Abstract
Progress in isolating stem cells from tissues, or generating them from adult cells by nuclear transfer, encourages attempts to use stem cells from affected individuals for gene correction and autologous therapy. Current viral vectors are efficient at introducing transgenic sequences but result in random integrations. Gene targeting, in contrast, can directly correct an affected gene, or incorporate corrective sequences into a site free from undesirable side effects, but efficiency is low. Most current targeting procedures, consequently, use positive-negative selection with drugs, often requiring ≥10 days. This drug selection causes problems with stem cells that differentiate in this time or require feeder cells, because the feeders must be drug resistant and so are not eliminated by the selection. To overcome these problems, we have developed a procedure for isolating gene-corrected stem cells free from feeder cells after 3-5 days culture without drugs. The method is still positive-negative, but the positive and negative drug-resistance genes are replaced with differently colored fluorescence genes. Gene-corrected cells are isolated by FACS. We tested the method with mouse ES cells having a mutant hypoxanthine phosphoribosyltransferase (Hprt) gene and grown on feeder cells. After 5 days in culture, gene-corrected cells were obtained free from feeder cells at a “purity” of >30%, enriched >2,000-fold and with a recovery of ≈20%. Corrected cells were also isolated singly for clonal expansion. Our FACS-based procedure should be applicable at small or large scale to stem cells that can be cultured (with feeder cells, if necessary) for ≥3 days.
Keywords: electroporation, flow cytometry, fluorescent protein, gene targeting, gene therapy
Hematopoietic stem cells (HSC) are among the longest known and best studied stem cells (1). They are characterized by their ability to fully repopulate the bone marrow of suitably prepared recipients and have been used therapeutically in the form of bone marrow transplants for several decades. HSC are, however, difficult to culture ex vivo without differentiating, and normal HSC have been induced to increase to only about six times the input number of HSC in vitro (2). The in vitro expansion of HSC from experimental animals has been enhanced (up to approximately forty times input) by the introduction of transgenes coding for MDR1 or HOXB4 (3, 4), but HSC cannot currently be clonally expanded in vitro. Their ability to self-replicate extensively in vivo is, however, undoubted because a single HSC is sufficient to repopulate the bone marrow of a recipient, which can then be used to repopulate the bone marrows of secondary recipients (5, 6).
The history of bone marrow transplantations clearly emphasizes that problems are likely to arise when there are histocompatibility differences between donors and recipients. Thus, the first successful bone marrow transplant was with an identical twin (genetically an autologous transplant) (7). Only the subsequent elucidation of histocompatibility antigens allowed success with allogeneic transplants (8). But, even now, the need for pretransplant myeloablation with cytotoxic drugs and/or irradiation and the need for posttransplant immune suppression limits the use of bone marrow transplantation to life-threatening conditions. Despite these limitations, bone marrow or cord blood transplantation is the only curative treatment for many malignancies of the hematopoietic system, for severe aplastic anemia, and for severe combined immunodeficiency. Beta zero thalassemia is the only common genetic disease for which bone marrow or cord blood transplantation is generally accepted, although it is a possibility for sickle cell anemia (9). In both these conditions, the availability of an HLA-compatible donor is an important consideration.
Because securing histocompatible donors is often difficult or impossible, and because of the high risk of graft-versus-host disease, considerable efforts have been made to develop ways of correcting the genetic defect in HSC from affected individuals and of using the corrected cells for autologous transplantation. Transfer into the HSC of a functional copy of the affected gene, typically with a viral vector, has received the most attention. The procedure is highly effective, with transfection frequencies approaching 20% (10, 11). Unfortunately, there is no control over the sites where virally transduced sequences incorporate into the genome, and this lack has led to the occurrence of leukemia in some patients with adenosine deaminase deficiency in whom the original defect had been corrected by retroviral gene therapy (12). Homologous recombination (gene targeting) is an alternative method of correcting genetic defects (13, 14). The affected gene can be corrected directly, or a functional copy of the gene can be introduced into a locus known to be free from significant side effects [such as the hypoxanthine phosphoribosyltransferase (Hprt) locus]. The low efficiency of gene targeting (of the order of 1 corrected cell in 105 treated cells, when tested in ES cells) and the low frequency of stem cells in total bone marrow (of the order of 1 stem cell per 104 to 105 nucleated cells) has currently precluded its successful application to HSC.
Gene correction by homologous recombination has, nevertheless, been demonstrated to occur in mouse hematopoietic progenitor cells (HPCs), which are more common in bone marrow (of the order of 2 HPC per 103 nucleated cells), and the frequency of recombination was indistinguishable from that obtained with ES cells (15). The gene defect corrected in the HPC experiments was a mutation in the Hprt gene that would cause Lesch-Nyhan disease in human males. Corrected cells were identified by their ability to form colonies in a selective medium that kills uncorrected cells. However, because this type of direct selection of targeted cells is possible for only a very few genes, indirect strategies for gene targeting have been developed. The positive-negative strategy devised by Mansour et al. (16) is most commonly used; it identifies targeted cells by their ability to form colonies resistant to two drugs. Its application in stem cells is limited to those that remain undifferentiated for the time required for uncorrected (drug-sensitive) cells to be killed and for corrected (drug-resistant) cells to form colonies (typically ≥10 days). Some stem cells, including HSC, are difficult to maintain undifferentiated in culture for this length of time, some do not form colonies, and some survive ex vivo only in the presence of accessory cells, which must be resistant to the selective drugs and so are not eliminated by them.
To overcome these limitations and restrictions, we have developed a FACS-based gene-targeting strategy that can be applied to any stem cells that can be cultured (with feeder cells, if necessary) for ≥3 days without differentiating. The stem cells do not have to be clonable, although the procedure can isolate cells singly for use with stem cells that can be clonally expanded. We have tested the procedure by correcting a mutation in the Hprt gene in ES cells. Gene-corrected cells were obtained in bulk with a recovery of ≈20%, enrichment >2,000, and at a “purity” of ≈30%.
Materials and Methods
Gene-Correcting Vectors. Two gene-correcting vectors were derived from an Hprt-targeting vector previously described (17). A green-positive, cyan-negative gene-correcting vector (Fig. 1A Middle) was made by inserting sequences coding for an EGFP (BD Clontech) between a BstBI and a BspEI site in the Hprt-vector, and inserting sequences coding for an enhanced cyan fluorescence protein (ECFP; BD Clontech) into the SalI site of the vector. A cyan-positive, green-negative gene-correcting vector (see Fig. 3C) was made by inserting the ECFP sequences between a BstBI and an AvrII site in the Hprt-targeting vector, and inserting sequences coding for a humanized Renilla GFP with a nuclear localizing sequence (hrGFP; Stratagene) into the SalI site. All fluorescence genes were driven by a human β-actin promoter together with its first intron (1.3 kb) on a 1,311-bp AvrII/AluI fragment from a 4.3-kb sequence that spans the promoter (18, 19) (accession no. AY582799).
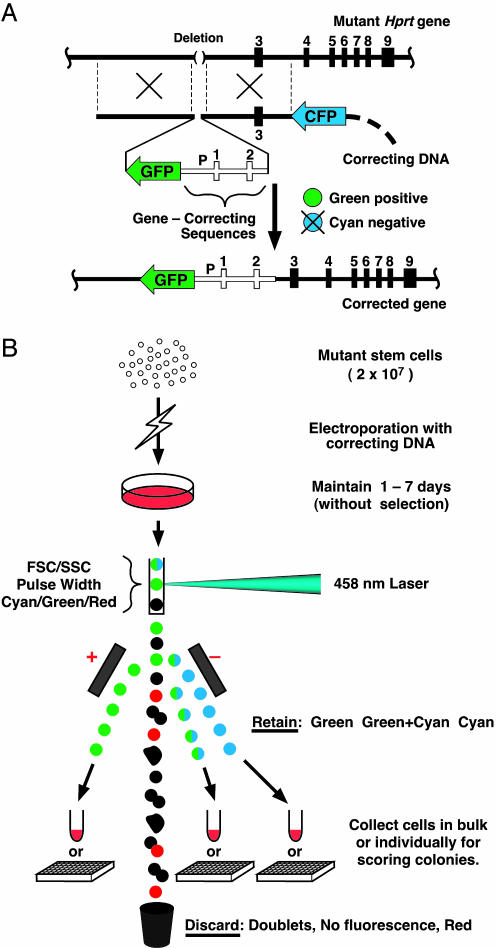
Fig. 1.
Gene correction by homologous recombination with vectors containing fluorescent protein genes. (A) A green-positive, cyan-negative targeting vector for correcting a gene mutation by homologous recombination with a defective Hprt gene as an example. (Top) The structure of the target gene, which has a deletion of ≈55 kb that removed the promoter and exons 1 and 2, but left exons 3 through 9. (Middle) The gene-correcting vector, which includes a GFP gene and the missing Hprt promoter P and exons 1 and 2 all flanked by the homologous sequences that control the recombination. A CFP gene, located outside the homologous regions, is also part of the targeting vector. (Bottom) The corrected gene with the GFP gene used for positive selection now adjacent to it. The CFP gene used for negative selection is excluded by the homologous recombination. The open boxes and line indicate the correcting DNA. The heavy horizontal lines and filled boxes represent endogenous sequences and exons in the target gene and homologous sequences in the correcting vector. The heavy dashed line indicates bacterial sequences. (B) Mutant stem cells are electroporated with correcting DNA and maintained on feeders for 1-7 days with passages but without drug selection. Cells are dispersed with trypsin and sorted by differences in their forward light scatter (FSC), side scatter (SSC), and pulse width and by their fluorescence after excitation with a 458-nm laser. Cells with green or green plus cyan, or cyan fluorescence are collected in bulk, or singly into the wells of a 384-well plates. Doublets, cells with no fluorescence, or cells fluorescing red are discarded. Colonies derived from sorted cells are counted before and after HAT selection.
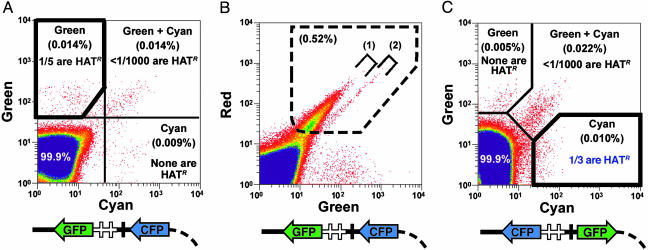
Fig. 3.
Sorting criteria for collecting gene-corrected cells. (A) Green and cyan fluorescence used to collect gene-corrected cells 7 days after electroporation introducing the green-positive, cyan-negative vector diagrammed at the bottom of the panel. The heavy polygon indicates the gating used for collecting green-positive, cyan-negative cells (0.014% of the total input); one in five of these cells produces HATR colonies in which the mutant gene is corrected. Less than 1 in 1,000 of green-positive, cyan-positive cells (0.014% of input cells) and none of the green-negative, cyan-positive cells (0.009% of input cells) are HATR. Single cells are shown in red. Yellow and blue indicate increasing numbers of cells. (B) Red and cyan fluorescence used to exclude cells with autofluorescence in experiments with the green-positive, cyan-negative vector by rejecting cells within the dashed polygon (0.52% of input cells). These include minor fractions of feeder cells and stem cells that autofluoresce, indicated by square brackets no. 1 and no. 2, respectively. (C) Green and cyan fluorescence used to collect gene-corrected cells 7 days after electroporation introducing the cyan-positive, green-negative vector diagrammed at the bottom of the panel in which the CFP and GFP genes of the earlier vector have been interchanged. The heavy polygon indicates the gating used to collect cyan-positive, green-negative cells (0.01% of input cells); 1 in 3 of these cells produce HATR colonies in which the mutant gene has been corrected. Less than 1 in 1,000 of cyan-positive, green-positive cells (0.022% of input cells) and none of the cyan-negative, green-positive cells (0.005% of input cells) are HATR. No significant autofluorescence encroaches on the cyan channel, and so the red channel is not needed with this vector, which is the one currently used.
Fluorescence-Activated Cell Sorting. Flow cytometry and sorting were with a MoFlo cell sorter (DakoCytomation, Fort Collins, CO). An argon-ion laser tuned to 458 nm (Coherent Innova-90) with output set to 150 mW was used to excite GFP and cyan fluorescent protein (CFP) (20). The CFP fluorescence was separated from the GFP fluorescence with a 500-nm short-pass dichroic mirror and collected with a 485/30-nm band-pass (BP) filter (Chroma Technology, Rockingham, VT). The GFP fluorescence was collected with a 510/AF23-nm BP filter (Omega Optical, Brattleboro, VT). Red fluorescence used for autofluorescence detection was separated with a 560-nm long-pass (LP) dichroic mirror (Chroma Technology) and collected with a 575-nm alpha long-pass (ALP) filter (Omega Optical). When sorting for GFP fluorescence only (Fig. 2), an argon-ion laser tuned to 488 nm was used. Sorting was conducted at room temperature (≈25°C).
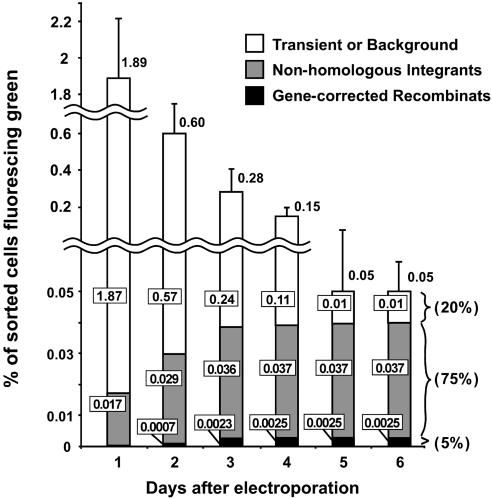
Fig. 2.
The time course of events after culture for 1-6 days after electroporation with the green-positive, cyan-negative vector diagramed in Fig. 1 A. The vertical axis shows the percentage of sorted cells fluorescing green. Open bars indicate cells that produce nonfluorescing colonies when expanded. Gray bars indicate cells that produce green-fluorescing HAT-sensitive colonies; these are cells that have incorporated the GFP gene into their genomes nonhomologously. Black bars indicate cells that produce green-fluorescing HAT-resistant colonies; these are the cells in which the mutant gene has been corrected by homologous recombination. The numbers in the boxes show the percentages of sorted cells falling into the three categories indicated by the shading of the bars. The error bars show the standard errors of the results from four experiments. Note that, although a green-positive, cyan-negative vector was used in this experiment, sorting was only for green fluorescence. Note also that the vertical axis of the figure is interrupted and has two scales.
Isolating Gene-Corrected Stem Cells. The mouse ES cells used in our experiments were a subclone (BK4) of E14TG2a, a cell line derived from 129/Ola mice with a deletion in the Hprt gene (21). The ES cells were grown and maintained on murine embryonic fibroblasts in DMEM supplemented with 15% heat-inactivated FBS, 10 μM 2-mercaptoethanol, and 2 mM l-glutamine. Electroporation was with ≈2 × 107 ES cells in the presence of 10 μg of the targeting DNA in a cuvette having a gap size of 4 mm and an area of 160 mm2 with a 1-s discharge from a 250-μF capacitor charged to 300 V. The ES cells were maintained for 1-7 days, with passages but without drug selection, on feeder layers of irradiated murine embryonic fibroblasts. Cells were then dispersed with trypsin and sorted by differences in their forward scatter (FSC), side scatter (SSC), and pulse width and by their fluorescence after excitation with a 458-nm laser. Cells with “green” or “green plus cyan” or “cyan” fluorescence were collected in bulk, or singly into the wells of 384-well plates in which feeder cells were already present. Doublets, cells with no fluorescence, or cells fluorescing “red” were discarded. To assess the frequency of gene correction, the sorted cells were plated for colony formation, and the numbers of colonies were counted with and without HAT selection (16 μg/ml hypoxanthine, 0.175 μg/ml aminopterin, and 4.8 μg/ml thymidine in the ES cell medium described above) for 7 days.
Assessing Transient Gene Expression. To assess the frequency and time course of transient expression (Fig. 2) of the electroporated gene-targeting construct, we used the green-positive, cyan-negative construct and the same general procedures as in a full-scale sorting experiment except that we sorted only by green fluorescence. Green-positive cells were collected in bulk at days 1-6 after electroporation. The percentage of cells in which gene correction had been achieved was determined by plating the collected cells for colony formation and by counting the numbers of resulting colonies with and without HAT selection. The colonies were also scored for fluorescence.
Results and Discussion
Gene-Targeting with Two Fluorescence Genes. To develop our FACS-based procedure, we used male-derived mouse ES cells having a deletion mutation in the X-linked Hprt gene, which enables easy detection of gene correction by testing for resistance to HAT. Our procedure retains the positive-negative strategy first described by Capecchi and his collaborators (16), but the positively selectable drug resistance gene (typically a neomycin phosphotransferase gene) is replaced with a gene coding for a GFP. This GFP gene is inserted into the target locus when homologous recombination occurs. Cells having the desired gene correction therefore fluoresce green. The conventional negatively selectable gene (typically a viral thymidine kinase gene) is replaced with a CFP gene located outside the sequences involved in the homologous recombination. This CFP gene is eliminated when homologous recombination occurs. Corrected cells therefore have no cyan fluorescence. Cells incorporating the CFP gene nonhomologously into their genomes fluoresce cyan. Cells in which gene correction has been achieved are consequently green-positive, cyan-negative (Fig. 1 A).
The overall procedure (summarized in Fig. 1B) uses electroporation to introduce a gene-correcting vector into the mutant stem cells. After the electroporation, the treated cells are cultured in nonselective medium for 1-7 days in the presence of feeder cells. The cultured cells are then dispersed with trypsin and sorted by their green and/or cyan fluorescence under a 458-nm excitation, with prior gating to discard debris, feeder cells, doublets, and autofluorescing cells. Sorting for green-positive, cyan-negative cells recovers corrected cells and eliminates any feeder cells that escape the prior gating because the feeder cells do not fluoresce green. Gene-corrected cells can be isolated in bulk, or singly for clonal expansion. (Details of the criteria for prior gating and for distinguishing green and cyan fluorescing cells are in Fig. 4, which is published as supporting information on the PNAS web site.)
Fates of Exogenous DNA. Gene-correcting DNA introduced into a cell can result in at least three nonexclusive outcomes. (i) The DNA may remain for a time in the cells and be transiently expressed but never integrated into the genome. (ii) The DNA, whole or in fragments, may be incorporated nonhomologously into the genome. (iii) The DNA may recombine homologously and correct the mutant gene. The relative frequency of these outcomes varies with the DNA construct and the target locus, but in general their frequencies are i >> ii > iii. Our initial experiments were therefore aimed primarily at determining the time course and relative frequencies of these three outcomes. For this purpose, we used the green-positive, cyan-negative vector shown in Fig. 1 A, but sorted positively only for green fluorescence together with prior gating to exclude feeder cells, debris, and cell doublets. Fig. 2 shows that, after 1 day in culture, 1.89% of the trypsinized cells fluoresced green. This percentage decreased progressively until, after 5 days, it stabilized at ≈0.05%.
To ascertain the types of events that had occurred in the fluorescing cells collected on the different days, we plated the sorted green-fluorescing cells onto feeder layers for culture with or without HAT in the medium. The resulting colonies were then scored for gene correction (HAT-resistant), or lack of correction (HAT-sensitive) and examined visually for their fluorescence. (Note that the HAT selection is used only as an assay for gene correction; it is not a part of the gene-targeting process. It can be replaced by other assays, such as PCR or Southern blotting.) Fig. 2 shows that the vast majority (1.87/1.89 = 99%) of the 1.89% of input cells that fluoresced green at day 1 give rise to nonfluorescing colonies when expanded (open bars in the figure), indicating that they expressed GFP only transiently (outcome i). The percentage of green-fluorescing cells decreased >10-fold over days 2 through 4 as transient expression decreased, so that, by day 5, and stably thereafter, only 0.05% of the input cells fluoresced green. Transient expression is unlikely to persist for 5 days, yet we found that 20% of the green-fluorescing cells collected at days 5 and 6 gave nonfluorescing colonies. We interpret these colonies as representing a small background percentage (0.01%) of unmodified cells mis-sorted as green-fluorescing during the initial FACS. This interpretation is supported by the results of Southern blotting DNA isolated from several of the nonfluorescing colonies; the DNA contained no detectable vector sequences (data not shown).
The percentage of sorted cells that gave rise to green-fluorescing colonies increased as integration of the GFP gene into the genome increased (gray plus black bars in Fig. 2) from 0.017% at day 1 to 0.037% at day 4 and thereafter. At day 1, all of the green-fluorescing colonies were HAT-sensitive (gray bars), showing that they are nonhomologous integrants (outcome ii). At day 4, 5% of the green-fluorescing colonies (derived from 0.0025% of the input cells) were HAT-resistant (solid black bars in Fig. 2), showing that the mutation had been corrected in these cells (outcome iii). By day 5, when the percentage of transiently expressing cells had decreased to an insignificant level and the proportion of cells in the three categories had stabilized, we find that 1 in 20 (5%) of the sorted green-fluorescing cells are gene-corrected; 75% are nonhomologous GFP integrants; and 20% are unmodified cells.
Isolation of Gene-Corrected Stem Cells by Multichannel-FACS Sorting. A 4-fold improvement in the efficiency of discarding noncorrected cells with the vector shown in Fig. 1 A was achieved by using red and cyan fluorescence in addition to green together with the prior gating to exclude feeder cells, debris and cell doublets (Fig. 3A). The addition of red fluorescence enables the detection and therefore exclusion of a quite minor but nonetheless significant proportion of feeder and stem cells with background red and green autofluorescence. These cells give a signal that encroaches on the green channel even in the absence of any vector DNA; they can be eliminated by excluding cells that fluoresce within the dashed polygon in Fig. 3B. (Square bracket no. 1 in the figure indicates autofluorescing feeder cells; square bracket no. 2 indicates autofluorescing stem cells.) The addition of cyan fluorescence allows exclusion of cells that have nonhomologously incorporated cyan-coding sequences into their genomes, alone or accompanied with green-coding sequences. The net result of using three fluorescent channels is that 1 in 5 (20%) of the collected cells are now gene-corrected instead of 1 in 20 when using green only.
Table 1 summarizes quantitative data obtained from several gene-correcting experiments with the green-positive, cyan-negative vector shown in Fig. 3 A and B Lower. When no sorting is used, <0.01% (0.0079 ± 0.0008%) of the electroporated cells are HAT-resistant (HATR) after 7 days in culture. When the cells are sorted positively for green fluorescence, ≈5% (4.9 ± 0.8%) are HATR. Sorting positively for green fluorescence and negatively for cyan and red fluorescence increases this to ≈20% (19 ± 2%) (1 in 5). Recovery after sorting is ≈20% (21 ± 3%), as judged by the number of HATR cells in the sorted populations (61 or 66) compared with the number in the unsorted population (316) after 7 days. Enrichment [(% of gene-corrected cells after sorting) ÷ (% of gene-corrected cells in the input)] is >2,000.
Table 1. Effects of sorting when using a green-positive, cyan-negative vector.
| Sorting colors
|
|||||
|---|---|---|---|---|---|
| Input cells | Green+ | Green+, cyan-, red- | Green+, cyan+, red- | Green-, cyan+, red- | |
| No. of cells | 2 × 107 | 5951 ± 177* | 1655 ± 48* | 1660 ± 84* | 1066 ± 82* |
| No. of colonies | 4 × 106 | 1239 ± 47 | 347 ± 24 | 315 ± 31 | 192 ± 14 |
| No. of HATR colonies | 316 ± 19 | 61 ± 13 | 66 ± 8 | 0 | 0 |
| Percentage that are HATR | 0.0079 ± 0.0008 | 4.9 ± 0.8 | 19 ± 2 | <0.1 | 0 |
| Percentage recovery | — | 19 ± 3† | 21 ± 3† | — | — |
| Fold enrichment of gene-corrected cells | — | 620 ± 56‡ | 2433 ± 401‡ | — | — |
Cell numbers are adjusted to an input of 2 × 107 cells per experiment and are presented as the mean of four experiments ± standard deviation
Percentage recovery = (no. of HATR colonies after sorting × 100) ÷ (no. of HATR colonies in the input cells)
Fold enrichment of gene-corrected cells = (% of HATR colonies after sorting) ÷ (% of HATR colonies in the input cells)
A further improvement was achieved by interchanging the GFP and CFP genes in the correcting DNA vector, as illustrated in Fig. 3C, with the result that now one in three of the sorted cyan-fluorescing cells is gene corrected. This improvement is largely because autofluorescence does not encroach on the cyan channel. Accordingly, switching the positive fluorescence to cyan and the negative to green eliminates the need to gate with red fluorescence to remove autofluorescing cells. This cyan-positive, green-negative vector is the one that we now use.
An obvious future improvement would flank the positive fluorescent gene with lox P sites to allow for its removal in gene-corrected stem cells. We also note that, if uncorrected stem cells are acceptable in the sorted cell population, gene-corrected cells can be isolated with no decrease in recovery by sorting after as few as 3 days in culture. This reduction in time is an important benefit with stem cells (such as HSCs) that are difficult to maintain undifferentiated in culture. An important feature of our method is that it allows the separation of individual cells into 384-well plates (as illustrated in Fig. 1B) for use with stem cells that can be clonally expanded. Correctly targeted clones can then be identified by PCR or other means, and pooled or further expanded. This cloning enables the recovery of stem cells free from any nonhomologous integrants. If feeder cells are required during the clonal expansion, they are easily removed by re-sorting.
General Considerations. Recent successes in isolating ES cells from human blastocysts or generating them from adult cells by nuclear transfer has catalyzed a tremendous surge in efforts to adapt these versatile cells for treating human diseases. Two recent experiments with partially differentiated ES cells illustrate some of the possibilities. In one experiment, embryoid bodies generated in vitro from cloned human ES cells were able to provide cells that acted as pacemakers after the embryoid bodies were injected into the left ventricle of pig hearts in which the atrioventricular bundle of His had been ablated (22). The recipient animals were treated with immunosupressive drugs to prevent immune rejection. In a different experiment, putative endodermal precursors generated in vitro from cloned mouse ES cells enabled the survival of factor IX-deficient hemophilic mice after successful engraftment after injection into the liver parenchyma (23). Immune reactions were not observed despite a complete MHC barrier, raising the interesting possibility that the partially differentiated ES cells induced immune tolerance. Mesenchymal stem cells (MSC) derived from adult bone marrow may also have properties enabling them to avoid allogeneic rejection (24). Nevertheless, the most certain way of avoiding histocompatibility problems with stem cells or cells derived from them is to use autologous grafts.
The fluorescence-based gene correction procedure we have described here should be applicable on a small or large scale to any type of stem cell for which culture conditions exist or can be developed that permit survival of corrected cells for ≥3 days. The procedure does not use drugs and can be used with stem cells that require feeder or stromal cells for their maintenance during culture or during ex vivo expansion, because they can be efficiently removed by FACS. Our findings should encourage further efforts to use gene targeting to correct or functionally replace a mutant gene in stem cells for use in autologous gene therapy.
Supplementary Material
Acknowledgments
We thank C. R. Bagnell, Jr., S. S. Boggs, C. J. S. Edgell, M. H. Edgell, R. Givens, S. Kirby, N. Maeda, and N. N. Malouf for their help and encouragement and V. J. Madden for technical assistance. The work was supported by National Institutes of Health Grants HL-37001 and HL-71266.
Author contributions: S.H., D.C., and O.S. designed research; S.H., L.W.A., T.H., J.E.C., D.C., and O.S. performed research; S.H., L.W.A., T.H., J.E.C., D.C., and O.S. contributed new reagents/analytic tools; S.H., L.W.A., T.H., J.E.C., D.C., and O.S. analyzed data; and S.H., L.W.A., D.C., and O.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: CFP, cyan fluorescent protein; HPRT, hypoxanthine phosphoribosyltransferase; HSC, hematopoietic stem cells; HAT, hypoxanthine/aminopterin/thymidine; HATR, HAT-resistant.
References
- 1.Shizuru, J. A., Negrin, R. S. & Weissman, I. L. (2005) Annu. Rev. Med. 56, 509-538. [DOI] [PubMed] [Google Scholar]
- 2.Sauvageau, G., Iscove, N. N. & Humphries, R. K. (2004) Oncogene 23, 7223-7232. [DOI] [PubMed] [Google Scholar]
- 3.Bunting, K. D., Galipeau, J., Topham, D., Benaim, E. & Sorrentino, B. P. (1998) Blood 92, 2269-2279. [PubMed] [Google Scholar]
- 4.Antonchuk, J., Sauvageau, G. & Humphries, R. K. (2002) Cell 109, 39-45. [DOI] [PubMed] [Google Scholar]
- 5.Boggs, D. R., Boggs, S. S., Saxe, D. F., Gress, L. A. & Canfield, D. R. (1982) J. Clin. Invest. 70, 242-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benveniste, P., Cantin, C., Hyam, D. & Iscove, N. N. (2003) Nat. Immunol. 4, 708-713. [DOI] [PubMed] [Google Scholar]
- 7.Thomas, E. D., Lochte, H. L., Jr., Cannon, J. H., Sahler, O. D. & Ferrebee, J. W. (1959) J. Clin. Invest. 38, 1709-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dausset, J., Rapaport, F., Ivanyi, P. & Colombani, J. (1965) in Histocompatibility Testing, eds. Balner, H., Cleton, F. & Eernisse, J. (Munksgaard, Copen-hagen), pp. 63-78.
- 9.Iannone, R., Ohene-Frempong, K., Fuchs, E. J., Casella, J. F. & Chen, A. R. (2005) Pediatr. Blood Cancer 44, 436-440. [DOI] [PubMed] [Google Scholar]
- 10.Cavazzana-Calvo, M., Hacein-Bey, S., de Saint Basile, G., Gross, F., Yvon, E., Nusbaum, P., Selz, F., Hue, C., Certain, S., Casanova, J. L., et al. (2000) Science 288, 669-672. [DOI] [PubMed] [Google Scholar]
- 11.Aiuti, A., Slavin, S., Aker, M., Ficara, F., Deola, S., Mortellaro, A., Morecki, S., Andolfi, G., Tabucchi, A., Carlucci, F., et al. (2002) Science 296, 2410-2413. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina, S., Von Kalle, C., Schmidt, M., McCormack, M. P., Wulffraat, N., Leboulch, P., Lim, A., Osborne, C. S., Pawliuk, R., Morillon, E., et al. (2003) Science 302, 415-419. [DOI] [PubMed] [Google Scholar]
- 13.Smithies, O., Gregg, R. G., Boggs, S. S., Koralewski, M. A. & Kucherlapati, R. S. (1985) Nature 317, 230-234. [DOI] [PubMed] [Google Scholar]
- 14.Thomas, K. R., Folger, K. R. & Capecchi, M. R. (1986) Cell 44, 419-428. [DOI] [PubMed] [Google Scholar]
- 15.Hatada, S., Nikkuni, K., Bentley, S. A., Kirby, S. & Smithies, O. (2000) Proc. Natl. Acad. Sci. USA 97, 13807-13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansour, S. L., Thomas, K. R. & Capecchi, M. R. (1988) Nature 336, 348-352. [DOI] [PubMed] [Google Scholar]
- 17.Bronson, S. K., Plaehn, E. G., Kluckman, K. D., Hagaman, J. R., Maeda, N. & Smithies, O. (1996) Proc. Natl. Acad. Sci. USA 93, 9067-9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leavitt, J., Gunning, P., Porreca, P., Ng, S. Y., Lin, C. S. & Kedes, L. (1984) Mol. Cell. Biol. 4, 1961-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frederickson, R. M., Micheau, M. R., Iwamoto, A. & Miyamoto, N. G. (1989) Nucleic Acids Res. 17, 253-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beavis, A. J. & Kalejta, R. F. (1999) Cytometry 37, 68-73. [DOI] [PubMed] [Google Scholar]
- 21.Hooper, M., Hardy, K., Handyside, A., Hunter, S. & Monk, M. (1987) Nature 326, 292-295. [DOI] [PubMed] [Google Scholar]
- 22.Kehat, I., Khimovich, L., Caspi, O., Gepstein, A., Shofti, R., Arbel, G., Huber, I., Satin, J., Itskovitz-Eldor, J. & Gepstein, L. (2004) Nat. Biotechnol. 22, 1282-1289. [DOI] [PubMed] [Google Scholar]
- 23.Fair, J. H., Cairns, B. A., Lapaglia, M. A., Caballero, M., Pleasant, W. A., Hatada, S., Kim, H. S., Gui, T., Pevny, L., Meyer, A. A., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 2958-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan, J. M., Barry, F. P., Murphy, J. M. & Mahon, B. P. (2005) J. Inflamm. (London) 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.