Abstract
Chanarin-Dorfman syndrome (CDS) is a rare autosomal recessive form of nonbullous congenital ichthyosiform erythroderma (NCIE) that is characterized by the presence of intracellular lipid droplets in most tissues. We previously localized a gene for a subset of NCIE to chromosome 3 (designated “the NCIE2 locus”), in six families. Lipid droplets were found in five of these six families, suggesting a diagnosis of CDS. Four additional families selected on the basis of a confirmed diagnosis of CDS also showed linkage to the NCIE2 locus. Linkage-disequilibrium analysis of these families, all from the Mediterranean basin, allowed us to refine the NCIE2 locus to an ∼1.3-Mb region. Candidate genes from the interval were screened, and eight distinct mutations in the recently identified CGI-58 gene were found in 13 patients from these nine families. The spectrum of gene variants included insertion, deletion, splice-site, and point mutations. The CGI-58 protein belongs to a large family of proteins characterized by an α/β hydrolase fold. CGI-58 contains three sequence motifs that correspond to a catalytic triad found in the esterase/lipase/thioesterase subfamily. Interestingly, CGI-58 differs from other members of the esterase/lipase/thioesterase subfamily in that its putative catalytic triad contains an asparagine in place of the usual serine residue.
Introduction
Chanarin-Dorfman syndrome (CDS [MIM 275630]), also referred to as “neutral-lipid–storage disease with ichthyosis,” is a rare autosomal recessive nonlysosomal inborn error of neutral-lipid metabolism. CDS is characterized by an intracellular accumulation of triacylglycerol (TG) droplets in most tissues. Since the first reports of the disorder (Rozenszajn et al. 1966; Dorfman et al. 1974; Chanarin et al. 1975), ∼30 patients have been described in the literature, most of them from Middle Eastern countries. The clinical phenotype involves multiple organs and systems. Ichthyosis is always present, and, more variably, liver steatosis with hepatomegaly, muscle weakness (or myopathy), ataxia, neurosensory hearing loss, subcapsular cataracts, nystagmus, strabismus, and mental retardation are sometimes present (Rozenszajn et al. 1966; Dorfman et al. 1974; Chanarin et al. 1975). The clinical aspect of ichthyosis is a nonbullous congenital ichthyosiform erythroderma (NCIE) with fine white scaling on an erythematous background. Patients are often born as collodion babies and may present bilateral ectropion and eclabion. No modifications of hair, nails, teeth, or mucosa have been reported. Most common blood tests, including those of serum lipids, are normal, but muscular and hepatic enzymes are often found to be moderately elevated (two- to threefold). A diagnosis is confirmed by a peripheral blood smear, which shows lipid droplets in granulocytes (Jordans anomaly; see Jordans et al. 1953). These lipid droplets have also been observed in multiple cell types, including basal keratinocytes of the epidermis. Electron microscopy shows multiple vacuoles in the cytoplasm, without a surrounding membrane (Miranda et al. 1979). Biochemical studies have excluded an abnormality in uptake, transport, or β-oxidation of fatty acids. Activities of various enzymes (lipase, carboxyesterase, and enzymes of glycerolipid synthesis) have been analyzed and found to be normal (Williams et al. 1988; Hilaire et al. 1995; Igal and Coleman 1996, 1998). Nevertheless, the TG content of lymphocytes, macrophages, or fibroblasts in culture from affected patients is 2- to 20-fold higher than that in normal cells, even in lipid-deficient medium (Williams et al. 1988; Hilaire et al. 1995). The specific defect has been postulated to be an abnormal recycling of TG-derived mono- or diacylglycerols to specific phospholipids (Igal and Coleman 1996, 1998) or to be a defect in the catabolism of long-chain fatty acids (Hilaire et al. 1995).
We previously localized a gene for NCIE (i.e., NCIE2, [MIM 604780]) to a 7.7-cM region on chromosome 3p21 (Fischer et al. 2000), which we identified as the CDS locus. We have narrowed this interval by analysis of additional families and have found mutations in a recently described gene, CGI-58 (Lai et al. 2000), in 13 patients from nine unrelated families. CGI-58 encodes a new protein, CGI-58, with homology to proteins of a large family characterized by an α/β hydrolase fold. The strongest homology was found for sequence motifs that characterize the esterase/lipase/thioesterase subfamily (Schrag and Cygler 1997). CGI-58 differs from other members of the esterase/lipase/thioesterase subfamily in that its putative catalytic triad contains an asparagine in place of the usual serine residue.
Subjects and Methods
Subjects and Specimens
We studied five of the six original families that we had reported in our gene-localization study (Fischer et al. 2000), as well as an additional four families for which a diagnosis of CDS had been clearly established. A total of 13 patients and 35 nonaffected family members from these nine families were analyzed. Three families were from Algeria, two families were from Tunisia, two families were from Turkey, one family was from France, and one family was from Morocco. All the parents were available. Seven families were known to be consanguineous because of marriages between first cousins. Two families were not known to be consanguineous, but, in one of these families, the parents came from the same village on the island of Djerba, in Tunisia. Signed, informed consent was obtained from each patient and each family member. A detailed medical record including dermatologic examination (performed by B.B., F.C., A.K., J.-L.V., or A.W.), as well as neurologic, ophthalmologic, and oto-rhino-laryngologic investigations, and pedigree information were established. DNA was extracted from whole blood by standard procedures. Lymphoblastoid cell lines were established for all patients and most of the parents. Peripheral blood smears were stained with the May-Grünwald-Giemsa and oil-red-O stains, to detect lipid droplets in neutrophils and in monocytes. Skin biopsies were also performed in four patients, and fibroblast cultures were initiated.
Genetic Analysis
Genotyping with fluorescent markers was performed as described elsewhere (Fischer et al. 2000). Haplotypes were constructed under assumption of the most-parsimonious linkage phase. Linkage programs were used under the assumptions of autosomal recessive inheritance, full penetrance, and a disease frequency, in the general population, of 1/1,000,000. Pairwise LOD scores were calculated by the MLINK program of the LINKAGE 5.1 package (Lathrop et al. 1985), with consanguineous loops, if known, being incorporated into the pedigree files. For linkage-disequilibrium analysis, the excess of the disease-associated alleles was calculated by the Pexcess equation: Pexcess=(Paffected-Pnormal)/(1-Pnormal), in which Paffected and Pnormal denote, respectively, the frequency of the disease-associated allele on disease-bearing chromosomes (15) and normal chromosomes (20) (Hästbacka et al. 1992). Chromosomes that either were shared by two sibs or were homozygous were counted only once. For χ2 estimation, we used the combined-allele method (Aksentijevich et al. 1993), corrected by Bonferroni’s procedure (Weir 1990).
Physical Map
The sequences from the initial interval were identified by the Genoscope working-draft utility. This tool identifies, by electronic PCR, human-genome working-draft sequences from the interval that contains one or more RHdb markers. The physical map, from Golden Path, of our 1.3-Mb interval consisted of 10 BAC or PAC clones (GenBank accession numbers AC022627, AC022985, AC006055, AC024719, AC073222, AC06058, AC024188, AC076962, AC069071, and AC018473). Sequences from these BACs were compared to sequences in databases (Ensembl and Golden Path) in which results of new and previously known gene structures were annotated by several gene-prediction programs, to identify candidate genes for mutation analysis.
Mutation Screening
Mutation analysis was performed in affected patients and in both parents in the nine families. Intronic oligonucleotide primers for acetyl-coenzyme acyltransferase-1 (ACAA1), sterol 12α-hydroxylase (CYP8B1), and CGI-58 were designed, by the Primer3 program, from the genomic sequences NT_006022, AF090320, and AC006055, respectively. The primer sequences for CGI-58 are shown in table 1. PCR was performed in a 45-μl mixture containing 50 ng of genomic DNA (in 10 μl), by standard procedures. After initial denaturation for 3 min at 96°C, Taq polymerase was added at 94°C (hot start), and 35 cycles of amplification were performed, consisting of 40 s at 94°C, 30 s at the optimal annealing temperature (55–60°C), and 30 s elongation at 72°C, followed by 10-min terminal elongation. For the amplification of exon 1, we used the Advantage-GC Genomic PCR kit (Clontech). One microliter of purified PCR product was added to 1 μl of sense or antisense primer (10 μM) and 3 μl of BigDye terminator mixture (PE Applied Biosystems). The linear amplification consisted of initial denaturation for 5 min at 96°C, 25 cycles of denaturation for 10 s at 96°C, and annealing/extension for 4 min at 60°C. The PCR products were purified and sequenced on an Applied Biosystems Sequencer 3700. Both strands from all patients and controls were sequenced for the entire coding region and for the intron/exon boundaries.
Table 1.
Primer Sequences for CGI-58
|
Sequence(5′→3′) |
||
| Forward | Reverse | |
| Exon 1 | ATAAGTCCCGGGCTTGCCCGCCGGCGGCT | GGTGGCTTATACAACAACGGGGCGGACCCTCC |
| Exon 2 | CCATGCTTTGTGCATGTTAG | AAACAAATCTCCTTGGGGTC |
| Exon 3 | AGAGAATGTCTGCCTTGTGG | TGAGGTAGGTCTTCCCCTTT |
| Exon 4 | GGGTTCAGGGTTTTCTTGTT | CGTGAAGGTTTTTGAAGGTG |
| Exon 5 | GACCTGGGGTCAGAAGTTCA | AATGTGTGCTTTTTCCCACC |
| Exon 6 | GTAGTTCACGGTTTGGACAT | CTTAGGTGCTGGAAAAGCTA |
| Exon 7 | TCAGAAATCACTTCCTAAATTGG | TTTAAATACAGTGGCTCTCACTT |
| RT-PCR | CACATGGTGCCCTACGTCTAT | TTTCCCATCAGAGCTTCAGTG |
Extension of Genomic DNA Sequence—and 5′ Rapid Amplification of cDNA Ends (RACE)
Long-range PCR was used to amplify the 5′ genomic sequence up to exon 3 (Takara LA kit). The 8-kb PCR product of intron 1 was purified on an agarose gel and was sheared on a capillary Hydroshear (Genemachines). After repair of extremities, by T4-DNA polymerase and ligation with BstXI adapters (Invitrogen), the product was fractionated and purified on agarose gel to obtain 2-kb fragments. Ligation with the vector pcDNA-2.1 (Invitrogen) was followed by cloning in the DH10B strain of Escherichia coli. After purification of DNA, sequencing was performed, by a LICOR 4200 sequencer, on both ends. Sequences were assembled by PHRED and PHRAP software. We isolated CGI-58 cDNA from four different human tissues—fetal small intestine, leukocyte, liver, and skeletal muscle (Marathon-Ready and Clontech)—by 5′ RACE PCR, by use of the Marathon cDNA amplification kit (Clontech) according to the manufacturer’s instructions (protocol PT1156-1), with antisense primers from the mRNA sequence (nucleotides 288–259 and 64–37; GenBank accession number AF151816).
Reverse-Transcriptase PCR (RT-PCR)
Isolation of RNA from cultured lymphoblastoid cell lines was performed by standard procedures. RT-PCR was performed using the RT-PCR kit (Life Technology) with the forward primer from exon 1 (nucleotides 75–96) and with the reverse primer from exon 7 (nucleotides 1074–1052) (table 1), both from the mRNA sequence AF151816, producing a 977-bp fragment. The RT-PCR conditions were 30 min at 50°C, for reverse transcription, and incubation for 15 min at 95°C, for inactivation of reverse transcriptase, followed by 35 cycles of 1 min at 94°C, 30 s at 60°C, and 50 s at 72°C and by final extension for 10 min at 72°C. RT-PCR fragments were electrophoresed on a 2% agarose gel, were stained with ethidium bromide, and were visualized under a UV transilluminator.
Results
Patients
We recently described a genetic localization, on chromosome 3p21, of an erythrodermic form of ichthyosis in 6 families from a collection of 51 consanguineous families (Fischer et al. 2000). In one patient from a Turkish family (family B4), a more severe erythematous phenotype was observed with ectropion, eclabion, and diffuse alopecia with eroded plaques on the scalp. Examination of a peripheral blood smear in this patient revealed the presence of lipid droplets in granulocytes, suggesting a diagnosis of CDS. Lipid droplets were also found in the other families, with the exception of the Algerian patient from family B5; this latter family was therefore excluded. We also collected four additional families (families B7–B10) for which a diagnosis of CDS had been established. Altogether, nine families from the Mediterranean basin were analyzed, and, in these families, there were 13 affected patients and 35 nonaffected family members.
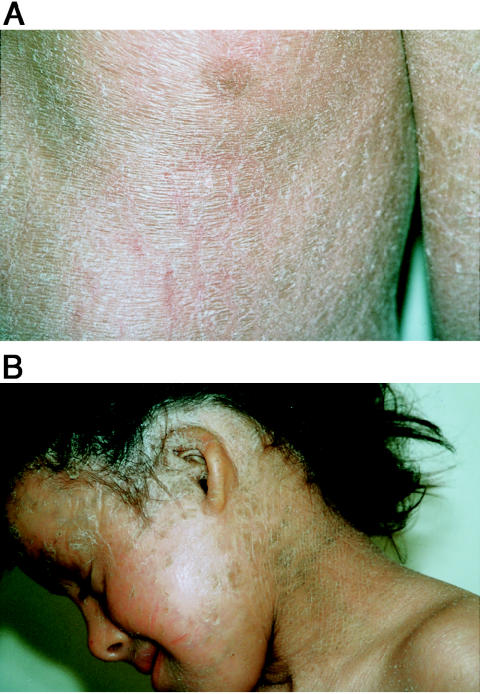
All the affected subjects presented with the clinical features of CDS, as summarized in table 2. Ichthyosiform erythroderma is characterized by fine white scales on an erythematous background, all over the body (fig. 1A)—with the exception of the extension sites of the limbs, where the scales were larger and grayish. Three of these families have been described elsewhere (Kaassis et al. 1998; Venencie et al. 1988; Wollenberg et al. 2000). The two main clinical features (i.e., NCIE and hepatomegaly or liver steatosis) were present in all patients available for liver examination, but with differing severities between families and between patients from the same family. The involvement of other organs and systems (CNS, eyes, ears, and muscle) was much more variable. We found, in six patients, small malformed thick external ears (fig. 1B) and, in two patients, diffuse alopecia with eroded plaques on the scalp, but these symptoms are nonspecific markers for several different types of ichthyosis (Traupe 1989).
Table 2.
Summary of Patients’ Clinical Data[Note]
|
Symptoms |
Vacuoles in |
||||||||
| Family/Patient (Sex) | Age(years) | Place ofOrigin | NCIE? | Liver Disease | Serum Enzyme(s)a (Level[s]) | Eyes and Ears | Other | Leukocytes? | Other Organs? |
| B2 (F) | 16 | Tunisia | Yes | NA | NA | NA | NA | Yes | No |
| B7 (M) | 18 | Tunisia | Yes | Hepatomegaly, liver steatosis | SGOT (<3-fold), SGPT (<2-fold), γGT (<3-fold), CPK (<3-fold), LDH (<2-fold) | Bilateral cataract, mild deafness | Growth retardation | Yes | Skin, liver, bone marrow |
| B3 (M) | 16 | Algeria | Yes | None | AP (<2-fold), CPK (<3-fold) | Ectropion, small ears | Mental retardation | Yes | Skin |
| B8: | |||||||||
| B8.1 (F) | 2 | Algeria | Yes | None | AP (>3-fold), SGOT (<3-fold), SGPT (<2-fold), CPK (<3-fold), LDH (<2-fold) | Small ears | None | Yes | No |
| B8.2 (F) | 8 | Algeria | Yes | None | AP (>3-fold), SGOT (<3-fold), SGPT (<3-fold), CPK (<3-fold) | Small ears | None | Yes | No |
| B6 (F) | 16 | Algeria | Yes | None | AP (>3-fold) | Bilateral sensory deafness | Mental retardation | Yes | No |
| B1: | |||||||||
| B1.1 (F) | 5 | Morocco | Yes | NA | NA | NA | NA | Yes | No |
| B1.2 (F) | 8 | Morocco | Yes | NA | NA | NA | NA | Yes | No |
| B10: | |||||||||
| B10.1 (M) | 16 | Turkey | Yes | Hepatomegaly | NA | Small ears, strabismus | Growth retardation | Yes | Skin |
| B10.2 (F) | 21 | Turkey | Yes | Liver steatosis | AP (<2-fold), CPK (<2-fold), LDH (<2-fold), aldolase (<2-fold) | Small ears | None | Yes | |
| B10.3 (M) | 17 | Turkey | Yes | Hepato-/splenomegaly, liver steatosis | AP (<2-fold), SGOT (<2-fold), CPK (>3-fold), LDH (<2-fold), aldolase (<3-fold) | Small ears | None | Yes | Skin |
| B4 (F) | 11 | Turkey | Yes | Hepatomegaly | AP (<2-fold), SGOT (<3-fold) | Ectropion, alopecia | None | Yes | No |
| B9 (F) | 16 | France | Yes | Hepatomegaly | SGOT (<3-fold), SGPT (<3-fold), AP (<3-fold), aldolase (<3-fold) | Ectropion, strabismus, alopecia | None | Yes | No |
Note.— Patients are listed in the same order as are families in figure 2. NA = not available.
SGOT = serum glutamate oxaloacetate transaminase; SGPT = serum glutamate pyruvate transaminase; γGT = γ-glutamyltransferase; AP = alkaline phosphatase; CPK = creatine phosphokinase; LDH = lactate dehydrogenase.
Figure 1.
A, Patient B3 at age 14 years, exhibiting typical phenotype of NCIE, with fine white scaling on erythrodermal background. B, Patient B8.2 at age 8 years, exhibiting small ears and abnormal pattern of hair distribution.
Linkage and Haplotype Analysis—and Refinement of the Interval
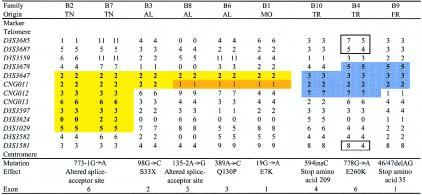
The original cosegregating region of 7.7 cM, on 3p21, was previously defined, in families B2 and B4, by loss of homozygosity in an affected child and was limited by the loci D3S3522 (telomeric) and D3S1581 (centromeric). We performed genotyping in the aforementioned region by use of 18 markers from public databases and of 3 new CA-repeat markers that we identified from a PAC clone (GenBank accession number AC006055). The order of the markers has changed from that described in our previous report (Fischer et al. 2000), and the telomeric limit of the homozygous region is now different. The haplotypes for 13 of the markers are presented in figure 2. The proband in consanguineous family B5, which we had excluded clinically, showed homozygous regions of 11 cM, on chromosome 3p21, and 18 cM, on chromosome 14 (containing the transglutaminase 1 gene, which is associated with lamellar ichthyosis [MIM 242300] [Huber et al. 1995; Russell et al. 1995]); linkage analysis for each localization showed maximum LOD scores of only 0.91 and 1.08, respectively, so we were not able to demonstrate linkage, in this family, to either of these loci. Data on additional families with CDS increased both the initial pairwise LOD score at θ=0, for the marker D3S3564, from 5.43 (after elimination of Algerian family B5) to 7.41 and the maximum LOD score at θ=0, for the marker CNG012, to 11.00. The multipoint LOD score at the same locus was 14.44. All the patients, including those in the two families in which there was no known consanguinity, showed homozygous alleles in the interval between the loci D3S3678 and D3S3582. We observed a common haplotype for seven markers in the two families of Tunisian origin, which further refined the interval between loci D3S3647 and D3S1029, to a region of ∼1 cM, or ∼1.3 Mb. The two Turkish families and the French family shared alleles for two markers (D3S3647 and CNG011), suggesting the possibility of a common ancestor (fig. 2). Results of linkage-disequilibrium analysis of the nine families with CDS were not statistically significant for any of the markers, when the χ2 test was used. After stratification of families according to their geographical origin, however, we observed a Pexcess of 1 for marker D3S3647 in the six families from North Africa, as well as for another allele of the same marker in the three families from Turkey and France. No statistical analysis was performed, because of an insufficient number of subjects.
Figure 2.
Common haplotypes in families from Tunisia (TN), Algeria (AL), Morocco (MO), Turkey (TR), and France (FR). Loss of homozygosity in family B4 is indicated by boxes. Bases are numbered beginning with the ATG initiation codon in exon 1 (A is at position 46 in cDNA sequence). Sequences for CNG (Centre National de Génotypage) microsatellite primers are available on request or from the Centre National de Génotypage website.
Identification of Candidate Genes
Because of the intracellular accumulation of lipid droplets in patients with CDS, we focused our candidate-gene search on enzymes that could be implicated in lipid metabolism (Gibbons et al. 2000). No mutations were found in two candidate genes, ACAA1 and CYP8B1, which were located within the previously defined interval of 7.7 cM. These two genes proved to be outside the reduced region.
The refined interval includes 10 known genes; 4 of them (ZNF197, ZFP, ZNF35, and DHHC1) encode a zinc-finger domain, 1 (HKLP2) encodes a kinesin-like protein, 1 (TGM4) encodes a prostate-specific transglutaminase, 1 (TNA) encodes tetranectin, and 3 (FLJ10375, FLJ22969, and CGI-58) have no known function. CGI-58 is close (150 kb) to marker D3S3647, which showed the maximum Pexcess of 1. CGI-58 attracted our attention because it contains an α/β hydrolase fold, which is also present in several enzymes—notably, lipases and esterases—implicated in lipid metabolism (Ollis et al. 1992; Cygler et al. 1993; Schrag and Cygler 1997). CGI-58 was recently identified by a comparative proteomic approach, in which the Caenorhabditis elegans proteome was used as an alignment template to match known human-gene transcripts from human expressed-sequence–tag nucleotide databases. From the 7,390 sequences evolutionarily conserved in C. elegans, 150 putative full-length human-gene transcripts (named “CGI,” for “comparative gene identification”) were assembled (Lai et al. 2000). Additional searches against the UniGene human-gene database provided information about expression patterns and chromosome localizations. CGI-58 is widely expressed in various tissues, including skin, lymphocytes, liver, skeletal muscle, and brain (SAGEmap). This broad pattern of expression is consistent with the CDS phenotype.
Gene Structure of CGI-58
The available genomic sequence of CGI-58 was incomplete. Five exons (exons 3–7) were contained in BAC clone AC006055, which did not include the first 139 bases of the CGI-58 mRNA (AF151816). A gap of ∼50 kb was present in the physical map. To get the genomic sequence of all the exons, BLAST searches, for either finished or working-draft sequences with the 139-base sequence, were performed and led to the isolation of a genomic DNA sequence (X87485), which matched with 53 bases (positions 1–53) of the mRNA sequence and corresponded to part of exon 1. This sequence also contained 332 bases of the first intron, as well as 29 bases upstream of the initiating methionine codon (instead of 6 bases upstream, as in the aforementioned mRNA). This methionine is embedded in a consensus sequence as described by Kozak (1996). The genomic sequence of exon 2 was not detected in the genomic DNA, but the mRNA sequence (positions 54–139) was considered to correspond to this second exon. We determined the size and the sequence of the two introns between exons 1 and 3, by long-range PCR, with primers from the putative exon 2 sequence and the available parts of introns 1 and 2. The size (45 bases) of the 5′ UTR mRNA was confirmed by 5′ RACE, and this 5′ extension was sequenced. The total length of the mRNA is 1,362 bases. The sequence of the upstream region was determined by sequence walking on the overlapping BAC clone, C-2338A22, from the CIT-D (Caltech Institute of Technology, segment D) library. The complete gene structure and the sizes of the exons and of the introns are shown in table 3. The sequences of the mRNA and of the gene, including the 5′-flanking region with 607 bases upstream of the putative transcription-initiation site, are available from GenBank (accession number AL606838).
Table 3.
Exon-Intron Organization CGI-58
|
Splice Site |
|||||
| Exon | Exon Length(bp) | Starting Position in cDNA | Acceptor | Donor | Intron Length(bp) |
| 1 | 92 | 1 | GGAGAGAG/gtaagcgc | 8,235 | |
| 2 | 86 | 93 | tgctctag/GTCAGGAT | GTTAAAAT/gtaaggct | 2,852 |
| 3 | 373 | 179 | ttcattag/GTGTGCCT | CCATCAAG/gtaagtgg | 9,112 |
| 4 | 155 | 552 | cccaacag/GGTTAATC | ACCCTTTG/gtgagtgc | 3,082 |
| 5 | 112 | 707 | ttaaatag/GTTTAAGT | ACTCCAAG/gtgagggt | 2,600 |
| 6 | 187 | 819 | cgttaaag/TGGTGAGA | AGACAATA/gtaagtgt | 585 |
| 7 | 357 | 1006 | ttatccag/GCTATTCT | ||
| Total mRNA | 1,362 | ||||
CGI-58 Mutations
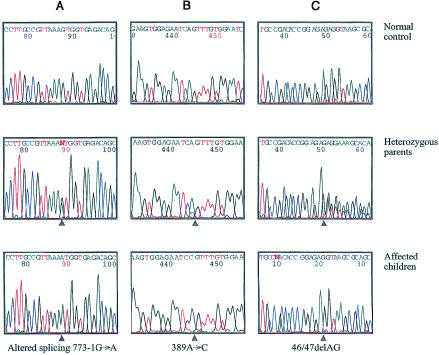
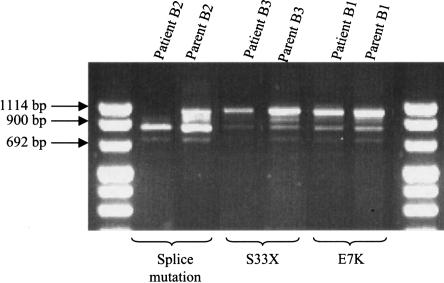
Mutation analysis of the seven coding exons and of the exon-intron boundaries of CGI-58 revealed eight different homozygous mutations in the nine families that we studied, corresponding to eight different haplotypes (fig. 2). The two families of Tunisian origin, which share a common haplotype, have the same splice-site mutation. Mutations were found in each exon, with the exception of exons 5 and 7. There were (a) four point mutations (amino acids 7, 33, 130, and 260), of which one (that at amino acid 33) creates a premature stop codon; (b) a deletion of two nucleotides (46/47delAG) and an insertion of one nucleotide (594insC), leading, in both cases, to a frameshift and a premature stop codon (at amino acid positions 35 and 209, respectively); and (c) two splice-site mutations affecting the invariant A or G of the acceptor–splice-site AG dinucleotide in exons 3 and 6 (figs. 2 and 3). These latter splice-site mutations are expected to lead to aberrant splicing, which was confirmed by RT-PCR (fig. 4).
Figure 3.
CGI-58 mutations in three patients with CDS and in their heterozygous parents. A, Mutation in acceptor splice site of exon 6. B, Point mutation at nucleotide 389, changing glutamine at position 130 to proline. C, Deletion of 2 bp at nucleotides 46/47, leading to frameshift and to premature stop codon at position 35. Arrowheads indicate the positions of the mutations.
Figure 4.
RT-PCR of cultured lymphoblastoid cell lines from three patients and from their heterozygous parents. Patient B2 has a homozygous splice-site mutation, which reduces the size of the mRNA fragment. Both normal and altered mRNA fragments are detected in the heterozygous parent. The size of the normal mRNA fragment is 997 bp.
Sequence Analysis of the CGI-58 Protein—and Identification of Conserved Residues
The CGI-58 open reading frame encodes a 349-amino-acid protein of ∼39 kD. No signal peptide was predicted by any of several sequence-analysis programs, all of which proposed a cytoplasmic localization for the protein. Comparison of the amino acid sequence with public nonredundant protein databases revealed strong homology with several members of a large and rapidly increasing family defined by an α/β hydrolase fold (amino acid residues 102–345) (Ollis et al. 1992; Heikinheimo et al. 1999). The strongest homology found was for the sequence motif (amino acid residues 76–178) bearing one of the three canonical residues that define the catalytic triad formed by the nucleophile/acid/histidine residues, which are present in this order in members of the esterase/lipase/thioesterase subfamily. An LLGHNLGG motif (the nucleophile component of which is included in amino acid residues 149–156), in which asparagine replaces serine, is close to a lipase consensus sequence (LLGHSLGG), in which serine defines the nucleophilic elbow (Schrag and Cygler 1997); although the amide of asparagine is not charged at neutral pH, serine and asparagine are both polar amino acids. For the two other motifs of the catalytic triad, slight sequence homology suggests that the histidine at residue 327, inside the motif GsmXHsmX (amino acid residues 324–328), where “sm” denotes a residue with a small side chain, forms the third element of the triad; the second of these two motifs, the acid of the active-site triad, could be either the aspartate residue at 301, inside the motif GARSCIDG (amino acid residues 295–302), or the glutamic acid residue at 260, which we found mutated in one family, inside the motif PSGETA (amino acid residues 257–262). The latter of these is the least conserved of the three motifs (Cygler et al. 1993; Schrag and Cygler 1997). A His-Gly dipeptide, located 72 residues N-terminal to the catalytic asparagine—and found in many, but not all, lipases—is also conserved and is predicted to define the margins of the lipid-binding pocket (Schrag and Cygler 1997).
Discussion
The principal difficulties encountered in gene discovery in rare disorders include a limited number of patients and families, often with diverse ancestral origins, and a poorly defined phenotype, which increases both clinical and genetic heterogeneity of the disorder. This was the situation in CDS, in which, to date, only ∼30 patients, mainly from the Mediterranean basin, have been reported. The clinical and genetic heterogeneity of ichthyoses is well known and, to a large extent, has been documented (see Traupe 1989). Clinically, patients with CDS present with an erythrodermic form of ichthyosis, with involvement of other organs, such as liver, CNS, eyes, and ears. A diagnosis of CDS is easily confirmed by a simple blood smear, in which the characteristic lipid droplets are observed in the cytoplasm of granulocytes. These droplets are also seen in leukocytes of heterozygous carriers of CDS (Wollenberg et al. 2000).
At least six loci and one gene (TGM1) for ichthyoses are now known. Other genes have been identified for lipid-metabolism disorders in which ichthyosis is part of a syndrome that involves other organs, such as liver, CNS, eyes, and ears. These disorders include the classic form of X-linked ichthyosis (MIM 308100)—due to a deficiency in steroid sulfatase (Webster et al. 1978), which leads to accumulation of cholesterol sulfate—and Refsum disease (MIM 266500)—in which phytanoyl-CoA hydrolase is defective and leads to the accumulation of phytanic-acid cholesterol esters (Jansen et al. 1997). Sjögren-Larsson syndrome (MIM 270200) is associated with mutations in the fatty–aldehyde dehydrogenase gene, FALDH (De Laurenzi et al. 1996). Conradi-Hünermann-Happle syndrome (or rhizomelic chondrodysplasia punctata type II [MIM 302960]) is due to a defect in the 3β-hydroxysteroid-Δ8,Δ7-isomerase (Braverman et al. 1999). One case of congenital hemidysplasia with ichthyosis and limb defects (CHILD) syndrome (MIM 308050) was also reported to be due to a defect in this enzyme (Grange et al. 2000). Mutations in the gene encoding the reduced-nicotinamide-adenine-dinucleotide steroid dehydrogenase–like protein are also known to cause CHILD syndrome (König et al. 2000). All of these syndromes are characterized by an accumulation of lipids, which leads to disorders of cornification (Melnik 1989). There are also lysosomal disorders, such as multiple sulfatase deficiency (MIM 272200) and Gaucher disease type 2 (MIM 230900), in which ichthyosis is part of the clinical picture.
In the nine families that we studied, we found eight different haplotypes, each of which corresponds to a different mutation. However, after stratification of the families according to their geographical origin, haplotype analysis helped us in refining the initial interval.
Progress in genome-sequencing projects has led to numerous interspecies-homology comparisons. In this way, Lai et al. (2000) identified 150 proteins from C. elegans that have homology to human proteins. We have identified mutations in 1 of these 150 proteins, CGI-58, as the cause of CDS.
CGI-58 belongs to a large family of proteins—most of which are enzymes but which also include other proteins, such as glutactin, vitellogenin, neurotactin, and thyroglobulin—characterized by an α/β-fold domain (Zhang et al. 1998; Nardini and Dijkstra 1999). The α/β-hydrolase-fold family is characterized by a catalytic triad composed of a nucleophile, an acid, and histidine that are distant from each other in sequence but close in three-dimensional structure (Zhang et al. 1998). The order of these residues in the sequence defines the various subfamilies. The esterase/lipase/thioesterase subfamily (InterPro accession number IPR000379) includes 94 genes and is the 31st-most-populous InterPro family in the human proteome (International Human Genome Sequencing Consortium 2001). The nucleophile in the catalytic triad is practically always serine, but it is occasionally replaced by cysteine or glutamic acid; it is embedded in a conserved consensus motif, GxSxG. This constitutes the first instance of which we are aware in which asparagine replaces serine in the nucleophile position. Although database searches have revealed an asparagine residue in this motif in various proteins, such proteins usually are hypothetical proteins but also may be proteins without an α/β hydrolase fold. The extended consensus motif LLGHSLGG (with serine instead of asparagine) is found in triglyceride lipases and in lecithin-cholesterol acyltransferase, in both prokaryotic and eukaryotic organisms (Schrag and Cygler 1997). The acid of the triad usually is aspartic acid but also may be glutamic acid; the motifs surrounding the acid and histidine residues are less conserved.
Proteins that are homologous to CGI-58 are present in most species that, to date, have been sequenced, including bacteria, yeast, Arabidopsis thaliana, C. elegans, drosophila, and mice. The highest homology is observed in species most closely related to humans, but, in most cases, the function of the protein is unknown. However, CGI-58 exhibits high homology with epoxide hydrolase and acetyltransferase, in C. elegans and various prokaryotes. There is, on human chromosome 14, a paralog, FLJ12816, that exhibits 56% homology, at the protein level, with CGI-58; this 38.8-kD protein shows remarkable similarity, in sequence and gene structure, to CGI-58, but it contains serine in its catalytic triad and also has an unknown function. Further studies will be needed to determine the exact function of these proteins.
Acknowledgments
We wish to thank the members of the families for their participation in this study. We would like to acknowledge the Généthon DNA Bank, for continuous technical support, and L. Cattolico, for help in sequencing. We are especially grateful to Susan Cure for help in writing the manuscript. This study was supported by the Centre National de Génotypage, Association Française contre les Myopathies, and Généthon.
Electronic-Database Information
The accession numbers and URLs for data in this article are as follows:
- BD Biosciences Clontech, http://www.clontech.com/index.shtml (for protocol PT1156-1)
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Centre National de Génotypage, http://www.cng.fr/ (for sequences of CNG microsatellite primers)
- Ensembl Genome Server, http://www.ensembl.org/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human CGI-58 mRNA [accession number AF151816], human genomic DNA [of chromosome 3 and clone NL1232R] [accession number X87485], human chromosome 3p22 contig 7 PAC RPCI4-672N11 [accession number AC006055], and CGI-58 mRNA and gene sequences [AL606838])
- Genoscope working-draft utility, http://www.genoscope.cns.fr/cgi-bin/WD.cgi/
- Human Genome Project Working Draft (“Golden Path”), http://genome.ucsc.edu/
- InterPro, http://www.ebi.ac.uk/interpro/ (for esterase/lipase/thioesterase subfamily [accession number IPR000379])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CDS [MIM 275630], NCIE2 [MIM 604780], lamellar ichthyosis [MIM 242300], X-linked ichthyosis [MIM 308100], Refsum disease [MIM 266500], Sjögren-Larsson syndrome [MIM 270200], Conradi-Hünermann-Happle syndrome [MIM 302960], CHILD syndrome [MIM 308050], multiple sulfatase deficiency [MIM 272200], and Gaucher disease type 2 [MIM 230900])
- SAGEmap, http://www.ncbi.nlm.nih.gov/SAGE/
- UniGene, http://www.ncbi.nlm.nih.gov/UniGene/
References
- Aksentijevich I, Pras E, Gruberg L, Shen Y, Holman K, Helling S, Prosen L, Sutherland GR, Richards RI, Dean M, Pras M, Kastner DL (1993) Familial Mediterranean fever (FMF) in Moroccan Jews: demonstration of a founder effect by extended haplotype analysis. Am J Hum Genet 53:644–651 [PMC free article] [PubMed] [Google Scholar]
- Braverman N, Lin P, Moebius FF, Obie C, Moser A, Glossmann H, Wilcox WR, Rimoin DL, Smith M, Kratz L, Kelley RI, Valle D (1999) Mutations in the gene encoding 3β-hydroxysteroid-Δ8,Δ7-isomerase cause X-linked dominant Conradi-Hünermann syndrome. Nat Genet 22:291–294 [DOI] [PubMed] [Google Scholar]
- Chanarin I, Patel A, Slavin G, Wills EJ, Andrews TM, Stewart G (1975) Neutral-lipid storage disease: a new disorder of lipid metabolism. Br Med J 1:553–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cygler M, Schrag JD, Sussman JL, Harel M, Silman I, Gentry MK, Doctor BP (1993) Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci 2:366–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V, Rogers GR, Hamrock DJ, Marekov LN, Steinert PM, Compton JG, Markova N, Rizzo WB (1996) Sjögren-Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nat Genet 12:52–57 [DOI] [PubMed] [Google Scholar]
- Dorfman ML, Hershko C, Eisenberg S, Sagher F (1974) Ichthyosiform dermatosis with systemic lipidosis. Arch Dermatol 110:261–266 [PubMed] [Google Scholar]
- Fischer J, Faure A, Bouadjar B, Blanchet-Bardon C, Karaduman A, Thomas I, Emre S, Cure S, Özgüc M, Weissenbach J, Prud'homme J-F (2000) Two new loci for autosomal recessive ichthyosis on chromosomes 3p21 and 19p12-q12 and evidence for further genetic heterogeneity. Am J Hum Genet 66:904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons GF, Islam K, Pease RJ (2000) Mobilisation of triacylglycerol stores. Biochim Biophys Acta 1483:37–57 [DOI] [PubMed] [Google Scholar]
- Grange DK, Kratz LE, Braverman NE, Kelley RI (2000) CHILD syndrome caused by deficiency of 3β-hydroxysteroid-Δ8,Δ7-isomerase. Am J Med Genet 90:328–335 [DOI] [PubMed] [Google Scholar]
- Hästbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E (1992) Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat Genet 2:204–211 [DOI] [PubMed] [Google Scholar]
- Heikinheimo P, Goldman A, Jeffries C, Ollis DL (1999) Of barn owls and bankers: a lush variety of α/β hydrolases. Struct Fold Des 7:141–146 [DOI] [PubMed] [Google Scholar]
- Hilaire N, Salvayre R, Thiers JC, Bonnafé MJ, Nègre-Salvayre A (1995) The turnover of cytoplasmic triacylglycerols in human fibroblasts involves two separate acyl chain length-dependent degradation pathways. J Biol Chem 270:27027–27034 [DOI] [PubMed] [Google Scholar]
- Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SP, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D (1995) Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science 267:525–528 [DOI] [PubMed] [Google Scholar]
- Igal RA, Coleman RA (1996) Acylglycerol recycling from triacylglycerol to phospholipid, not lipase activity, is defective in neutral lipid storage disease fibroblasts. J Biol Chem 271:16644–16651 [DOI] [PubMed] [Google Scholar]
- ——— (1998) Neutral lipid storage disease: a genetic disorder with abnormalities in the regulation of phospholipid metabolism. J Lipid Res 39:31–43 [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:850–92111237008 [Google Scholar]
- Jansen GA, Ofman R, Ferdinandusse S, Ijlst L, Muijsers AO, Skjeldal OH, Stokke O, Jakobs C, Besley GT, Wraith, JE, Wanders RJ (1997) Refsum disease is caused by mutations in the phytanoyl-CoA hydroxylase gene. Nat Genet 17:190–193 [DOI] [PubMed] [Google Scholar]
- Jordans GHW (1953) The familial occurrence of fat-containing vacuoles in the leukocytes diagnosed in two brothers suffering from dystrophia musculorum progressiva (Erb). Acta Med Scand 145:419–423 [DOI] [PubMed] [Google Scholar]
- Kaassis C, Ginies JL, Berthelot J, Verret JL (1998) Le syndrome de Dorfman-Chanarin. Ann Dermatol Venereol 125:317–319 [PubMed] [Google Scholar]
- König A, Happle R, Bornholdt D, Engel H, Grzeschik KH (2000) Mutations in the NSDHL gene, encoding a 3β-hydroxysteroid dehydrogenase, cause CHILD syndrome. Am J Med Genet 90:339–346 [PubMed] [Google Scholar]
- Kozak M (1996) Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome 7:563–574 [DOI] [PubMed] [Google Scholar]
- Lai CH, Chou CY, Ch'ang LY, Liu CS, Lin W (2000) Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res 5:703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1985) Multilocus linkage analysis in humans: detection and estimation of recombination. Am J Hum Genet 37:482–498 [PMC free article] [PubMed] [Google Scholar]
- Melnick B (1989) Epidermal lipids and the biochemistry of keratinization. In: Traupe H (ed) The ichthyoses: a guide to clinical diagnosis, genetic counseling, and therapy. Springer, Heidelberg, pp 14–42 [Google Scholar]
- Miranda A, DiMauro S, Eastwood A, Hays A, Johnson WG, Olarte M, Whitlock R, Mayeux R, Rowland LP (1979) Lipid storage myopathy, ichthyosis, and steatorrhea. Muscle Nerve 1:1–13 [DOI] [PubMed] [Google Scholar]
- Nardini M, Dijkstra BW (1999) α/β Hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 6:732–737 [DOI] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A (1992) The α/β hydrolase fold. Protein Eng 3:197–211 [DOI] [PubMed] [Google Scholar]
- Rozenszajn L, Klajman A, Yaffe D, Efrati P (1966) Jordans' anomaly in white blood cells: report of case. Blood 28:258–265 [PubMed] [Google Scholar]
- Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, Bale SJ (1995) Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet 9:279–283 [DOI] [PubMed] [Google Scholar]
- Schrag JD, Cygler M (1997) Lipases and α/β hydrolase fold. Methods Enzymol 284:85–107 [DOI] [PubMed] [Google Scholar]
- Traupe H (1989) The ichthyoses: a guide to clinical diagnosis, genetic counseling, and therapy. Springer, Heidelberg [Google Scholar]
- Venencie PY, Armengaud D, Foldès C, Vieillefond A, Coulombel L, Hadchouel M (1988) Ichthyosis and neutral lipid storage disease (Dorfman-Chanarin syndrome). Pediatr Dermatol 5:173–177 [DOI] [PubMed] [Google Scholar]
- Webster D, France JT, Shapiro LJ, Weiss R (1978) X-linked ichthyosis due to steroid-sulphatase deficiency. Lancet 1:70–72 [DOI] [PubMed] [Google Scholar]
- Weir BS (1990) Genetic data analysis. Sinauer, Sunderland, MA [Google Scholar]
- Williams ML, Monger DJ, Rutherford SL, Hincenbergs M, Rehfeld SJ, Grunfeld C (1988) Neutral lipid storage disease with ichthyosis: lipid content and metabolism of fibroblasts. J Inherit Metab Dis 11:131–143 [DOI] [PubMed] [Google Scholar]
- Wollenberg A, Geiger E, Schaller M, Wolff H (2000) Dorfman-Chanarin syndrome in a Turkish kindred: conductor diagnosis requires analysis of multiple eosinophils. Acta Dermatol Venereol 80:39–43 [DOI] [PubMed] [Google Scholar]
- Zhang L, Godzik A, Skolnick J, Fetrow JS (1998) Functional analysis of the Escherichia coli genome for members of the α/β hydrolase family. Fold Des 3:535–548 [DOI] [PubMed] [Google Scholar]