Abstract
Fifty-four epidemiologically unrelated multidrug-resistant Salmonella enterica serovar Typhimurium isolates, collected between 1992 and 2000 in Italy, were analyzed for the presence of integrons. Strains were also tested for Salmonella genomic island 1 (SGI1), carrying antibiotic resistance genes in DT104 strains. A complete SGI1 was found in the majority of the DT104 strains. Two DT104 strains, showing resistance to streptomycin-spectinomycin and sulfonamides, carried a partially deleted SGI1 lacking the flost, tetR, and tetA genes, conferring chloramphenicol-florfenicol and tetracycline resistance, and the integron harboring the pse-1 gene cassette, conferring ampicillin resistance. The presence of SGI1 was also observed in serovar Typhimurium strains belonging to other phage types, suggesting either the potential mobility of this genomic island or changes in the phage-related phenotype of DT104 strains.
Salmonella enterica is one of the most common pathogens causing food-borne infections in Italy (10). The epidemic strain serovar Typhimurium definitive phage type 104 (DT104) has been identified as a major cause of salmonellosis in humans and animals, in both Europe (13, 21, 32) and the United States (1, 12, 30).
The majority of the DT104 isolates are characterized by resistance to six drugs: ampicillin (A), chloramphenicol (C), streptomycin (S), spectinomycin (Sp), sulfonamides (Su), and tetracycline (T) (23, 31). Between 80 and 90% of isolates of DT104 show this resistance type (33).
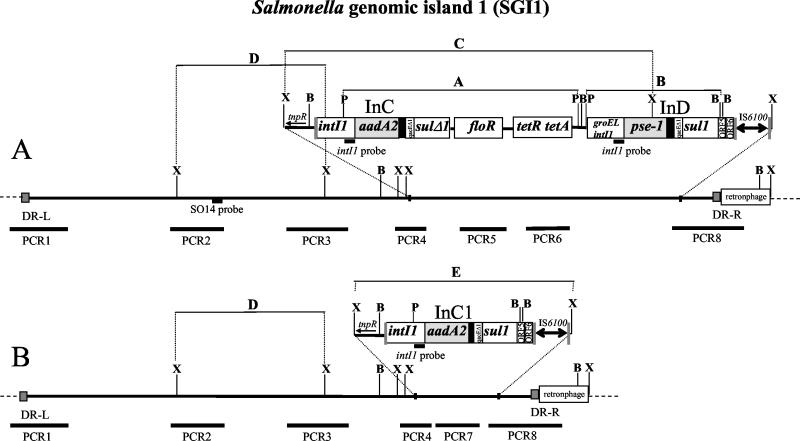
The resistance mechanism in ACSSpSuT-type DT104 has been recently elucidated, and resistance genes have been described, all located within the same chromosomal locus, designated Salmonella genomic island 1 (SGI1) (2, 3). SGI1 is a 43-kb genomic island, showing 44 coding sequences, most of them encoding hypothetical proteins. SGI1 shows two 18-bp direct repeats at the external boundaries, which strongly supports the hypothesis that site-specific recombination events may have driven the insertion of the island within the serovar Typhimurium chromosome (2, 3).
The ACSSpSuT resistance genes have been identified within a 14-kb region in SGI1, carried by two class 1 integrons, InC and InD, encoding the aminoglycoside resistance gene aadA2 and the β-lactamase pse-1 gene, respectively (4). The intervening region, encompassing the two integrons, contains the floR gene, conferring resistance to florfenicol-chloramphenicol, and the tetR and tetA (class G) genes, conferring tetracycline resistance (2, 3, 4). InC and InD show a peculiar structure with respect to other class 1 integrons (19, 20, 29). InC carries the qacEΔ1 gene, conferring resistance to disinfectants, and a truncated, nonfunctional sul1 gene in the 3′-conserved segment (3′-CS). InD shows a deletion of the integrase gene (intI1) which is linked to the groEL gene in the 5′-conserved segment (5′-CS) (2, 3, 4); the qacEΔ1 gene, the sul1 gene, conferring sulfonamide resistance, and the two open reading frames of unknown function, ORF5 and ORF6, are contained in the 3′-CS followed by the IS6100 element (20). The multidrug resistance region in SGI1 is bounded by characteristic 25-bp inverted repeats and flanked by a direct duplication of 5 bp of the target sequence, indicating that it was inserted by a transpositional mechanism (EMBL accession no. AF261825) (19, 20).
The aim of our study was to analyze the molecular basis of antibiotic resistance by searching for, identifying, and characterizing integrons and other resistance genes in multidrug-resistant serovar Typhimurium isolated in Italy from humans and food animals. Strains were also analyzed for the presence and conservation of SGI1.
MATERIALS AND METHODS
Bacterial strains.
Fifty-four apparently epidemiologically unrelated serovar Typhimurium isolates of human and animal origin, representative of frequent phage types and recurrent multidrug resistance profiles in Italy (10), were chosen from the collections of the Istituto Zooprofilattico delle Venezie (Padua, Italy) and of the Istituto Superiore di Sanità (Rome, Italy). Serotypes were determined with anti-O and anti-H antisera obtained from Behringwerke AG (Marburg, Germany). Phage types were determined according to the work of Callow (5), and antibiotic resistance was determined by the disk diffusion assay on Mueller-Hinton agar with commercial antimicrobial susceptibility disks (Oxoid, Basingstoke, United Kingdom; Becton Dickinson Microbiological Systems, Cockeysville, Md.), according to the recommendations of the National Committee for Clinical Laboratory Standards (18). The following antibiotics were tested: ampicillin, ceftazidime, chloramphenicol, streptomycin, spectinomycin, sulfonamides, tetracycline, trimethoprim (Tp), kanamycin (K), gentamicin (G), tobramycin (N), amikacin, nalidixic acid, and ciprofloxacin. Strain ST30 was assigned to PT (phage type) U302 by John E. Threlfall (Public Health Laboratory Service, Colindale, London, United Kingdom).
Preparation of total DNA and Southern blot hybridization.
Small-scale DNA preparations were made with 3 ml of bacterial liquid cultures grown overnight, as described by Ezaki and colleagues (9). Four micrograms of total DNA was digested with PvuII-BamHI or XbaI restriction enzymes. Restricted fragments were separated by 1% agarose gel electrophoresis and transferred onto positively charged nylon membranes (Roche Diagnostics, Monza, Italy) by standard methods (24). Southern blot hybridization was carried out under high-stringency conditions (24). A specific probe for the intI1 gene was obtained as previously described (6). The S014 and the PCR8 probes were obtained by PCR amplification with S014-FW-S014-RV and S044-FW-DR-RV primer pairs, respectively (Table 1). DNA probes were [α-32P]dCTP labeled with a random priming kit (Life Technologies, Milan, Italy).
TABLE 1.
Primers used in this study
| Primer | DNA sequence | Amplicon in Fig. 1 | EMBL accession no., nucleotide positions |
|---|---|---|---|
| DR-FW | 5′-GGGCAAAGCGCAGCTATTAG-3′ | AF261825, 13-32 | |
| S004-RV | 5′-CCCGCAGGGTAAGTAATG-3′ | PCR1 | AF261825, 3275-3258 |
| S011-FW | 5′-CGCCGGCTCCAAAGGAAATGG-3′ | AF261825, 11764-11784 | |
| S014-RV | 5′-AATTTCCTCATCGTCTAGC-3′ | PCR2 | AF261825, 14859-14842 |
| S014-FW | 5′-AGTCTGTGGCATGAAGAA C-3′ | AF261825, 14521-14539 | |
| S022-FW | 5′-CGCTGCAAGCACAATGATGA-3′ | AF261825, 18301-18320 | |
| S024-RV | 5′-GGTACGGTATCGCCTAAGTG-3′ | PCR3 | AF261825, 21930-21911 |
| S026-FW | 5′-TCGGGTAATCTCAGCAGAGC-3′ | AF261825, 25021-25040 | |
| int-RV | 5′-GGGCATGGTGGCTGAAGGACC-3′ | PCR4 | AF261825, 27266-27246 |
| 5′-CS | 5′-GGCATCCAAGCAGCAAG-3′ | AF261825, 27892-27908 | |
| 3′-CS | 5′-AAGCAGACTTGACCTGA-3′ | AF261825, 28900-28884 | |
| flo-FW | 5′-ATGACCACCACACGCCCCG-3′ | AF261825, 30482-30500 | |
| flo-RV | 5′-CTAGACGACTGGCGACTTC-3′ | PCR5 | AF261825, 31696-31678 |
| tetR-FW | 5′-CTGCTGATCGTGGGTCT-3′ | AF261825, 32657-32673 | |
| tetA-RV | 5′-TTGCGAATGGTCTGCGT-3′ | PCR6 | AF261825, 333797-33781 |
| aadA2-FW | 5′-GAGCGCCATCTGGAATCAACG-3′ | AF261825, 28058-28078 | |
| ORF5-RV | 5′-CCGAACGTTCGGAGGCTCCT-3′ | PCR7 | AF261825, 39750-39731 |
| S044-FW | 5′-ACCAGAGAGAGTTATCGAGC-3′ | AF261825, 41911-41930 | |
| DR-RV | 5′-CACGAAAAGGAGACGATGAGA-3′ | PCR8 | AF261825, 44483-44463 |
| strAF | 5′-AGCAGAGCGCGCCTTCGCTG-3′ | NC_001740.1, 761-780 | |
| strAR | 5′-CCAAAGCCCACTTCACCGAC-3′ | NC_001740.1, 1464-1445 |
PCR amplification, cloning, and sequencing.
Standard PCR amplifications were performed with primers listed in Table 1 and 2.5 U of Taq DNA polymerase (Roche Diagnostics), according to the manufacturer's recommendations. All PCR amplifications were run at 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min, for a total of 30 cycles. Amplification products were sequenced (26) with fluorescent dye-labeled dideoxynucleotides and a 373 automatic DNA sequencer (Perkin-Elmer, Foster City, Calif.). Comparative analysis of nucleotide sequences was performed by the advanced BLAST search program 2.0 within the QBLAST system at the National Center for Biotechnology Information site (www.ncbi.nlm.nih.gov/blast/).
Pulsed-field gel electrophoresis (PFGE).
Preparation of total DNA was performed as described by R. K. Gautom (11) except that 0.5 mg of proteinase K per ml was dissolved in the bacterial suspension before addition of 1% melted agarose containing 1% sodium dodecyl sulfate. DNA plugs were digested with XbaI (Roche Diagnostics). Electrophoresis was performed in a CHEF-DRII electrophoresis system (Bio-Rad Laboratories) at 14°C with 0.5× Tris-borate-EDTA running buffer and 1% pulsed-field certified agarose (Bio-Rad Laboratories). Electrophoresis conditions were as follows: phase 1, initial switch time, 0.5 s; final switch time, 60 s for 24 h, with 6 V cm−1. For visualization, gels were stained in ethidium bromide, destained in water, and photographed.
RESULTS
Detection of integrons in multidrug-resistant serovar Typhimurium isolated in Italy.
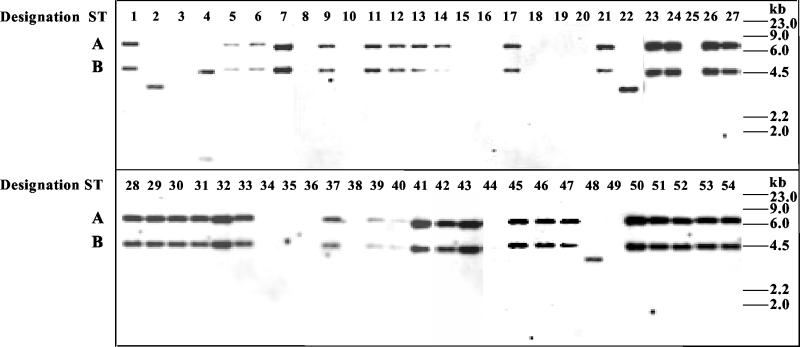
Forty-one isolates of DT104, 33 of resistance type ACSSpSuT and 8 with different resistance profiles, and 13 resistant isolates of other phage types (Table 2) were analyzed for the presence of class 1 integrons. The integron search was performed by Southern blot hybridization with the intI1 gene probe (6), on total DNA digested with PvuII-BamHI restriction enzymes. This hybridization is expected to produce, in DT104 strains carrying SGI1, two integrase-positive restriction fragments of 7,738 bp (band A) and 4,402 bp (band B) (Fig. 1A). Band A contains InC and the floR, tetA, and tetR resistance genes, from the PvuII site at position 27276 to the PvuII site at position 35014 (EMBL accession no. AF261825). Band B contains InD from the PvuII site at position 35546 to the BamHI site at position 39948 in the SGI1 DNA sequence (Fig. 1A). As expected, all ACSSpSuT DT104 strains showed both bands, indicating that the SGI1 region containing the two integrons was well conserved (Fig. 2). Interestingly, the A and B bands were also observed in a strain belonging to DT1 (ST9) and in a PT U302 strain (ST30), both showing the ACSSpSuT resistance profile (Fig. 2).
TABLE 2.
Characteristics of the 54 serovar Typhimurium isolates tested in this studya
| Source, yr | No. | Phage type | Antibiotic resistance | Integron-borne gene cassette(s) | strA gene | SGI1 | Designation(s) |
|---|---|---|---|---|---|---|---|
| Human, 1992 | 2 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST7, ST41 |
| Human, 1993 | 4 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST23, ST24, ST26, ST28 |
| Human, 1994 | 3 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST29, ST32, ST33 |
| Human, 1994 | 1 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | SGI1 partially deleted | ST37 |
| Animal, 1994 | 2 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST27, ST31 |
| Human, 1995 | 3 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST11, ST39, ST40 |
| Human, 1996 | 6 | DT104 | ACSSpSuT | aadA2, pse-1 | Pos | Pos | ST5, ST12, ST13, ST14, ST17, ST21 |
| Human, 1997 | 1 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST1 |
| Animal, 1998 | 1 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST6 |
| Human, 1999 | 1 | DT104 | ACSSpSuT | aadA2, pse-1 | Pos | Pos | ST43 |
| Animal, 1999 | 3 | DT104 | ACSSpSuT | aadA2, pse-1 | Pos | Pos | ST42, ST47, ST52 |
| Human, 2000 | 2 | DT104 | ACSSpSuT | aadA2, pse-1 | Pos | Pos | ST45, ST46 |
| Animal, 2000 | 4 | DT104 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST50, ST51, ST53, ST54 |
| Animal, 1999 | 1 | DT104 | ASSpSuT | aadA1 | Pos | Neg | ST48 |
| Human, 1997 | 1 | DT104 | SSpSu | aadA2 | Pos | SGI1 partially deleted | ST2 |
| Animal, 1998 | 1 | DT104 | SSpSu | aadA2 | Pos | SGI1 partially deleted | ST22 |
| Human, 1994 | 1 | DT104 | ASSuT | Neg | Pos | Neg | ST38 |
| Animal, 1999 | 2 | DT104 | ASSuT | Neg | Pos | Neg | ST44, ST49 |
| Human, 1995 | 1 | DT104 | AST | Neg | Pos | Neg | ST10 |
| Animal, 1993 | 1 | DT104 | SSuTp | Neg | Pos | Neg | ST25 |
| Animal, 1994 | 1 | U302 | ACSSpSuT | aadA2, pse-1 | Neg | Pos | ST30 |
| Human, 1995 | 1 | DT1 | ACSSpSuT | aadA2, pse-1 | Pos | Pos | ST9 |
| Human, 1997 | 1 | RDNC | ASSpSuGKTp | aadB, oxaI-aadAI | Neg | Neg | ST4 |
| Human, 1995 | 1 | DT1 | S | Neg | Neg | Neg | ST8 |
| Human, 1996 | 1 | DT12 | ASSuTTp | Neg | Pos | Neg | ST15 |
| Human, 1994 | 1 | DT179 | AST | Neg | Pos | Neg | ST35 |
| Human, 1994 | 1 | DT194 | AST | Neg | Pos | Neg | ST34 |
| Human, 1996 | 1 | DT194 | AST | Neg | Pos | Neg | ST20 |
| Human, 1997 | 1 | DT208 | ASSuT | Neg | Pos | Neg | ST3 |
| Human, 1996 | 1 | DT208 | AST | Neg | Pos | Neg | ST19 |
| Human, 1994 | 1 | DT208 | ASTTp | Neg | Pos | Neg | ST36 |
| Human, 1996 | 1 | UT | AST | Neg | Pos | Neg | ST18 |
| Human, 1996 | 1 | UT | SGNK | Neg | Pos | Neg | ST16 |
RDNC, reacts but does not conform to any standard phage type; UT, phage untypeable; Neg, negative; Pos, positive.
FIG. 1.
Schematic representation of the entire and partially deleted SGI1. (A) XbaI (X), BamHI (B), and PvuII (P) restriction map was deduced from the DT104 DNA sequence released under EMBL accession no. AF261825. (B) Restriction map of the partially deleted SGI1 was experimentally determined. White boxes represent intI1, qacEΔ1, sul1, floR, tetR, tetA, ORF5, and ORF6 genes. The IS6100 position is indicated between two arrows. The shaded boxes represent gene cassettes inserted into the InC and InD integrons. Black boxes are the 59-base element sites, and DR-R and DR-L boxes represent the direct repeat junctions of SGI1 in the Salmonella chromosome (dotted lines). Thin bars represent Southern blot hybridization bands A, B, C, and D. Heavy lines represent PCR amplicons (PCR1 to PCR8). The positions of the intI1 gene and S014 probes are indicated.
FIG. 2.
Southern blot hybridization of serovar Typhimurium strains by the integrase gene. Total DNAs restricted with BamHI-PvuII were separated by 1% agarose gel electrophoresis, blotted onto nylon membranes, and hybridized with an intI1 gene-specific probe (6). Numbers above lanes indicate the designation number for each strain. The positions of bands A and B are indicated. Molecular size standards are shown on the right.
Three DT104 strains showing resistance types SSpSu (ST2 and ST22) and ASSpSuT (ST48) were positive for a PvuII-BamHI fragment of approximately 3,700 bp (Fig. 2). This band indicates the presence of an integron located within a different genetic structure from that of the DT104 SGI1. Integron-positive plasmids were not found in these strains, suggesting that integrons carried by ST2, ST22, and ST48 are located on the bacterial chromosome.
Strain ST4, showing the ASSpSuGKTp resistance type, was positive for two bands of approximately 4,300 and 800 bp, suggesting the presence of two integrons. All the other serovar Typhimurium strains were negative for the presence of integrons (Fig. 2).
Antibiotic resistance genes.
Integron-borne gene cassettes were amplified with the 5′-CS and 3′-CS primer pair (16). All DT104 strains showing the ACSSpSuT resistance profile produced two PCR products of 1,008 and 1,133 bp. By DNA sequencing, these bands corresponded to the well-characterized aadA2 and pse-1 gene cassettes of InC and InD as previously described (Table 2) (25). The same gene cassettes were amplified and sequenced from strains ST9 (DT1) and ST30 (U302).
Strains ST2, ST22, and ST48 produced an amplicon of about 1,000 bp. These amplicons were fully sequenced. The nucleotide sequence of the ST48 integron revealed the presence of the aadA1 gene cassette, encoding streptomycin-spectinomycin resistance. This gene cassette was 99.9% identical, without amino acid changes, to the aadA1 gene cassette identified within the In2 integron carried by the Tn21 transposon (EMBL accession no. AF071413) (17).
Nucleotide sequences of amplicons obtained from both ST2 and ST22 strains showed the presence of the aadA2 gene cassette, encoding streptomycin-spectinomycin resistance. These gene cassettes were 100% (ST2) and 99.9% (ST22) identical to the aadA2 gene cassette carried by InC in SGI1 (EMBL accession no. AF261825). In the ST22 DNA sequence, a single point mutation at position 28211 was identified, resulting in an amino acid change (from glutamic acid to lysine) in the deduced protein sequence. The same substitution was previously described for the DT104 strain H3380 (EMBL accession no. AF071555) (4).
Two PCR products of 2,000 and 850 bp were obtained from strain ST4. The DNA sequence of the 850-bp amplicon revealed the presence of the aadB gene cassette, conferring resistance to kanamycin and gentamicin, while the DNA sequence of the 2,000-bp amplicon revealed the integron-borne oxa1 and aadA1 gene cassettes, conferring ampicillin and streptomycin-spectinomycin resistance, respectively.
Serovar Typhimurium strains were then tested for strA, a streptomycin resistance gene that is an alternative to the integron-borne aadA gene cassette (28). Positive PCR amplifications with the strAF-strAR primer pair (Table 1) were obtained from 21 of the 54 strains (Table 2), including the integron-negative DT104 strains ST25, ST38, ST10, ST44, and ST49 and the ampicillin-, tetracycline-, and chloramphenicol-susceptible DT104 strains ST2, ST22, and ST48.
Analysis of SGI1.
In order to gather more information on the presence and conservation of SGI1 in the DT104 strains, Southern blot hybridization and PCR amplification experiments were performed.
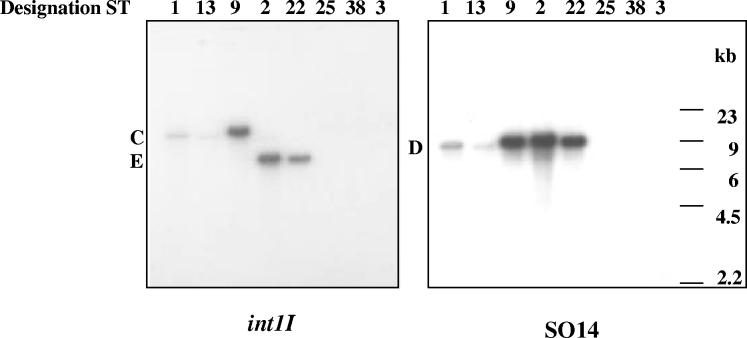
SGI1 was initially sought by using Southern blot hybridization with the intI1 probe on total DNA restricted with XbaI, in DT104 strains ST2 and ST22 lacking the A and B PvuII-BamHI bands, and in the ST9 strain (DT1), which tested positive for the presence of SGI1. ST1 and ST13 were used as positive controls and ST25, ST38, and ST3 were used as negative controls in these experiments.
As shown in Fig. 3, ST1 and ST13 produced an integrase-positive band of 11,758 bp (band C in Fig. 1A), as expected from the XbaI sites located at positions 26038 and 37796 in the SGI1 DNA sequence. This band was also observed in ST9, while ST2 and ST22 showed a different band of approximately 7,000 bp (band E in Fig. 3). As expected, band C was missing in the three strains lacking integrons (ST25, ST38, and ST3).
FIG. 3.
Southern blot hybridization of the SGI1 island. Total DNAs restricted with XbaI were separated by 0.8% agarose gel electrophoresis, blotted onto nylon membranes, and hybridized with the intI1 gene probe and with the S014 probe. Numbers above lanes indicate the designation number for each strain. The positions of bands C, D, and E are indicated. Molecular size standards are shown on the right of the figure.
The XbaI Southern blot was then hybridized with the S014 probe, located in the left arm of SGI1. This probe recognized the 8,921-bp XbaI restriction fragment (band D in Fig. 1A) in both ST2 and ST22 as well as in ST1, ST13, and ST9 (Fig. 3), revealing that this region of SGI1 was well conserved in these strains. Finally, ST2, ST22, and ST1 BamHI-restricted DNAs were hybridized with the PCR8 amplification product as probe (Fig. 1). This probe recognized in both ST2 and ST22 as well as the ST1 positive control strain the 4,699-bp BamHI restriction fragment containing ORF5, ORF6, IS6100 of InD, and part of the retron phage (data not shown).
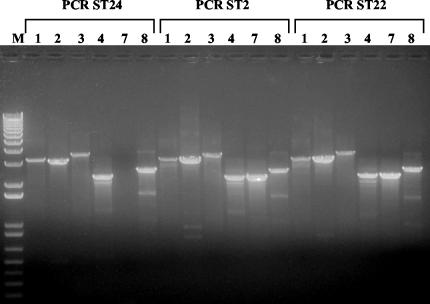
To fully characterize the conservation and structure of SGI1 in ST2 and ST22, PCR was performed with primer pairs listed in Table 1 (PCR amplifications relative to the SGI1 map are illustrated in Fig. 1). PCR experiments performed on the left (PCR1) and right (PCR8) direct repeat junctions of SGI1 and on other SGI1 regions (PCR2, PCR3, and PCR4) demonstrated that ST2 and ST22 produced indistinguishable bands with respect to the ST24 control strain (Fig. 4). In particular, PCR4, obtained with primers S026-FW and int-RV, demonstrated that integrons in ST2 and ST22 were located within SGI1, linked to the S026 DNA sequence. However, PCR amplifications with flo-FW-flo-RV and tetA-FW-tetR-RV primer pairs (PCR5 and PCR6) were negative in ST2 and ST22, demonstrating that these strains do not carry the floR and the tetA and tetR genes, respectively (data not shown). Finally, ST2 and ST22 produced a PCR product of approximately 2,000 bp (Fig. 4) with the aadA2-FW-ORF5-RV primer pairs designed on the aadA2 gene cassette and on the ORF5 DNA sequence, respectively (PCR7, Fig. 1B). This amplicon is about 10 kb shorter than the expected amplicon for complete SGI1 (ca. 12 kb). Southern blot hybridization and PCR amplification results demonstrate that ST2 and ST22 harbor only one of the two integrons of SGI1; this integron (InC1 in Fig. 1) carries the same aadA2 gene cassette of InC and shows a 3′-CS identical to that of InD, carrying qacEΔ1, conferring resistance to disinfectants; the entire sul1 gene, conferring sulfonamide resistance; ORF5; ORF6; and the IS6100 element (Fig. 1B).
FIG. 4.
PCR analysis of SGI1. PCR1 to PCR8 were performed on the ST2 and ST22 strains. Strain ST24 was used as DT104 positive control. The amplification product PCR7 (ca. 12 kb) was not obtained from the ST24 DT104 control strains, since no long-run PCR conditions were used in these experiments. A 1-kb marker (KiloBase DNA marker; Pharmacia Biotech, Milan, Italy) was used as a standard.
PCR amplification (PCR1 to PCR8) was then performed on the remaining isolates, demonstrating that, with the exception of strain ST37, all ACSSpSuT DT104 strains carried an intact SGI1 (Table 2). Strain ST37 was negative for PCR1 and PCR2 but positive for the other PCR amplifications performed on SGI1, suggesting the partial deletion of the left arm of the genomic island.
An intact SGI1 was identified in ST9 (DT1) and in ST30 (U302), while it was completely absent in ST48 (DT104), which harbors the aadA1 integron-borne gene cassette.
PFGE of macrorestricted genomic DNA.
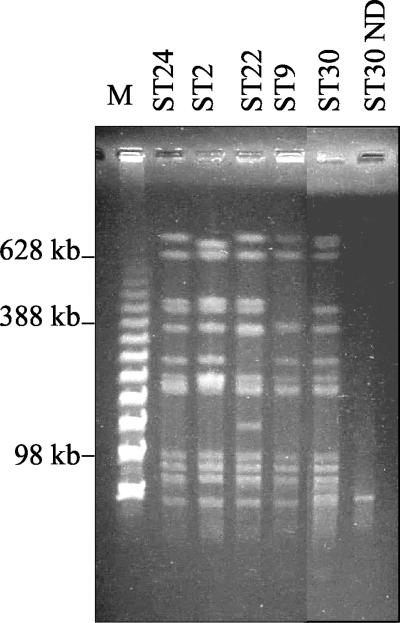
Strains ST2 (DT104), ST22 (DT104), ST9 (DT1), and ST30 (U302) were analyzed by PFGE following digestion with XbaI. Strain ST24 was used as the DT104 reference control strain. The results showed that the three DT104 strains were highly related at the chromosomal level. ST30 (U302) showed a DT104-related PFGE pattern differing by three bands (Fig. 5). PT U302 has been previously shown to be closely related to DT104, and phage conversion of DT104 to U302 has been observed, possibly by the acquisition of plasmids (15, 34). The presence of a plasmid in ST30 was revealed by PFGE of undigested DNA (Fig. 5), suggesting that this plasmid could have caused a DT104-U302 phage conversion in this strain. In contrast, ST9 (DT1) was divergent at the chromosomal level from DT104, differing by at least four bands in the PFGE pattern (Fig. 5). In this case the presence of the DT104 resistance traits could be explained by the horizontal transfer of SGI1.
FIG. 5.
Analysis by PFGE. Serovar Typhimurium DNAs were digested by XbaI and separated by PFGE on a 1% agarose gel. Designation numbers of tested strains are reported above each lane. Lane M, the molecular size lambda ladder ranged from 48.5 to 1,000 kb (Roche Diagnostics). ND, not digested.
DISCUSSION
Several studies have focused on resistance genes in DT104 strains, and new molecular methods for detection and analysis of the DT104 clone have been proposed elsewhere (8, 14). Most of these methods are based on PCR amplification of DT104-related traits, such as the 1.0- and 1.2-kb integrons or the floR resistance gene located within the resistance gene cluster (22).
More recently, the cloning and sequencing of the entire genomic island from multidrug-resistant DT104 have opened new possibilities for investigating genetic characteristics of this important and widely diffused Salmonella clone. However, complete information is not available for the conservation of SGI1 among strains circulating in animals and humans, and the frequency of transfer of the genomic island to serovar Typhimurium strains of other phage types has not been quantified. This information could be important to understanding the origin and evolution of the genomic island and potential horizontal mobility of SGI1.
To study antibiotic resistance genes carried by integrons and to investigate the presence of SGI1, multidrug-resistant serovar Typhimurium strains of different phage types of both human and animal origin were analyzed by both PCR amplification and Southern blot hybridization. All DT104 strains showing the ACSSpSuT resistance profile harbor the two integrons InC and InD, located within SGI1. Two DT104 strains, isolated in Italy in 1997 (ST2) and in 1998 (ST22) and showing the SSpSu resistance profile, revealed the presence of a partially deleted SGI1. These isolates lack the region of the island encoding resistance to chloramphenicol-florphenicol and tetracycline and also the InD integron carrying the pse-1 ampicillin resistance gene. These two DT104 strains carried the InC1 integron, which shows a 3′-CS, including the qacEΔ1, sul1, ORF5, ORF6, and IS6100 DNA sequences. This integron was found in an XbaI integrase gene-positive band smaller than that observed in DT104 control strains.
The ST2 and ST22 resistance island may represent the precursor of the DT104 resistance gene cluster in which InC1 is the unique integron harbored by the genomic island. Our findings suggest that the assembly of the resistance gene cluster within SGI1 could be due to sequential acquisition of resistance determinants. In fact, InC could derive from InC1 by deletion of part of the 3′-CS caused by the insertion of the floR and tet genes. The second integron, InD, could also have been acquired through an independent integration event, leading to the full assembly of the resistance gene cluster. The deleted SGI1 could then represent the precursor of the genomic island before the assembly of InD, floR, and tet genes. However, the genetic structure observed in ST2 and ST22 could also be explained by homologous recombination between InC and InD in the 3′-CS. Recombination between the two sul1 genes could lead to the deletion of a 10-kb DNA region containing the floR, tet, intI1, and pse-1 genes and to the reconstruction of InC1 carrying the 3′-CS of InD.
A different deletion in the genomic island was observed in the ST37 ACSSpSuT DT104 strain, lacking the left arm of SGI1. These results demonstrate that several rearrangements could occur in SGI1, suggesting a constant and dynamic evolution of this genetic trait.
In another DT104 strain (ST48) an integron that carried the aadA1 gene cassette was found to be located in the chromosome. This strain lacked SGI1, demonstrating that integrons other than InC and InD can be acquired by DT104 strains. Most of the DT104 strains showing different resistance profiles than the ACSSpSuT resistance type lack SGI1 but contain the strA resistance gene, conferring streptomycin resistance. ST2, ST22, and ST48, as well as many DT104 strains harboring the entire SGI1, also carried the strA gene, suggesting that multiple mechanisms of resistance can be simultaneously present in these strains.
Of note, the entire SGI1 was found in the chromosome of ST9 (DT1) and in the ST30 (U302) isolate, suggesting the potential capacity of mobilization of SGI1. However, recent studies have demonstrated that strains of U302, DT120, and DT12 of the ACSSpSuT resistance type are probably derived from DT104 by a change of phage sensitivity (15). This possibility should be taken into consideration. The presence of the DT104 resistance genes has also been previously reported for several serovar Agona strains (2, 7). However, the multiresistant serovar Agona strains lacked the retron phage at the DR-R boundary of SGI1, while results with serovar Typhimurium strains ST2 and ST22 indicate that in these strains SGI1 is located between the thdf gene and the retron phage, a situation similar to that described for the multiresistant DT104 strain. There is no experimental evidence demonstrating the molecular mechanism of the SGI1 horizontal transfer among Salmonella strains, although transduction experiments with a P22-like phage demonstrated a facilitated transduction of resistance genes to a susceptible Salmonella strain (27).
Multidrug resistance in serovar Typhimurium is now a cause of great concern in both clinical and veterinary medicine. Studies of evolution and dissemination of resistance determinants may help us to better understand the origin of such strains and the transmission from food animals to humans.
Acknowledgments
We are grateful to John E. Threlfall for encouraging discussions, helpful suggestions, and characterization of the ST30 strain. We thank Paul D. Fey for critical reading of the manuscript; Sergio Arena, Susanna Mariotti, and Maria Giovanna Daga for technical assistance; and F. Riccobono for DNA sequencing.
This research was supported by grants to Alessandra Carattoli on the “Progetto Antibiotico Resistenza 2000” and to Ida Luzzi and Antonia Ricci on the “Ricerca Corrente 1999 e 2000” from the Italian Ministry of Health.
REFERENCES
- 1.Besser, T. E., M. Goldoft, L. C. Pritchett, R. Khakhria, D. D. Hankock, D. H. Rices, J. M. Gay, W. Johnson, and C. C. Gay. 2000. Multiresistant Salmonella Typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol. Infect. 124:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D. A., G. A. Peters, L. K. Ng, and M. R. Mulvery. 2000. Partial characterisation of a genomic island associated with the multidrug resistant region of Salmonella enterica Typhimurium DT104. FEMS Microb. Lett. 189:285-291. [DOI] [PubMed] [Google Scholar]
- 4.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callow, B. 1959. A new phage typing scheme for Salmonella typhimurium. J. Hyg. (Cambridge) 57:346-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli, A., L. Villa, C. Pezzella, E. Bordi, and P. Visca. 2001. Expanding drug-resistance through integron acquisition by IncFI plasmids of Salmonella enterica serotype Typhimurium. Emerg. Infect. Dis. 7:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloeckaert, A., K. S. Boumedine, G. Flaujac, H. Imberechts, I. D'Hooghe, and E. Chaslus-Dancla. 2000. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob. Agents Chemother. 44:1359-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebner, P. D., and A. G. Mathew. 2001. Three molecular methods to identify Salmonella enterica serotype Typhimurium DT104: PCR fingerprinting, multiplex PCR and rapid PFGE. FEMS Microbiol. Lett. 205:25-29. [DOI] [PubMed] [Google Scholar]
- 9.Ezaki, T., N. Takeuchi, S. L. Liu, A. Kai, H. Yamamoto, and E. Yabuuchi. 1988. Small-scale DNA preparation for rapid genetic identification of Campylobacter species without radioisotope. Microbiol. Immunol. 32:141-150. [DOI] [PubMed] [Google Scholar]
- 10.Fantasia, M., E. Filetici, S. Arena, and S. Mariotti. 1998. Serotype and phage type distribution of salmonellas from humans and non-human sources in Italy in the period 1973-1995. Eur. J. Epidemiol. 14:701-710. [DOI] [PubMed] [Google Scholar]
- 11.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glynn, M. K., C. Bopp, W. Dewitt, P. Dabney, M. Mokhtar, and F. J. Angulo. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333-1338. [DOI] [PubMed] [Google Scholar]
- 13.Hancock D., T. Besser, J. Gay, D. Rice, M. Davis, and C. Gay. 2000. The global epidemiology of multiresistant Salmonella enterica serovar Typhimurium DT104, p. 217-243. In C. Brown and C. Bolin (ed.), Emerging diseases of animals. ASM Press, Washington, D.C.
- 14.Khan, A. A., M. S. Nawaz, S. A. Khan, and C. E. Cerniglia. 2000. Detection of multidrug-resistant Salmonella typhimurium DT104 by multiplex polymerase chain reaction. FEMS Microbiol. Lett. 182:355-360. [DOI] [PubMed] [Google Scholar]
- 15.Lawson, A. J., M. U. Dassama, L. R. Ward, and E. J. Threlfall. 2002. Multiply resistant (MR) Salmonella enterica serotype Typhimurium DT 12 and DT 120: a case of MR DT 104 in disguise? Emerg. Infect. Dis. 8:434-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levesque, C., L. Pichè, C. Larose, and P. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1992. Performance standards for antimicrobial disc susceptibility tests. M2A2. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 19.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poppe, C., N. Smart, R. Khakhria, W. Johnson, J. Spika, and J. Prescot. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559-565. [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchett, L. C., M. E. Konkel, J. M. Gay, and T. E. Besser. 2000. Identification of DT104 and U302 phage types among Salmonella enterica serotype Typhimurium isolates by PCR. J. Clin. Microbiol. 38:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridley, A., and E. J. Threlfall. 1998. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT104. Microb. Drug Resist. 4:113-118. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J. E., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1997. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157:177-181. [DOI] [PubMed] [Google Scholar]
- 26.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmieger, H., and P. Schicklmaier. 1999. Transduction of multiple drug resistance of Salmonella enterica serovar Typhimurium DT104. FEMS Microbiol. Lett. 170:251-256. [DOI] [PubMed] [Google Scholar]
- 28.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 29.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 30.Tauxe, R. V. 1999. Salmonella Enteritidis and Salmonella Typhimurium DT104: successful subtypes in the modern world, p. 37-52. In W. M. Scheld, W. A. Craig, and J. M. Hughes (ed.), Emerging infections 3. ASM Press, Washington, D.C.
- 31.Threlfall, E. J., L. R. Ward, and B. Rowe. 1998. Multiresistant Salmonella typhimurium DT104 and Salmonella bacteraemia. Lancet 352:287-288. [DOI] [PubMed] [Google Scholar]
- 32.Threlfall, E. J., J. A. Frost, L. R. Ward, and B. Rowe. 1994. Epidemic in cattle and humans of Salmonella typhimurium DT 104 with chromosomally integrated multiple drug resistance. Vet. Rec. 134:577. [DOI] [PubMed] [Google Scholar]
- 33.Threlfall, E. J., J. A. Skinner, A. Graham, L. R. Ward, and H. R. Smith. 2000. Resistance to ceftriaxone and cefotaxime in non-typhoidal Salmonella enterica in England and Wales, 1998-99. J. Antimicrob. Chemother. 46:860-862. [DOI] [PubMed] [Google Scholar]
- 34.Walker, R. A., E. Lindsay, M. J. Woodward, L. R. Ward, and E. J. Threlfall. 2001. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype Typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb. Drug Resist. 7:13-21. [DOI] [PubMed] [Google Scholar]