressing CT-CagA (or GFP-CT-CagA) elongated, extending basal pseudopodia that advance and retract between and below adjacent cells (Fig. 4 A and C; and see Movies 3 and 4, which are published as supporting information on the PNAS web site). However, unlike CagA, this fragment was not localized to the junctions or cell membrane and remained diffusely distributed within the cell cytoplasm. Furthermore, despite inducing cell elongation, the formation of protrusive pseudopodia, CT-CagA did not disrupt the distribution of junctional proteins (Fig. 4A), cause changes in cell polarity (Fig. 4E), cause the loss of cell–cell adhesion (Fig. 4C), or induce cell migration (Fig. 7). The C terminus of CagA was sufficient, however, to induce MMP activity because its expression induced the formation of pseudopodia that were able to penetrate basement membranes in 8% (n = 234) of CT-CagA-expressing cells (versus 0.6% of control cells (n = 1,890, t test P < 0.0001).
Previous studies have also shown that association of H. pylori with junctional proteins depends on CagA delivery into epithelial cells but does not depend on CagA phosphorylation (1, 20). We expressed the N-terminal two-thirds of the CagA protein (NT-CagA or GFP-NT-CagA, amino acids 1–877), lacking the EPIYA tyrosine phosphorylation motifs, and found that this region is sufficient to target CagA to the junctions and the plasma membrane (Fig. 4B). We confirmed this finding by expressing three other constructs of different sizes that lack EPIYA motifs, as well as a mutant form of CagA with the EPIYA tyrosines mutated into serines (EPISA) (see Fig. 5). Like full-length CagA, NT-CagA was localized to sites of junction formation in preconfluent monolayers (Fig. 4D), but, unlike the full-length protein, NT-CagA did not cause detectable redistribution of junctional proteins to the basolateral membranes (Fig. 4B), nor did it disrupt cell polarity (Fig. 4F). Furthermore, it did not induce morphological changes, cell migration, or invasive behavior (Fig. 3C). Together, these findings demonstrate that the signaling triggered by the EPIYA motifs of CagA is necessary to cause cellular elongation and pseudopodial activity but is not sufficient to induce all of the events resembling the EMT that are induced by the full-length protein. Furthermore, they suggest that localization of CagA to the junctions, mediated by the N terminus, is necessary to activate these events.
To test whether the activities of the N and C termini of CagA act independently of each other, we tagged NT-CagA with monomeric red fluorescent protein (13) (RFP-NT-CagA), coexpressed it with GFP-CT-CagA, and monitored the behavior of cells expressing the two domains in trans. An unexpected finding was that when both peptides were expressed in the same cell, they colocalized near the plasma membrane (Fig. 4G), whereas when expressed individually, GFP-CT-CagA was diffuse in the cytoplasm (Fig. 4A). This colocalization of the cotransfected peptides suggests that the two domains may interact with each other by means of noncovalent associations or as part of a complex with host cell proteins and that the N terminus is dominant in determining CagA's subcellular localization. In support of this interpretation, immunoprecipitation of a CT-CagA fragment tagged with a FLAG tag also coprecipitated the NT-CagA fragment when expressed in trans (Fig. 10, which is published as supporting information on the PNAS web site).
Discussion
In this study, we showed that CagA, a cancer-associated effector protein of H. pylori, is sufficient to disrupt the mechanisms that maintain normal epithelial differentiation, including cell adhesion, cell polarity, and the inhibition of migration. These effects appear to be controlled by different domains in the CagA protein, some of which are phosphorylation-dependent, and involve RTK-like signaling, whereas others are phosphorylation independent and relate to the apical junctions. We suggest that CagA subdomains work in concert in the vicinity of the apical junctional complex to orchestrate CagA's effects on cell behavior. We propose that targeting CagA to the apical junctional complex through the N terminus of the molecule confers specificity to the RTK-like signaling induced by the phosphorylated C terminus.
The cellular behavior induced by CagA is reminiscent of oncogenes that disrupt cytoskeletal signaling and induce EMTs. CagA is known to interact with a number of potential oncogenes including the RTK c-Met (10), the tyrosine phosphatase SHP-2 (7), and the adaptor protein Grb2 (8). Although CagA has no amino acid homology with eukaryotic proteins involved in these signaling pathways, it may have structural similarities to proteins that can link and activate these pathways. For example, CagA may share some of the functional properties of Gab proteins, docking molecules that serve as scaffolds to mediate the activation of multiple signals downstream of RTK (22). CagA and Gab proteins have been implicated in the activation of the Ras/MAPK (23) and PI3-kinase (10) pathways, both of which have been linked to EMTs (24, 25). The carcinogenic effects of CagA have also been recently correlated with activation of the β-catenin pathway (26), which has important roles in epithelial differentiation and the induction of EMTs (27).
Unlike known oncogenes, CagA is a noninheritable signal delivered to the host by infecting microorganisms residing on the cell membrane. How can a signal that disrupts cell–cell and cell–matrix interactions contribute to carcinogenesis? Our findings point to the concept that disrupting cell–cell adhesion, apicobasal polarity, and cell–matrix interactions of epithelia may lead to defects of cellular differentiation and signaling that can act very early in the process of tumor initiation/promotion (28–31). In fact, disruption of cell–matrix interactions by ectopic expression of MMPs has been shown to be enough to induce carcinomas in animal models (30).
Because carcinogenesis is a multistep process, it is also possible that CagA's effects in vivo become pathologic only in the context of other acquired mutations or in specific cell types. For example, CagA itself may select for cells that can survive in its presence, resulting in an aberrant population of cells with intrinsic or acquired resistance to apoptotic signals induced by the loss of junctions and polarity (32). Furthermore, a recent observation indicates that chronic Helicobacter infection in mice results in recruitment of bone marrow-derived mesenchymal stem cells into the gastric epithelium and that these cells can become the target of malignant transformation (33). Although this model did not test the role of CagA in carcinogenesis, it is possible that, in the presence of CagA, the behavior and differentiation programs of gastric precursor cells is disrupted, and their ability to self-renew, resist apoptosis, and undergo EMTs could make these cells more susceptible to long-term pathogenic effects of CagA. Whether gastric stem cells are targets for CagA delivery in vivo is an important question for the future. Interestingly, a recent report by Oh et al. (34) suggests that H. pylori have the capacity to directly interact with gastric precursor cells.
Supplementary Material
Acknowledgments
We thank S. Falkow, R. Rappuoli, A. Muller, D. Monack, and R. Vogelmann for critical reading of the manuscript and helpful discussions. W. J. Nelson shared many reagents and time-lapse microscopy equipment. We also thank S. Yamada for sharing microscopy expertise; N. Pacchiani, G. Corsi, and M. Pentecost for assistance in generating the figures; A. Kawale for cell culture help; and S. Censini and M. Stein for sharing sera and clones. This work was supported by National Institutes of Health Grants AI38459 (to M.R.A. and L.T.) and DDC DK56339 (to M.R.A.).
Author contributions: F.B., L.T., A.C., and M.R.A. designed research; F.B., L.B., and M.R.A. performed research; and F.B., A.C., and M.R.A. analyzed data and wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MDCK, Madin–Darby canine kidney; GFP, enhanced green fluorescent protein; RTK, receptor tyrosine kinase; EMT, epithelial-to-mesenchymal transition; MMP, matrix metalloproteinase.
References
- 1.Amieva, M. R., Vogelmann, R., Covacci, A., Tompkins, L. S., Nelson, W. J. & Falkow, S. (2003) Science 300, 1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segal, E. D., Cha, J., Lo, J., Falkow, S. & Tompkins, L. S. (1999) Proc. Natl. Acad. Sci. USA 96, 14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahi, M., Azuma, T., Ito, S., Ito, Y., Suto, H., Nagai, Y., Tsubokawa, M., Tohyama, Y., Maeda, S., Omata, M., et al. (2000) J. Exp. Med. 191, 593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backert, S., Ziska, E., Brinkmann, V., Zimny-Arndt, U., Fauconnier, A., Jungblut, P. R., Naumann, M. & Meyer, T. F. (2000) Cell. Microbiol. 2, 155-164. [DOI] [PubMed] [Google Scholar]
- 5.Odenbreit, S., Puls, J., Sedlmaier, B., Gerland, E., Fischer, W. & Haas, R. (2000) Science 287, 1497-1500. [DOI] [PubMed] [Google Scholar]
- 6.Stein, M., Rappuoli, R. & Covacci, A. (2000) Proc. Natl. Acad. Sci. USA 97, 1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashi, H., Tsutsumi, R., Muto, S., Sugiyama, T., Azuma, T., Asaka, M. & Hatakeyama, M. (2002) Science 295, 683-686. [DOI] [PubMed] [Google Scholar]
- 8.Mimuro, H., Suzuki, T., Tanaka, J., Asahi, M., Haas, R. & Sasakawa, C. (2002) Mol. Cell 10, 745-755. [DOI] [PubMed] [Google Scholar]
- 9.Stein, M., Bagnoli, F., Halenbeck, R., Rappuoli, R., Fantl, W. J. & Covacci, A. (2002) Mol. Microbiol. 43, 971-980. [DOI] [PubMed] [Google Scholar]
- 10.Churin, Y., Al-Ghoul, L., Kepp, O., Meyer, T. F., Birchmeier, W. & Naumann, M. (2003) J. Cell Biol. 161, 249-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higashi, H., Nakaya, A., Tsutsumi, R., Yokoyama, K., Fujii, Y., Ishikawa, S., Higuchi, M., Takahashi, A., Kurashima, Y., Teishikata, Y., et al. (2004) J. Biol. Chem. 279, 17205-17216. [DOI] [PubMed] [Google Scholar]
- 12.Xiang, Z., Censini, S., Bayeli, P. F., Telford, J. L., Figura, N., Rappuoli, R. & Covacci, A. (1995) Infect. Immun. 63, 94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A. & Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. USA 99, 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amieva, M. R., Salama, N. R., Tompkins, L. S. & Falkow, S. (2002) Cell. Microbiol. 4, 677-690. [DOI] [PubMed] [Google Scholar]
- 15.Shore, E. M. & Nelson, W. J. (1991) J. Biol. Chem. 266, 19672-19680. [PubMed] [Google Scholar]
- 16.Pollack, A. L., Runyan, R. B. & Mostov, K. E. (1998) Dev. Biol. 204, 64-79. [DOI] [PubMed] [Google Scholar]
- 17.Gumbiner, B. & Simons, K. (1987) Ciba Found. Symp. 125, 168-186. [DOI] [PubMed] [Google Scholar]
- 18.Thiery, J. P. (2002) Nat. Rev. Cancer 2, 442-454. [DOI] [PubMed] [Google Scholar]
- 19.Boghaert, E. R., Chan, S. K., Zimmer, C., Grobelny, D., Galardy, R. E., Vanaman, T. C. & Zimmer, S. G. (1994) J. Neurooncol. 21, 141-150. [DOI] [PubMed] [Google Scholar]
- 20.El-Etr, S. H., Mueller, A., Tompkins, L. S., Falkow, S. & Merrell, D. S. (2004) J. Infect. Dis. 190, 1516-1523. [DOI] [PubMed] [Google Scholar]
- 21.Kitadai, Y., Sasaki, A., Ito, M., Tanaka, S., Oue, N., Yasui, W., Aihara, M., Imagawa, K., Haruma, K. & Chayama, K. (2003) Biochem. Biophys. Res. Commun. 311, 809-814. [DOI] [PubMed] [Google Scholar]
- 22.Hatakeyama, M. (2003) Microbes Infect. 5, 143-150. [DOI] [PubMed] [Google Scholar]
- 23.Meyer-ter-Vehn, T., Covacci, A., Kist, M. & Pahl, H. L. (2000) J. Biol. Chem. 275, 16064-16072. [DOI] [PubMed] [Google Scholar]
- 24.Grande, M., Franzen, A., Karlsson, J. O., Ericson, L. E., Heldin, N. E. & Nilsson, M. (2002) J. Cell Sci. 115, 4227-4236. [DOI] [PubMed] [Google Scholar]
- 25.Bakin, A. V., Tomlinson, A. K., Bhowmick, N. A., Moses, H. L. & Arteaga, C. L. (2000) J. Biol. Chem. 275, 36803-36810. [DOI] [PubMed] [Google Scholar]
- 26.Franco, A. T., Israel, D. A., Washington, M. K., Krishna, U., Fox, J. G., Rogers, A. B., Neish, A. S., Collier-Hyams, L., Perez-Perez, G. I., Hatakeyama, M., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 10646-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birchmeier, C., Birchmeier, W. & Brand-Saberi, B. (1996) Acta Anat. (Basel) 156, 217-226. [DOI] [PubMed] [Google Scholar]
- 28.Lochter, A., Galosy, S., Muschler, J., Freedman, N., Werb, Z. & Bissell, M. J. (1997) J. Cell Biol. 139, 1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, S. & Ingber, D. E. (1999) Nat. Cell Biol. 1, E131-E138. [DOI] [PubMed] [Google Scholar]
- 30.Sternlicht, M. D., Lochter, A., Sympson, C. J., Huey, B., Rougier, J. P., Gray, J. W., Pinkel, D., Bissell, M. J. & Werb, Z. (1999) Cell 98, 137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingber, D. E. (2002) Differentiation (Berlin) 70, 547-560. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi, M., Hirose, F., Inoue, Y. H., Ohno, K., Yoshida, H., Hayashi, Y., Deak, P. & Matsukage, A. (2004) Cancer Sci. 95, 436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houghton, J., Stoicov, C., Nomura, S., Rogers, A. B., Carlson, J., Li, H., Cai, X., Fox, J. G., Goldenring, J. R. & Wang, T. C. (2004) Science 306, 1568-1571. [DOI] [PubMed] [Google Scholar]
- 34.Oh, J. D., Karam, S. M. & Gordon, J. I. (2005) Proc. Natl. Acad. Sci. USA 102, 5186-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.