Abstract
Background
The identification of sequence innovations in the genomes of mammals facilitates understanding of human gene function, as well as sheds light on the molecular mechanisms which underlie these changes. Although gene duplication plays a major role in genome evolution, studies regarding concerted evolution events among gene family members have been limited in scope and restricted to protein-coding regions, where high sequence similarity is easily detectable.
Results
We describe a mammalian-specific expansion of more than 20 rapidly-evolving genes on human chromosome Xq22.1. Many of these are highly divergent in their protein-coding regions yet contain a conserved sequence motif in their 5' UTRs which appears to have been maintained by multiple events of concerted evolution. These events have led to the generation of chimaeric genes, each with a 5' UTR and a protein-coding region that possess independent evolutionary histories. We suggest that concerted evolution has occurred via gene conversion independently in different mammalian lineages, and these events have resulted in elevated G+C levels in the encompassing genomic regions. These concerted evolution events occurred within and between genes from three separate protein families ('brain-expressed X-linked' [BEX], WWbp5-like X-linked [WEX] and G-protein-coupled receptor-associated sorting protein [GASP]), which often are expressed in mammalian brains and associated with receptor mediated signalling and apoptosis.
Conclusion
Despite high protein-coding divergence among mammalian-specific genes, we identified a DNA motif common to these genes' 5' UTR exons. The motif has undergone concerted evolution events independently of its neighbouring protein-coding regions, leading to formation of evolutionary chimaeric genes. These findings have implications for the identification of non protein-coding regulatory elements and their lineage-specific evolution in mammals.
Background
Discriminating mutations arising during the evolution of mammals which were selectively neutral from those which were adaptive is an important challenge for the current genomic era. In the main, beneficial mutations in mammalian genomes appear to have been gene duplication, rapid sequence divergence, and alteration in gene expression levels [1-4].
An additional lineage-specific mutational process which is also a substrate for selection is concerted evolution, via either unequal crossing-over or gene conversion [5,6]. Non-allelic gene conversion occurs during non-reciprocal homologous recombination when sequence-similar paralogues are misaligned. Converting sequences are often short, in the order of hundreds of basepairs, and when frequent and sustained can lead to the homogenisation of multigene family sequences [7], as observed for mammalian histone [8] and Hsp70 [9] genes. During gene conversion, repair at mismatched positions appears to be biased towards retention of G or C bases which leads to elevation in G+C nucleotide content [10].
Non-allelic gene conversion thus results in phylogenetic trees which display significantly greater proximity between a species' gene paralogues than for gene orthologues of a sister species [11]. Such phylogenetic relationships, however, are also indicative of lineage-specific gene duplication events. Nevertheless, when the affected genes are widely-spread on the genome these relationships are usually indicative of non-allelic gene conversion. This is because gene duplication most frequently results in tandem consecutive genes along the chromosome.
Gene conversion events are expected to occur mostly between protein-coding sequences of genes. This expectation arises from sequence conservation being, in general, highest in protein-coding regions, intermediate in untranslated regions (UTRs), and lowest within introns and intergenic regions [12]. Gene conversion thus is not expected between sequence-dissimilar and non-homologous genes.
Here, we describe genes whose evolution defies these expectations. We present evidence for gene conversion events between mammalian-specific genes which encode sequence-dissimilar and possibly non-homologous, proteins. These conversion events occurred not within their protein-coding or 3' UTR sequences, but rather within their 5' UTRs and upstream regions. We suggest that the occurrence of concerted evolution events during mammalian evolution led to multiple chimaeric genes, with 5' UTR and protein-coding sequences possessing different evolutionary pedigrees.
These proposed events occurred within and between genes from three separate families ('brain-expressed X-linked' [BEX], WWbp5-like X-linked [WEX] and G-protein-coupled receptor-associated sorting protein [GASP]), all of which contain single protein-coding exons. Bex1, -2 and -3 are 'brain-expressed X-linked genes' whose intracellular products bind protein [13]. BEX1 and BEX2 bind the olfactory marker protein (OMP) [14,15] whereas BEX3 binds the p75 neurotrophin receptor (p75NTR) [16], and the second mitochondria-derived activator of caspase (Smac) [17,18], as well as self-associating [16]. WEX proteins include WWbp5 which is a poorly-understood WW domain binding protein [19]. GASP-1 and -2 are G-protein-coupled receptor (GPCR) associated sorting proteins (GASPs) which bind to the COOH-termini of various GPCRs and modulate their endocytic sorting to lysosomes [20,21].
Genes from these three families are all tightly-clustered within a mammal-specific ~2 Mb region of human chromosome Xq22.1-q22.2. We find that these genes all arose during early eutherian evolution and have experienced substantial sequence divergence thereafter. Their localisation to brain tissues, and their unusual and rapid evolution, are thus consistent with their involvement in the evolution of innovative brain cortical structures among eutherian mammals.
Results and discussion
A 2.3 Mb region of human Xq22.1-q22.2 is specific to placental mammals
Our studies focused on a 2.3 Mb region of human and mouse X chromosomes that encode a collinear arrangement of multiple genes from BEX, WEX and GASP families (Figure 1; Table 1). This entire region has a counterpart in neither the chicken nor the metatherian (marsupial) Monodelphis domestica. Orthologues of two genes that flank the region, Gla (encoding α-galactosidase; NM_013463) and Glra4 (encoding glycine receptor subunit α4; NM_010297), are separated by less than 4 Kb on chicken chromosome 4, and 10 Kb on Monodelphis scaffold 13561 (which is expected to lie within the long arm of its X chromosome [22]). These two non-eutherians' intergenic regions are both devoid of sequences homologous to BEX, WEX and GASP genes, and of assembly gaps greater than 100 b.
Figure 1.
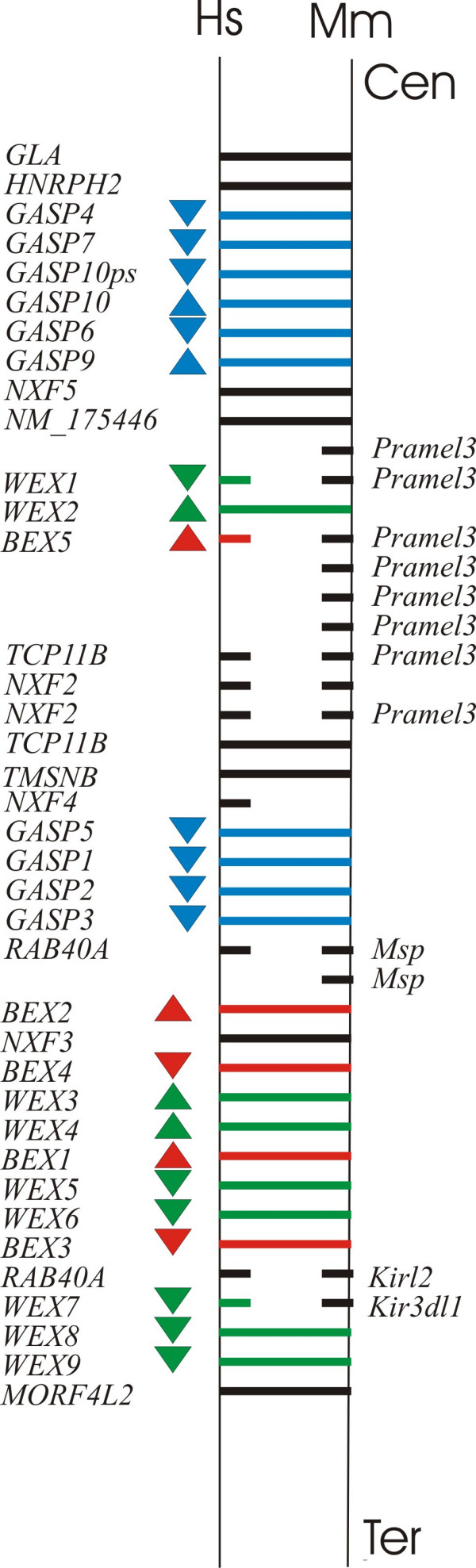
Protein-coding gene order in human chromosome Xq22.1-q22.2 (~2.3 Mb) and in its mouse orthologous region. Transcriptional orientations are indicated by filled arrow heads. Human- or mouse- specific genes are indicated by short bars. Abbreviations: Cen, Centromere; Ter, Terminal. Gene names are abbreviated as in Table 1.
Table 1.
Gene and transcript annotation of protein-coding genes located on human chromosome Xq22.1-q22.2. Human WEX1 has arisen from duplication of WEX2, and human BEX5 is absent from the mouse genome. a Levels of transcript expression in the brain are based on Microarray gene hybridization results [57]. Accession codes and expression tags are shown in parentheses. A gene expression in the brain is absent (-), present (+), or high relative to other tissues (++). Abbreviations: GLA, Galactosidase, alpha; HNRPH2, Heterogeneous nuclear ribonucleoprotein H2; GASP, G protein-coupled receptor-associated sorting; NXF, Nuclear RNA export factor; PRAMEL3, Preferentially expressed in melanoma like 3; WEX, WWbp5-like gene family; BEX, Brain-expressed X-linked; TCP11B, T-complex 11 B; TMSNB, Thymosin, beta, identified in neuroblastoma; RAB40A, Ras oncogene family member; MSP, Microsomal signal peptidase 23 kDa subunit (SPase 22 kDa subunit); KIRL2, Killer immunoglobulin-like receptor-like 2; KIR3DL1, Killer immunoglobulin-like receptor 3DL1; MORF4L2, Mortality factor 4 like 2;
| RefSeq Gene | RefSeq Protein | Synonyms | Gene name | Gene Start | Gene End | Transcriptional orientation | Expression in the braina (accession/expression-tag) | BGW element |
| NM_000169 | NP_000160 | GLA | 99424654 | 99434808 | - | + (GLA/214430_at) | ||
| NM_019597 | NP_062543 | HNRPH2 | 99435140 | 99440975 | + | + (HNRPH2) | + | |
| NM_152583 (BC036206) | NP_689796 | Q8K2R3 | GASP4 | 99512319 | 99560303 | + | + (gnf1h06491_at) | + |
| NM_016608 | NP_057692 | Alex1/Q9P291 | GASP7 | 99577371 | 99581530 | + | + (ARMCX1) | + |
| GASP10ψ | 99624346 | 99624903 | + | N/A | + | |||
| NM_019007 | NP_061880 | Q9NWJ3 | GASP10 | 99641972 | 99644848 | - | + (FLJ20811) | + |
| NM_016607 | NP_057691 | Alex3, Q9UH62 | GASP6 | 99649976 | 99654688 | + | + (ARMCX3) | + |
| NM_177949 | NP_808818 | Alex2, O60267 | GASP9 | 99682129 | 99686692 | - | + (ARMCX2) | + |
| NM_032946 | NP_116564 | NXF5 | 99858942 | 99884406 | - | N/A | ||
| AK094141 | XP_040486 | KIAA1789 | 99910469 | 99958857 | - | N/A | ||
| NM_080390 (AL540086) | NP_525129 | my048 | WEX1 | 100152572 | 100154538 | + | ++ (AF063606) | + |
| BC071675 | N/A | WEX2 | 100167309 | 100169228 | - | N/A | + | |
| BC042818 | N/A | BEX5 | 100180542 | 100182792 | - | N/A | + | |
| NM_017809 (BX647232) | NP060279 | NXF2 | 100242137 | 100353489 | + | + (NXF2/220257_x_at) | ||
| NM_017809 (BX647232) | NP060279 | NXF2 | 100387175 | 100498589 | - | + (NXF2/220257_x_at) | ||
| NM_021992 | NP_068832 | TMSNB | 100540461 | 100543504 | - | ++ Fetal brain (205347_s_at) | ||
| NM_207512 | NP_997395 | NXF4 | 100576750 | 100598478 | + | N/A | ||
| NM_022838 | NP_073749 | Q9H969 | GASP5 | 100626014 | 100630940 | + | + (FLJ12969/219335_at) | |
| NM_014710 | NP_055525 | Q43168 | GASP1 | 100678299 | 100685235 | + | ++ (GASP/204793_at) | |
| NM_138437 (AK123832) | NP_612446 | Q96D09 | GASP2 | 100739148 | 100744517 | + | N/A | |
| NM_030639 | NP_085142 | Q9BE11 | GASP3 | 100747511 | 100779225 | + | + (KIAA1701/213709_at) | |
| Rab40A-like | 100964038 | 100965103 | + | ++ (108_g_at) | ||||
| NM_018476 (BX098763) | NP_060946 | BEX1, HBEX2 | BEX2 | 101089445 | 101090956 | - | ++ (BEX1/218332_at) | + |
| NM_022052 | NP_071335 | NXF3 | 101102606 | 101119879 | - | - (NXF3/220110_s_at) | ||
| AK000959 | N/A | Bex4 | BEX4 | 101241922 | 101243977 | + | ++ (BEXL1/ 215440_s_at, 40916_at**) | |
| NM_153333 (AL709034) | NP_699164 | Wex3 | WEX3 | 101279783 | 101281945 | - | N/A | + |
| AL550633 | N/A | Wex4 | WEX4 | 101300507 | 101303513 | - | ++ (gnf1h07374_s_at, gnf1h07373_at) | + |
| NM_032621 (AL520932) | NP_116010 | Bex2, mouseBex1 | BEX1 | 101336132 | 101337663 | - | N/A | |
| NM_152278 (H09952) | NP_689491 | Wex5 | WEX5 | 101357022 | 101359108 | + | ++ (gnf1h06733_at) | + |
| NM_016303 (BF028506) | NP_057387 | Wex6 | WEX6 | 101383280 | 101385238 | + | + (WBP5 / 217975_at) | + |
| NM_206915 | NP_996798 | Ngfrap1 | BEX3 | 101403125 | 101404856 | + | ++ (NGFRAP1/217963_s_at) | + |
| NM_080879 | NP_543155 | RAB40A | 101526538 | 101546274 | - | N/A (RAB40A, 217589_at, 33477_at) | ||
| NM_024863 (AK024827) | NP_079139 | Wex7 | WEX7 | 101603016 | 101614300 | + | ++ (FLJ21174) | + |
| NM_032926 | NP_116315 | Wex8 | WEX8 | 101634691 | 101636711 | + | ++ (MGC15737) | + |
| NM_004780 | NP_004771 | TCEAL1 | WEX9 | 101655759 | 101657725 | + | ++ (TCEAL1) | + |
| NM_012286 | NP_036418 | MORF4L2 | 101702290 | 101713453 | - | + MORF4L2 |
Human Xq22.1-q22.2 thus appears to be an innovation of eutherian (placental) mammals. This is surprising since chromosome Xq initially arose after the divergence of the lineages leading to modern birds and mammals, but prior to the metatherian-eutherian split [23]. The observation however, provides an excellent opportunity to investigate issues underlying sequence innovation and functional innovation. We were interested in three interrelated questions: (i) Which evolutionary processes led to the origin of these genes? (ii) Are these genes' functions similar, and are they related to physiological or anatomical innovations in placental mammals? and, (iii) Why are these newly-acquired genes restricted to this one chromosomal region, rather than being dispersed throughout the remainder of the X chromosome or elsewhere?
BEX and WEX proteins are diverged homologues
We investigated the evolutionary origins of BEX, WEX and GASP genes using database searches of known protein and nucleotide sequences. We concluded that mammalian X-linked GASP genes are members of an ancient family that are discernible in early-branching bony vertebrates such as teleost fish (FLJ20811; NM_212853). By contrast, from the results of BLAST and PSI-BLAST [24] searches, BEX or WEX gene homologues appear to be absent outside of eutherian mammals. The order of BEX, WEX and GASP genes is conserved between human, rodent and canine X chromosomes and thus must have been present in their common ancestor.
Nevertheless, despite their sequence divergence we find that BEX and WEX protein sequences are homologous. Using COMPASS [25], significant similarity (Smith-Waterman score 82; E = 4.4 × 10-8) between a multiple sequence alignment of 25 sequences from the BEX protein family, and an alignment of 18 WEX sequences was observed. We thus predict that WEX and BEX proteins arose from a single ancestral gene early in eutherian evolutionary history, but diverged as separate families thereafter by numerous events of gene duplication. This is also consistent with BEX and WEX gene families possessing a common gene structure, namely three exons, with the most 3' of these always containing the entire protein-coding sequence.
5' UTR sequences of BEX, WEX and GASP genes are homologous
Despite the lack of evidence for homology between BEX or WEX, and GASP proteins, we were surprised to observe highly-similar sequences within the 5' UTRs of BEX and GASP genes. For example, a 383 b sequence (human chrX:101090862–101091244) which overlaps and extends the first non-coding exon of BEX2 is 91% identical to a region straddling the first exon of human GASP4.
Using MEME [26] we searched the first 5' UTR exons of human, mouse and rat BEX, WEX and GASP gene sequences for conserved DNA motifs. A highly significant DNA motif (E-value = 5.8 × 10-390, width = 57 bases) was identified by MEME in 37 of these 50 sequences. Using BLASTn searches of genomic sequences, we extended the width and breadth of these motifs, which we call BGW (Bex/GASP/Wex) elements (Figure 2). The program MAST [27] and the position-specific matrix of the MEME BGW motif was then used to search the NCBI non-redundant DNA database for significantly scoring DNA alignments. This search yielded statistically significant hits (E < 10-7) for 18 genomic sequences on human, mouse or rat chromosomes X, all of which represent the 5' UTRs of BEX, WEX and GASP genes. No significant alignments were observed to non-mammalian sequence or to mammalian sequence present outside of the X chromosome.
Figure 2.
Multiple sequence alignment of a conserved region of BGW sequence elements. Shaded if columns are conserved in at least 80% of sequences. Abbreviations: Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; NA, sequences not 5' to an identifiable gene structure.
These BGW motifs are not randomly positioned with respect to coding sequences: 18 of the 23 human homologues occur within the 5' UTRs of BEX or WEX or GASP family genes (Table 1). Of the remaining 5, 2 are homologous to other BGWs that are upstream of BEX or WEX or GASP family genes, as assessed by whole genome BLASTn searches (p < 0.01); 1 is 5' to a neighbouring gene HNRPH2 (and is highly conserved in orthologous sequences in dog, rat and mouse); and, 2, including 1 5' of GASP10ψ, are not upstream of known coding transcripts, so might represent false positives, pseudogenic copies or longer-range regulators. We conclude that the BGW element is a conserved non-coding sequence motif shared by BEX, WEX and GASP genes, which is restricted to eutherian chromosome Xq21-22.
Concerted evolution events within and between the 5' UTRs of BEX, WEX and GASP genes
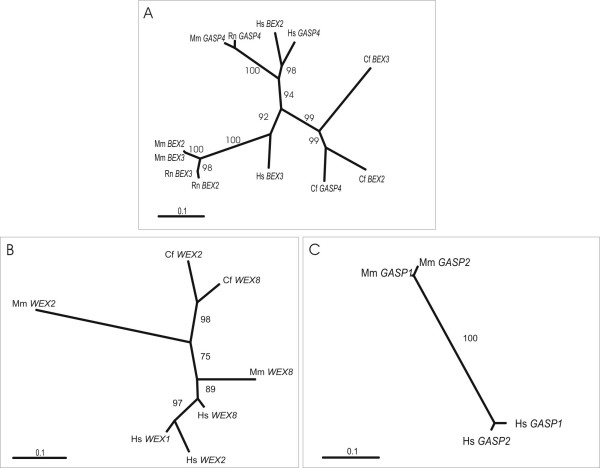
Further investigation demonstrated that the evolutionary histories of portions of BEX, WEX and GASP genes were not identical to the relationships expected from the species' phylogenetic tree. The first exon of the 5' UTR of BEX2, BEX3 and GASP4 genes encompasses the BGW motif and exhibits greater sequence similarities between paralogues than it does between orthologues (Figure 3A). This finding is indicative of independent concerted evolution events [7,9,11,28,29]. Bootstrap values are sufficiently high to indicate with confidence that concerted evolution events have occurred among these genes in all mammalian lineages examined (human, mouse, rat and dog).
Figure 3.
Concerted evolution events among BEX, WEX and GASP genes. Maximum likelihood phylogenetic trees for A) the first exon of the 5'UTR of BEX2, BEX3 and GASP4 genes B) the first exon of the 5' UTR of WEX2, WEX8 and WEX1 genes, and C) the exonic 5' UTR of GASP1 and GASP2 genes. 5' UTR sequences were extracted either from sequence transcripts or from homologous genomic regions. Maximum likelihood tree topologies and branch lengths were obtained using the program BASEML [54], for a given DNA sequence alignment. Each alignment was bootstrapped a 1000 times using the neighbour-joining method, and bootstrap values were overlaid the maximum likelihood tree. Abbreviations: Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Cf, Canis familiaris.
Notably, some of these concerted evolution events are lineage-specific. Bootstrap values (Figure 3A) support such events between human or dog BEX2 and GASP4, but not between mouse or rat BEX2 and GASP4, and between mouse, rat or dog BEX2 and BEX3, but not human BEX2 and BEX3.
Similarly, phylogenetic analysis of WEX2 and WEX8 genes indicates that concerted evolution events occurred recently, between the first 5' UTR exons, in the carnivore and primate lineages (Figure 3B). Finally, concerted evolution also occurred between the 5' UTRs of human or mouse GASP1 and GASP2 (Figure 3C).
Some of these concerted evolution events appear to have occurred relatively recently. In particular, concerted evolution between the 5' UTRs of mouse Bex2 and Bex3 (or rat Bex2 and Bex3) genes must have occurred very recently because they exhibit no substitutions of nucleotides within their BGW motifs (Figure 2).
Chimaerism among BEX, WEX and GASP genes
Concerted evolution events between BEX2, BEX3 and GASP4 are restricted to the non-coding regions of their genes. The GASP4 protein exhibits no discernible sequence similarity to either BEX2 or BEX3 proteins; moreover, BEX2 and BEX3 amino acid sequences are relatively divergent (42% identity). Thus, these three genes appear to be chimaeric: their 5' UTRs are highly similar and exhibit a recent ancestry, whereas their protein coding sequences are more distantly related.
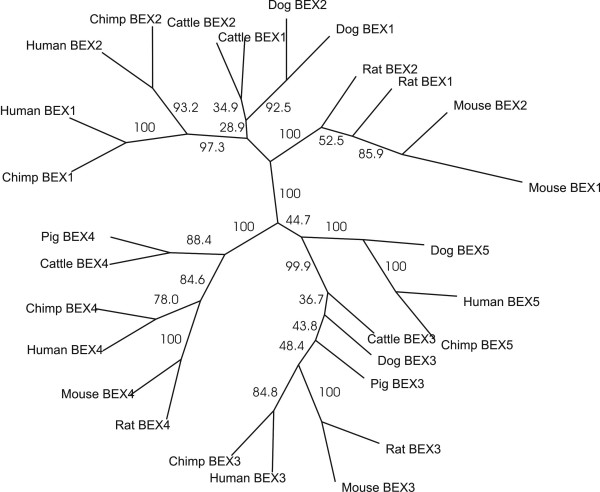
This surprising conclusion is supported by a phylogenetic tree of BEX protein coding sequences (Figure 4). With high reliability the protein coding regions of human BEX1 and BEX2 paralogues were found to be more similar than they are to their rodent (mouse and rat), ruminant (cattle) or carnivore (dog) orthologues; similarly mouse BEX1 and BEX2 are most closely related, as are dog BEX1 and BEX2 genes. By contrast, the predicted evolutionary relationships of the three other paralogues, BEX3, BEX4 and BEX5, recapitulate the expected species tree [30].
Figure 4.
Concerted evolution events among BEX1 and BEX2 protein-coding sequences. A maximum likelihood phylogenetic tree for the BEX family members' protein-coding regions was constructed using the program BASEML [54] and a DNA sequence alignment. The alignment was bootstrapped a 1000 times using the neighbour-joining method, and provided bootstrap values, which were overlaid onto the maximum-likelihood tree.
These results thus demonstrate distinct and complex evolutionary histories for different regions of genes: concerted evolution events in the 5' UTRs are supported among BEX2, BEX3 and GASP4, while concerted evolution events in protein coding regions are supported only between BEX1 and BEX2. Among WEX proteins, a similar analysis indicates that WEX2, WEX4 and WEX8 coding sequences have experienced concerted evolution events (data not shown).
A pseudogene of human GASP10 (GASP10ψ), which is positioned upstream of GASP10 on chromosome X, appears to be converting with GASP10 in primate (human), rodent (mouse) and carnivore (dog) lineages. Bootstrap analysis supports that GASP10 and GASP10ψ are significantly more similar to each other than they are to their orthologues from dog or mouse (bootstrap value = 100%, data not shown). Therefore, although GASP10ψ does not encode a functional protein, it may function as a redundant copy for facilitating gene conversion events to its neighbouring GASP10 gene, as has occurred elsewhere in the human genome [31,32].
Mechanism and mode of concerted evolution
These and similar inferences of concerted evolution (collated in Additional file 1) could have arisen due to unequal crossing-over, gene duplication or gene conversion. Unequal crossing-over and multiple gene duplications are unlikely mechanisms for concerted evolution of these genes because their orders and transcriptional orientations along their X chromosomes have suffered no rearrangements, inversions (except for one, involving rat WEX2) or large deletions when human, dog and mouse genomes are compared (Table 1; Figure 1; data not shown).
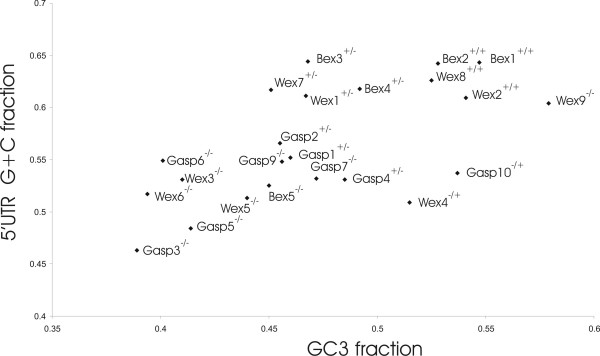
The findings also could have arisen from exon shuffling, where exons are duplicated and inserted into different gene contexts [33]. However, inter-allelic gene conversion is distinguished from exon shuffling in that it often occurs within regions possessing high G+C nucleotide compositions [8-10,34]. Indeed, from G+C fractions for BEX, WEX and GASP 5' UTRs and for the third positions of their protein coding regions ("GC3"), we found that sequences with evidence for concerted evolution possess significantly higher G+C fractions than sequences without such evidence (paired T-test, p(5' UTR) < 10-3, p(GC3) = 10-3) (Figure 5, Additional file 1). This indicates strongly that multiple events of interlocus gene conversion, rather than de novo sequence duplication or exon shuffling, have occurred among these genes.
Figure 5.
G/C levels are elevated in human genes that undergo concerted evolution events. 5' UTR G+C and protein-coding GC3 properties of human BEX, WEX and GASP genes are indicated. (+) or (-) symbols represent presence or absence of a concerted evolution event in 5' UTR (first/left symbol) or in protein-coding (second/right symbol) regions.
Evidence for sustained gene conversion events
Our findings indicate that the vertical transmission of BEX, WEX and GASP genes has been interrupted frequently by horizontal acquisitions of non-coding, as well as coding, sequences from genes that are closely-linked on human chromosome Xq22.1-q22.2. These genes' sequences appear to have been homogenized by multiple episodes of interlocus gene conversion. Because gene conversion is thought to proceed via formation of heteroduplexes between highly-similar sequences [35], this raises an interesting conundrum: how has recent gene conversion occurred between sequence-dissimilar genes drawn from different gene families?
We resolve this question by proposing that BEX and WEX proteins all arose from a common ancestral GASP-like gene and rapidly diverged in sequence thereafter (Figure 6). We further propose that sequence similarity has been preserved between these homologues' 5' UTRs by recurrent episodes of gene conversion, despite rapid divergence of sequences elsewhere in their genes.
Figure 6.
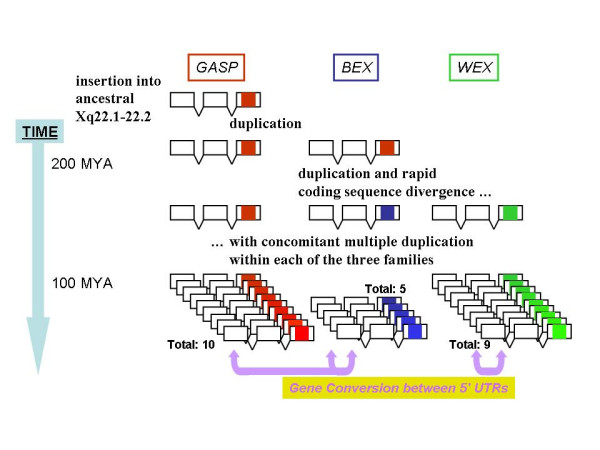
Proposed evolutionary events leading to BEX, WEX and GASP genes on human chromosome Xq22.1-q22.2. BEX and WEX genes arise due to duplication of a GASP-like gene early in eutherian mammalian history. These genes undergo multiple duplication events thereafter, but prior to the divergence of human and dog lineages. Multiple events of gene conversion, either between 5' UTRs or between coding sequences (Additional file 1), occur with the remaining coding sequence diverging rapidly due to relaxed selective constraints and/or adaptive evolution.
This scenario is supported by three observations: that BEX and WEX genes are homologous, that all three families utilize only a single exon to code for protein, and that BEX, WEX and GASP protein-coding sequences evolved rapidly. For the latter observation, we note that KA/KS values of these genes are high (median of 0.34) relative to a median value of 0.10 for all Ensembl human-mouse single orthologues (data not shown). These genes might then have arisen initially from duplication of GASP10, a gene that contains the BGW element and whose orthologues are known in earlier-diverging vertebrates, including fish (hypothetical protein FLJ20811; NM_212853). Unlike other GASP genes, GASP10 contains a three exon structure, similarly to these observed in BEX and WEX genes.
Selection of gene conversion events
To our knowledge there is only a single documented occurrence of gene conversion between paralogues' 3' UTRs [36], and a single observation of gene conversion between paralogues' 5' UTRs [37]. However, multiple conversion events that differentiate between coding and non-coding sequences, as well as sequence conversion between genes that otherwise are not demonstrably homologous, are completely unexpected.
We attribute this singular evolutionary scenario to placental mammal-specific functional innovation. BEX and WEX genes appear, from the absence of homologues among other vertebrates, to have arisen during mammalian evolution by rapid sequence divergence from a common ancestor. In other analogous situations, such as the evolution of the caseins and histatins from a single ancestral gene [38], rapid gene duplication and sequence diversification is causally linked to innovation in physiology and behaviour [2]. In this case, gene function is associated with binding to brain-specific receptors, as seen for BEX and GASP proteins [15,20,21].
On the basis of frameshifts, stop codons, the lack of an initiating methionine codon or introns, all BEX and WEX paralogues outside of the Xq22.1-q22.2 region appear to be retrotransposed processed pseudogenes. This suggests that there is selection for retention of these genes as a closely-linked group on the X chromosome, and perhaps points either to gene conversion being a necessary requirement for long-term sustenance and evolution of their functions, or an X-linked factor that is necessary for their proper gene functions.
Function prediction
The X chromosome contains a disproportionate number of genes related to mental functions which has been linked to the male preponderance of mental retardation cases [39,40]. However, of 9 X-linked genes that, when mutated, lead to mental impairment all possess orthologues in fish or even earlier-branching eukaryotes [40]. When expression information is available, all BEX, WEX and GASP genes are found to be expressed in the brain (Table 1). These eutherian-specific genes are thus possible candidates for the adaptive evolution of the neocortex, a region of the forebrain which is unique to mammals [41].
The presence of a conserved BGW element within the 5' UTRs of BEX, WEX and GASP genes is suggestive of its participation in regulation of translation. This is because translation rates have been shown previously to be affected by regulatory sequences, which include the start site consensus sequence, secondary structures, upstream AUGs, internal ribosome entry sites (IRES) and sequence specific recognition site for regulatory factors, such as protein or RNA [42,43]. Translational control of BEX, WEX and GASP genes might indicate that their proteins are utilized under specific physiological conditions [44], at developmental stages [45] or in subcellular compartments [46,47]. Another possible role for the BGW element might be to regulate alternative splicing. Although these genes possess only single protein coding exons there are several examples of transcripts that exhibit alternative splicing within their 5' UTR exons (e.g. WEX2 (mRNA BQ068054)) and others that exclude the protein-coding exon altogether (e.g. GASP5 (mRNA BC022066)).
Conclusion
We have described the evolutionary history of a large region of human chromosome X, which appears to be an innovation of placental mammals. This region encompasses three previously unrelated protein-coding gene families, BEX, WEX and GASP, which have been the product of multiple gene duplications and large protein-coding sequence diversification since the earliest eutherian mammal. Despite the lack of protein-coding sequence similarity between many genes, we were able to identify a mammalian conserved DNA motif in their exonic 5'UTR, suggesting that they are derived of a common single ancestor, probably a GASP-like gene, found in early-branching bony vertebrates.
We have shown that the evolution of these paralogous genes has been affected by multiple events of gene conversion acting to homogenize among 5'UTR sequences, protein-coding sequences or both. Events of gene conversion in these regions have led to the occurrence of chimaeric genes, where their 5' UTRs are highly similar and exhibit a recent ancestry, but their protein coding sequences are more distantly related. We showed that the composition of sequences undergoing concerted evolution is enriched with G and C nucleotides, suggesting that biased gene conversion has been the underlying mechanism rather than exon shuffling.
BEX, WEX and GASP genes are found to be expressed in the brain (Table 1), suggesting that these eutherian-specific genes are possible candidates for the adaptive evolution of the neocortex, a region of the forebrain which is unique to mammal. The presence of a conserved BGW element within the 5' UTRs of BEX, WEX and GASP genes is suggestive of its participation in regulation of translation, possibly resulting in different spatio-temporal localization of these genes products or in different alternative splicing forms. These findings thus hint at hitherto unappreciated modes of 5' UTR evolution. The identification of such 5' UTRs elsewhere in the genome thus will be required as a contribution to the delineation of all human sequence under selection.
Methods
Genome assemblies and gene models
The July 2003 human (based on NCBI Build 34), the October 2003 mm4 Mus musculus genome assembly (based on NCBI Build 32), the rn3 June 2003 Rattus norvegicus genome assembly (based on version 3.1), the canFam1 July 2004 Canis familiaris whole genome shotgun (WGS) assembly v1.0, the 13 Nov. 2003 chimpanzee (Pan troglodytes) Arachne assembly – NCBI Build 1 Version 1, and the February 2004 chicken (Gallus gallus) draft assembly were analysed. Gene annotations were extracted from the UCSC genome browser [48], or were predicted using Genewise [49] and available transcriptional information.
BGW element
The ENSEMBL web browser blast application [50] and the BlastN program [24] (without optimization for identical hits) were used to perform a sequence similarity search between a region 5' to Alex2 (chrX:99686693–99686992) and the entire human genome assembly. Multiply aligned genomic hits that were predicted (p < 0.01) to be homologous to this region were positioned on human X chromosome, only within an area containing BEX, WEX, and GASP family members. A similar search was done in mouse, rat, zebrafish and chicken genomes. Human, mouse and rat sequences were thereafter multiply aligned, and used to further search the human genomic area (chrX:99850000–100250000) using an HMM [51]. Additional similar sequences (E < 0.1) were added to generate the final multiple alignment (Figure 2).
Tree construction and bootstrap analysis
Protein-coding and exonic 5' UTR phylogenetic trees were both constructed from DNA sequence alignments. Sequences were aligned using ClustalW [52] or MULAN [53], and alignments were subjected to a neighbour-joining bootstrapping process (n = 1000). Non protein-coding branch length estimations were calculated using a maximum likelihood approach (BASEML [54]) as implemented by the molecular evolution package DAMBE [55]. In order to assign bootstrap values for the branch lengths, neighbour-joining bootstrap values were superimposed on the ML tree.
G+C and GC3 content analysis
G+C proportions contained within the exonic-5' UTR or the entire 5' UTR were based on genomic sequences. GC3 content was calculated from examining the GC fraction of the third nucleotide codon positions of protein-coding sequences.
Exonic 5' UTR sequence comparisons
The longest 5'-UTR sequence of each gene was chosen using all available mRNA and EST transcript information (Table 1). In all cases, sequences were further extended by 300 bases, using genomic data, to account for foreshortened transcript evidence. UTR sequences were then aligned using BLASTN and considered to be homologous when p < 0.001.
KA/KS analysis
Ratios of KA (the number of nonsynonymous substitutions per nonsynonymous site) to KS (the number of synonymous substitutions per synonymous site) were calculated using the yn00 method of Yang and Nielsen [56].
Authors' contributions
EEW identified the shared homology between the 5'UTR of BEX and GASP genes and generated much of the analysis. CPP identified the BGW motif, and assisted in data interpretation. EEW and CPP both contributed to the drafting of the manuscript.
Supplementary Material
Evidence for gene conversion events among mammalian BEX, WEX and GASP genes. Sequences were grouped according to the gene region they encompass: exonic 5' UTR, 5' UTR (exons and introns) and protein-coding regions. Validation method: (a) A phylogenetic tree indicating significantly greater proximity between gene paralogs in a particular genome than for gene orthologous of different organisms' genomes. Neighbour-joining bootstrap values for the tree topology were considered if they were greater than 85% (b) Sequence identity levels between paralogs are substantially higher than sequence identity levels between orthologs (>85% compared with <65%, respectively). Abbreviations: N/D, not determined.
Acknowledgments
Acknowledgements
We would like to thank the Medical Research Council UK for funding.
Contributor Information
Eitan E Winter, Email: eew@sanger.ac.uk.
Chris P Ponting, Email: chris.ponting@anat.ox.ac.uk.
References
- Copley RR, Goodstadt L, Ponting C. Eukaryotic domain evolution inferred from genome comparisons. Curr Opin Genet Dev. 2003;13:623–628. doi: 10.1016/j.gde.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Emes RD, Goodstadt L, Winter EE, Ponting CP. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Hum Mol Genet. 2003;12:701–709. doi: 10.1093/hmg/ddg078. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. London , George Allen and Unwin; 1970. [Google Scholar]
- Carrington M, Cullen M. Justified chauvinism: advances in defining meiotic recombination through sperm typing. Trends Genet. 2004;20:196–205. doi: 10.1016/j.tig.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Dover GA. Evolution of genetic redundancy for advanced players. Curr Opin Genet Dev. 1993;3:902–910. doi: 10.1016/0959-437X(93)90012-E. [DOI] [PubMed] [Google Scholar]
- Li WHGD. Fundamentals of Molecular Evolution. Sinauer Associates Inc. Sunderland, MA.; 1997. [Google Scholar]
- Galtier N. Gene conversion drives GC content evolution in mammalian histones. Trends Genet. 2003;19:65–68. doi: 10.1016/S0168-9525(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Kudla G, Helwak A, Lipinski L. Gene conversion and GC-content evolution in mammalian Hsp70. Mol Biol Evol. 2004;21:1438–1444. doi: 10.1093/molbev/msh146. [DOI] [PubMed] [Google Scholar]
- Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol Biol Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Slightom JL, Chang LY, Koop BF, Goodman M. Chimpanzee fetal G gamma and A gamma globin gene nucleotide sequences provide further evidence of gene conversions in hominine evolution. Mol Biol Evol. 1985;2:370–389. doi: 10.1093/oxfordjournals.molbev.a040357. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, Dodgson JB, Chinwalla AT, Cliften PF, Clifton SW, Delehaunty KD, Fronick C, Fulton RS, Graves TA, Kremitzki C, Layman D, Magrini V, McPherson JD, Miner TL, Minx P, Nash WE, Nhan MN, Nelson JO, Oddy LG, Pohl CS, Randall-Maher J, Smith SM, Wallis JW, Yang SP, Romanov MN, Rondelli CM, Paton B, Smith J, Morrice D, Daniels L, Tempest HG, Robertson L, Masabanda JS, Griffin DK, Vignal A, Fillon V, Jacobbson L, Kerje S, Andersson L, Crooijmans RP, Aerts J, van der Poel JJ, Ellegren H, Caldwell RB, Hubbard SJ, Grafham DV, Kierzek AM, McLaren SR, Overton IM, Arakawa H, Beattie KJ, Bezzubov Y, Boardman PE, Bonfield JK, Croning MD, Davies RM, Francis MD, Humphray SJ, Scott CE, Taylor RG, Tickle C, Brown WR, Rogers J, Buerstedde JM, Wilson SA, Stubbs L, Ovcharenko I, Gordon L, Lucas S, Miller MM, Inoko H, Shiina T, Kaufman J, Salomonsen J, Skjoedt K, Wong GK, Wang J, Liu B, Wang J, Yu J, Yang H, Nefedov M, Koriabine M, Dejong PJ, Goodstadt L, Webber C, Dickens NJ, Letunic I, Suyama M, Torrents D, von Mering C, Zdobnov EM, Makova K, Nekrutenko A, Elnitski L, Eswara P, King DC, Yang S, Tyekucheva S, Radakrishnan A, Harris RS, Chiaromonte F, Taylor J, He J, Rijnkels M, Griffiths-Jones S, Ureta-Vidal A, Hoffman MM, Severin J, Searle SM, Law AS, Speed D, Waddington D, Cheng Z, Tuzun E, Eichler E, Bao Z, Flicek P, Shteynberg DD, Brent MR, Bye JM, Huckle EJ, Chatterji S, Dewey C, Pachter L, Kouranov A, Mourelatos Z, Hatzigeorgiou AG, Paterson AH, Ivarie R, Brandstrom M, Axelsson E, Backstrom N, Berlin S, Webster MT, Pourquie O, Reymond A, Ucla C, Antonarakis SE, Long M, Emerson JJ, Betran E, Dupanloup I, Kaessmann H, Hinrichs AS, Bejerano G, Furey TS, Harte RA, Raney B, Siepel A, Kent WJ, Haussler D, Eyras E, Castelo R, Abril JF, Castellano S, Camara F, Parra G, Guigo R, Bourque G, Tesler G, Pevzner PA, Smit A, Fulton LA, Mardis ER, Wilson RK. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Brown AL, Kay GF. Bex1, a gene with increased expression in parthenogenetic embryos, is a member of a novel gene family on the mouse X chromosome. Hum Mol Genet. 1999;8:611–619. doi: 10.1093/hmg/8.4.611. [DOI] [PubMed] [Google Scholar]
- Baldisseri DM, Margolis JW, Weber DJ, Koo JH, Margolis FL. Olfactory marker protein (OMP) exhibits a beta-clam fold in solution: implications for target peptide interaction and olfactory signal transduction. J Mol Biol. 2002;319:823–837. doi: 10.1016/S0022-2836(02)00282-6. [DOI] [PubMed] [Google Scholar]
- Behrens M, Margolis JW, Margolis FL. Identification of members of the Bex gene family as olfactory marker protein (OMP) binding partners. J Neurochem. 2003;86:1289–1296. doi: 10.1046/j.1471-4159.2003.01940.x. [DOI] [PubMed] [Google Scholar]
- Mukai J, Shoji S, Kimura MT, Okubo S, Sano H, Suvanto P, Li Y, Irie S, Sato TA. Structure-function analysis of NADE: identification of regions that mediate nerve growth factor-induced apoptosis. J Biol Chem. 2002;277:13973–13982. doi: 10.1074/jbc.M106342200. [DOI] [PubMed] [Google Scholar]
- Kimura MT, Irie S, Shoji-Hoshino S, Mukai J, Nadano D, Oshimura M, Sato TA. 14-3-3 is involved in p75 neurotrophin receptor-mediated signal transduction. J Biol Chem. 2001;276:17291–17300. doi: 10.1074/jbc.M005453200. [DOI] [PubMed] [Google Scholar]
- Yoon K, Jang HD, Lee SY. Direct interaction of Smac with NADE promotes TRAIL-induced apoptosis. Biochem Biophys Res Commun. 2004;319:649–654. doi: 10.1016/j.bbrc.2004.05.043. [DOI] [PubMed] [Google Scholar]
- Jolliffe CN, Harvey KF, Haines BP, Parasivam G, Kumar S. Identification of multiple proteins expressed in murine embryos as binding partners for the WW domains of the ubiquitin-protein ligase Nedd4. Biochem J. 2000;351 Pt 3:557–565. doi: 10.1042/0264-6021:3510557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin F, Karcher P, Boeuf JJ, Matifas A, Kieffer BL. Identification of a novel family of G protein-coupled receptor associated sorting proteins. J Neurochem. 2004;89:766–775. doi: 10.1111/j.1471-4159.2004.02411.x. [DOI] [PubMed] [Google Scholar]
- Whistler JL, Enquist J, Marley A, Fong J, Gladher F, Tsuruda P, Murray SR, Von Zastrow M. Modulation of postendocytic sorting of G protein-coupled receptors. Science. 2002;297:615–620. doi: 10.1126/science.1073308. [DOI] [PubMed] [Google Scholar]
- Glas R, Marshall Graves JA, Toder R, Ferguson-Smith M, O'Brien PC. Cross-species chromosome painting between human and marsupial directly demonstrates the ancient region of the mammalian X. Mamm Genome. 1999;10:1115–1116. doi: 10.1007/s003359901174. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadreyev R, Grishin N. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J Mol Biol. 2003;326:317–336. doi: 10.1016/S0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Liao D. Concerted evolution: molecular mechanism and biological implications. Am J Hum Genet. 1999;64:24–30. doi: 10.1086/302221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenoi M, Mita K, Ichimura S, Kawano A. Higher frequency of concerted evolutionary events in rodents than in man at the polyubiquitin gene VNTR locus. Genetics. 1998;148:867–876. doi: 10.1093/genetics/148.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci U S A. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor M, Parker KL, Globerman H, New MI, White PC. Mutation in the CYP21B gene (Ile-172----Asn) causes steroid 21-hydroxylase deficiency. Proc Natl Acad Sci U S A. 1988;85:1600–1604. doi: 10.1073/pnas.85.5.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler J, Curnutte JT, Rae J, Barrett D, Patino P, Chanock SJ, Goerlach A. Recombination events between the p47-phox gene and its highly homologous pseudogenes are the main cause of autosomal recessive chronic granulomatous disease. Blood. 2000;95:2150–2156. [PubMed] [Google Scholar]
- Patthy L. Genome evolution and the evolution of exon-shuffling--a review. Gene. 1999;238:103–114. doi: 10.1016/S0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb BC. The properties of meiotic gene conversion important in its effects on evolution. Heredity. 1984;53 ( Pt 1):113–138. doi: 10.1038/hdy.1984.68. [DOI] [PubMed] [Google Scholar]
- Bargelloni L, Scudiero R, Parisi E, Carginale V, Capasso C, Patarnello T. Metallothioneins in antarctic fish: evidence for independent duplication and gene conversion. Mol Biol Evol. 1999;16:885–897. doi: 10.1093/oxfordjournals.molbev.a026178. [DOI] [PubMed] [Google Scholar]
- Bobba A, Marra E, Lattanzio P, Iolascon A, Giannattasio S. Characterization of the CYP21 gene 5' flanking region in patients affected by 21-OH deficiency. Hum Mutat. 2000;15:481. doi: 10.1002/(SICI)1098-1004(200005)15:5<481::AID-HUMU14>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci U S A. 2003;100:4060–4065. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH. X-linked genes and mental functioning. Hum Mol Genet. 2005;14 Spec No 1:R27–32. doi: 10.1093/hmg/ddi112. [DOI] [PubMed] [Google Scholar]
- Zechner U, Wilda M, Kehrer-Sawatzki H, Vogel W, Fundele R, Hameister H. A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution? Trends Genet. 2001;17:697–701. doi: 10.1016/S0168-9525(01)02446-5. [DOI] [PubMed] [Google Scholar]
- Allman J. Evolving Brains. New York , Scientific American Library; 2000. [Google Scholar]
- Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5'- and 3'-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kuhn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci U S A. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Slack FJ. Micro-RNAs: small is plentiful. J Cell Biol. 2002;156:17–21. doi: 10.1083/jcb.200111033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Palacios IM, St Johnston D. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu Rev Cell Dev Biol. 2001;17:569–614. doi: 10.1146/annurev.cellbio.17.1.569. [DOI] [PubMed] [Google Scholar]
- UCSC Genome Bioinformatics Site http://genome.cse.ucsc.edu
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl Web Browser Blast Application http://www.ensembl.org/Multi/Blastview?species=Homo_sapiens
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, Miller W. Mulan: Multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- GNF Gene Expression Atlas http://symatlas.gnf.org/SymAtlas
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evidence for gene conversion events among mammalian BEX, WEX and GASP genes. Sequences were grouped according to the gene region they encompass: exonic 5' UTR, 5' UTR (exons and introns) and protein-coding regions. Validation method: (a) A phylogenetic tree indicating significantly greater proximity between gene paralogs in a particular genome than for gene orthologous of different organisms' genomes. Neighbour-joining bootstrap values for the tree topology were considered if they were greater than 85% (b) Sequence identity levels between paralogs are substantially higher than sequence identity levels between orthologs (>85% compared with <65%, respectively). Abbreviations: N/D, not determined.