Abstract
Arginine:glycine amidinotransferase (AGAT) catalyzes the first step of creatine synthesis, resulting in the formation of guanidinoacetate, which is a substrate for creatine formation. In two female siblings with mental retardation who had brain creatine deficiency that was reversible by means of oral creatine supplementation and had low urinary guanidinoacetate concentrations, AGAT deficiency was identified as a new genetic defect in creatine metabolism. A homozygous G-A transition at nucleotide position 9297, converting a tryptophan codon (TGG) to a stop codon (TAG) at residue 149 (T149X), resulted in undetectable cDNA, as investigated by reverse-transcription PCR, as well as in undetectable AGAT activity, as investigated radiochemically in cultivated skin fibroblasts and in virus-transformed lymphoblasts of the patients. The parents were heterozygous for the mutant allele, with intermediate residual AGAT activities. Recognition and treatment with oral creatine supplements may prevent neurological sequelae in affected patients.
The creatine/creatine-phosphate system plays an important role in the storage and transmission of phosphate-bound energy (fig. 1). In humans, creatine is synthesized mainly in the liver and pancreas, in a process involving two reactions catalyzed by arginine:glycine amidinotransferase and by guanidinoacetate methyltransferase (Walker 1979). Creatine is transported via the blood, and its accumulation in the main target organs—the muscle and brain—is provided by an active transport process (Guimbal and Kilimann 1993) mediated by the creatine transporter protein (Sora et al. 1994). Guanidinoacetate methyltransferase (GAMT [MIM 601240]) deficiency is the first inborn error of creatine metabolism (von Figura et al. 2000), originally recognized in two unrelated patients with mental retardation, epilepsy, and extrapyramidal symptoms (Stöckler et al. 1996b). GAMT deficiency is characterized by accumulation of guanidinoacetate, which is the substrate for the activity of the deficient enzyme (Ganesan et al. 1997; Schulze et al. 1997; Stöckler et al. 1997; Leuzzi et al. 2000; van der Knaap et al. 2000), as well as by severe brain creatine deficiency, which is reversible by means of oral creatine substitution (Stöckler et al. 1994, 1996a). Recently, a second inborn error of creatine metabolism, the X-linked creatine transporter (MIM 300036) defect, has been described in a boy with mental retardation (Salomons et al. 2001). In the patients with the X-linked creatine transporter defect, brain creatine deficiency is not reversible by oral creatine substitution (Cecil et al. 2001).
Figure 1.
Metabolic pathway of creatine/phospho-creatine. CRTR, creatine transporter; CK, creatine kinase.
Bianchi et al. (2000) reported two female siblings, aged 4 and 6 years, with mental retardation and severe creatine deficiency in the brain. At the time of the present study, the molecular cause of the creatine deficiency syndrome in these two siblings remained unidentified: GAMT deficiency was excluded by the presence of normal enzyme activity, as measured in virus-transformed lymphoblasts (table 1), and X-linked creatine transporter deficiency was unlikely because of a prompt increase of brain creatine by means of oral creatine substitution. Retrospective investigation of the patients' biological material finally disclosed extremely low levels of urinary guanidinoacetate excretion (table 1), which, together with the absent resonance of creatine/creatine phosphate in the brain, as shown by in vivo proton magnetic resonance spectroscopy (Bianchi et al. 2000), is a diagnostic indicator of arginine:glycine amidinotransferase (AGAT [MIM 602360]) deficiency. AGAT deficiency is a novel disorder of creatine synthesis, which, until now, has not been described in humans. AGAT catalyzes the first of the two reactions in creatine biosynthesis, including the transfer of the amidino group from arginine to glycine, to form ornithine and guanidinoacetate, the immediate precursor of creatine. The human AGAT gene is localized on chromosome 15q15.3, and its genomic sequence has recently been identified (AGAT genomic DNA [GenBank accession number 1432573]). The AGAT genomic DNA is 16,858 bp long and consists of nine exons.
Table 1.
AGAT and GAMT activity and Urinary Guanidinoacetate Concentrations in Two Female Siblings with AGAT Deficiency[Note]
|
Activity(nmol/hr/mg protein) |
|||
| AGAT | GAMT | GuanidinoacetateExcretion in Urine(μmol/g creatinine,μmol/liter) | |
| Patient 1 | <.3 | .53 | 16, 13 |
| Patient 2 | <.3 | .64 | 17, 9 |
| Father | 4.9 | ND | ND |
| Mother | 6.3 | ND | ND |
| Controls | 12.6–23.4a | .61–.84b | 60–850, 50–500 |
Note.— Enzyme extracts were obtained from virus-transformed lymphoblasts by filtration (Millipore, cutoff 10,000 Daltons) of cell homogenate supernatants in 0.1 M Tris/HCl pH 8.0, 2 mM DTT, 0.1 mM EDTA, and incubated for 6 h with 100 mM arginine and 14C-glycine (3 μCi/ml). The formation of guanidinoacetate was measured by determination of radioactivity (14C) in the guanidinoacetate fraction after separation from glycine by high-performance liquid chromatography (Hypersil ODS reverse-phase column, 250×4 mm, 5 μm, Hewlett-Packard). The formation of guanidinoacetate was proportional to incubation time (provided that incubation time did not exceed 6 h) and to the amount of sample extract (provided that substrate turnover was not >10%). GAMT activities were determined as described by Ilas et al. (2000). Guanidinoacetate concentrations were measured in 24-h urine samples, by GC-MS (Hunneman and Hanefeld 1997) and by stable-isotope dilution (Struys et al. 1998). ND = not determined.
n=5.
n=8.
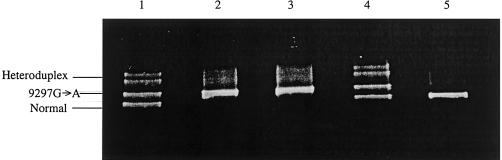
To further test the hypothesis that the creatine deficiency in both of these patients is due to AGAT deficiency, we first screened exon 1 by direct sequencing and screened exons 2–9 and adjoining introns of the human AGAT gene by denaturing gradient gel electrophoresis (DGGE) (for primer sequences and DGGE conditions, see table 2). Investigation of the PCR product from exon 3 of the AGAT gene in both patients revealed a single band whose position on DGGE differed from that of the wild-type allele (fig. 2, lanes 2, 3, and 5), whereas the bands representing the PCR products from the remaining exons did not differ from the wild-type alleles (data not shown). DGGE analysis in DNA samples from both parents showed two bands: one characteristic of the wild-type allele and the other identical to the mutant allele found in both index patients (fig. 2, lanes 1 and 4). Sequencing of all putative sequence variations as pointed out by the DGGE analysis confirmed patients 1 and 2 to be homozygous for a G→A transition at nucleotide position 9297, which converts a tryptophan (TGG) codon to a stop (TAG) codon at residue 149 (T149X) (fig. 3a). This nucleotide change predicts a severely truncated protein that lacks the active-site cysteine residue 407 on exon 9 (Humm et al. 1997). Sequence analysis of the parents' DNA showed heterozygosity for the wild-type and the mutant nucleotide sequence (fig. 3c).
Table 2.
Oligonucleotide Primers for the Amplification of Exons 1–9 of the AGAT Gene and Respective DGGE Conditions[Note]
|
Primer Sequencea |
||||||
| Fragment | Forward | Reverse | Length ofAmplifiedSegment(bp) | Denaturant Range(%) | Melting Point(°C) | PCR AnnealingTemperature(°C) |
| Exon 1 | 5′-CACCACCTGTTCCCGGCAGC-3′ | 5′-TCTCGTGCAATTCCGGCTAG-3′ | 381 | NAb | 85 | 64 |
| Exon 2 | 5′-CTGATAGAGCAATGCAATAC-3′ | 5′-(GC)AAGCAGTCAGAGGGTAGCAG-3′ | 337 | 25–65 | 76 | 55 |
| Exon 3a, 5′ end | 5′-TAAGTATTGTGTTTATTTGC-3′ | 5′-(GC)CTTCAATGACCAGTCAATGG-3′ | 240 | 15–55 | 68 | 50 |
| Exon 3b, 3′ end | 5′-ATTGAAGAAATGTGCAATAT-3′ | 5′-(GC) AGCTTTCCCCTTACACATTA-3′ | 220 | 15–55 | 69 | 50 |
| Exon 4a, 5′ end | 5′-(GC)ACACTACTGAAGTTGTCATG-3′ | 5′-TAAAGCTCATCAGCCATTGT-3′ | 263 | 25–65 | 73 | 55 |
| Exon 4b, 3′ end | 5′-TCACGCTTCTTTGAGTACCG-3′ | 5′-(GC)ACTCTTTGTGGATCATTTAG-3′ | 213 | 25–65 | 72 | 60 |
| Exon 5 | 5′-(GC)TTCTCATGATCCAGTGCATT-3′ | 5′-TTTTAATTTGGAAGTAAAGA-3′ | 289 | 25–65 | 72 | 50 |
| Exon 6 | 5′-(GC)AATATTTATCGATCGTATGT-3′ | 5′-TTAAAATAAAAGTGGCAATT-3′ | 314 | 25–65 | 71 | 50 |
| Exon 7 | 5′-(GC)TGATAAGGGCTATTAGAGAC-3′ | 5′-AAGCACCACTTCACACCTTA-3′ | 317 | 15–55 | 67 | 60 |
| Exon 8 | 5′-TCTTCACTTGTAAATCCTTG-3′ | 5′-(GC)TAACTTGGTGCCTCTAAAAG-3′ | 360 | 15–55 | 67 | 55 |
| Exon 9a, 5′ end | 5′-GAACCACAGGACTCCTCCAA-3′ | 5′-(GC)TAGGACTGTAAGGTGCCTCG-3′ | 238 | 15–55 | 69 | 60 |
| Exon 9b, 3′ end | 5′-TTCGTAATGCCAATTCCCTG-3′ | 5′-(GC)AAAGCACTACAGTTCATGCA-3′ | 230 | 25–65 | 75 | 60 |
Note.— GC clamp = 5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-3′.
Primer sequences according to human genomic AGAT gene sequence. All primers are intronic, with the exception of reverse primers of exons 3a, 4a, 9a and forward primers of exons 3b, 4b, 9b, which were placed on exon regions.
NA = not analyzed by DGGE, but directly sequenced.
Figure 2.
Identification of AGAT nonsense mutation in lymphoblasts of two female siblings and leukocytes of their parents, by DGGE analysis of exon 3. Lane 1, Father, genotype T149X/N. Lane 2, Patient 1, genotype T149X/T149X. Lane 3, Patient 2, genotype T149X/T149X. Lane 4, Mother, genotype T149X/N. Lane 5, Wild-type DNA from unaffected control, genotype N/N. DNA extraction from EBV-transfected lymphoblasts and cultivated fibroblasts was performed using guanidinhydrochloride/proteinase K (Nucleospin C+T kit [Machery Nagel]). PCR primers were designed to cover exons 1–9 and the adjoining introns of the AGAT gene. PCR reactions were carried out over 37 cycles with 2.5 U of Ampli-Taq Gold DNA polymerase (Perkin Elmer) in a Perkin-Elmer-Cetus DNA Thermal Cycler. DGGE analysis of PCR products was essentially performed as described by Greber-Platzer et al. (1997), after heteroduplex formation by loading 20 μl of sample onto an 8% polyacrylamide gel containing a 15%–55% or 25%–65% linearly increasing denaturing gradient (100% = 7 M urea and 40% (vol/vol) deionized formamide). Electrophoresis was performed at 160 V for 6 h in a 1× TAE buffer at 60°C, using a DCode system (Biorad). After electrophoresis, gels were stained in ethidium bromide and examined under ultraviolet illumination. DGGE analysis of exon 1 was not possible, because of its high melting temperature; therefore, direct sequencing of exon 1 was performed. In the patients, no aberrations from the normal sequence were found on exon 1.
Figure 3.
Direct sequencing of PCR products showing putative sequence variations, as pointed out by DGGE analysis, was performed by double-stranded automated cycle sequencing of the PCR products on a ABI 370 sequencer (Applied Biosystems). a, Patients 1 and 2. b, Control. c, Parents.
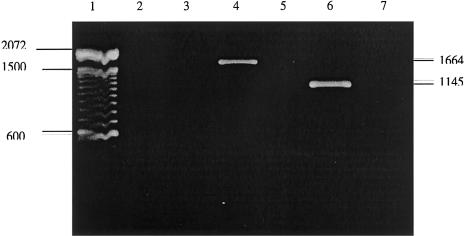
To investigate the effect of the genomic 9297 G→A nucleotide change at the cDNA level, we analyzed total lymphoblast RNA of both patients and of an unaffected control sample, by means of PCR following reverse transcription (RT-PCR). When primers flanking the entire cDNA sequence of the AGAT gene were used (AGAT mRNA [GenBank accession number S68805]), no transcript was detected at the cDNA level in the patients, whereas a normal-sized 1,664-bp cDNA sequence was detected in the unaffected control sample (fig. 4, lanes 2, 3, and 4). To exclude the possibility that 40 rounds of RT-PCR did not yield enough product from both patients, 35 additional rounds of PCR were performed, using a second pair of primers nested within the first primer pair. This again revealed no detectable cDNA in the patients, as well as the expected 1,145-bp cDNA in the unaffected control sample (fig. 4, lanes 5, 6, and 7). The finding of no detectable cDNA by RT-PCR suggests either complete absence of the AGAT gene or the presence of unstable RNA transcripts in the affected patients, which are subject to nonsense-mediated decay (Osborn and Upadhyaya 1999). To investigate the effect of the gene mutation at the translational level, we measured the biological activity of AGAT enzyme protein in cultivated fibroblasts and in virus-transformed lymphoblasts from both patients and their parents, by a radiochemical assay based on the separation, by high-performance liquid chromatography (HPLC), of radioactively labeled substrate from reaction product. Whereas AGAT activity was clearly measurable in normal control cell lines, no detectable activity was found in the cell extracts from the patients (table 1). Cell extracts from both parents showed intermediate residual activity, as would be expected in the heterozygous state.
Figure 4.
RT-PCR analysis to examine the expression of the T149X allele in lymphoblasts of both siblings. Lane 1, 100-bp DNA ladder. Lane 2, RT-PCR, patient 1. Lane 3, RT-PCR, patient 2. Lane 4, RT-PCR, unaffected control. Lane 5, Nested PCR, patient 1. Lane 6, Nested PCR, unaffected control. Lane 7, Nested PCR, patient 2. Total RNA extraction was performed from EBV-transfected lymphoblasts (Molecular Research Center, Inc.), using RNAzolB. A one step RT-PCR was performed using the Qiagen kit (Qiagen, Inc.). Two micrograms of the extracted RNA in aqueous solution was reverse transcribed in a volume of 50 μl containing 800 nM each of the AGAT specific primers on the 5′-UTR and 3′-UTR of the total RNA (forward primer: 5′-GAGCGACGCGGCCCAGAGGCCAGGAACATT-3′; reverse primer: 5′-CCTTTTGGAAAGAAAATTAAAAACAGAGGT-3′), 400 μM of each dNTP, 10 U of RNase inhibitor, 10 μl of 5× Qiagen buffer, and 2 μl RT-PCR enzyme mix, at 50°C for 30 min. A PCR activation step at 95°C for 15 min and 40 cycles of PCR consisting of denaturation at 94°C for 1 min, annealing at 65°C for 1 min, extension at 72°C for 1 min, followed by a final elongation at 72°C for 10 minutes were performed (PCR product 1,664 bp). In a second round of PCR, 35 cycles were performed in a PCR mixture containing 0.5 μl of PCR product from the first round of amplification, 400 nM of each nested primer on exons 1 and 9 (forward primer: 5′-ATGCTGCGGGTGCGGTGTCTGCGCGGCGGG-3′; reverse primer: 5′-ATCTTTTGAATTGGAACTTCATTGGCATCC-3′), 10 mM Tris-HCl, pH 8.3, 1.5 mM KCl, 2 mM MgCl2, 200 μM of each deoxyribonucleotide, and 2.5 U of AmpliTaq Gold DNA polymerase in a volume of 50 μl (PCR product 1,145 bp) . RT-PCR products were run on a 1% agarose gel and were visualized by ethidium bromide staining.
In addition to GAMT and creatine transporter deficiency, AGAT deficiency has been predicted as the third inborn error of creatine metabolism causing cerebral creatine deficiency syndromes (Stöckler-Ipsiroglu 1997). In the present report on two siblings with mental retardation, we have finally demonstrated AGAT deficiency as the third inborn error of creatine synthesis. In both patients, we were able to show by RT-PCR that the homozygous mutation resulting in a stop codon on exon 3 results in complete absence of AGAT or in unstable transcripts without detectable functional activity at the enzyme protein level. These findings confirm that the genomic mutation found in both patients fulfills the disease-causing criteria as described elsewhere (Cotton and Scriver 1998). Heterozygosity for the mutant allele and intermediate residual AGAT activities in the clinically asymptomatic parents indicate autosomal recessive inheritance of AGAT deficiency.
In both patients, deficient guanidinoacetate production was clearly reflected by extremely low plasma and urinary guanidinoacetate concentrations. Therefore, assessment of guanidinoacetate concentrations in body fluids may represent a valuable tool for identification of patients at risk. Care must be taken to choose a method that is sensitive enough to safely detect values below the normal range. The gas chromatographic–mass spectrometric (GC-MS) stable-isotope dilution method (Struys et al. 1998), as applied in these patients, as well as isotope dilution electrospray tandem mass spectrometry (Bodamer et al. 2001), may represent such methods. In contrast, accumulation of glycine and arginine, which are substrates for activity of the deficient enzyme, are alternatively metabolized. Therefore, significant accumulation of these amino acids does not occur, and amino acid analysis will not identify patients at risk for AGAT deficiency. Plasma creatine concentrations were reported to be normal in both patients (Bianchi et al. 2000); therefore, determination of plasma creatine concentrations may not be useful as a screening test for AGAT deficiency.
To provide a noninvasive diagnosis of human AGAT deficiency, we have developed a sensitive enzyme assay for the determination of AGAT activity in cultivated fibroblasts and in virus-transformed lymphoblasts, by radioactive-substrate labeling of glycine and separation, by HPLC, of radioactivity in the guanidinoacetate fraction. By this method, the radioactivity in the guanidinoacetate fraction of the reaction samples from normal controls was ∼30 times higher than the radioactivity found in the respective blank samples. This high sensitivity was the prerequisite for reliable measurements in fibroblasts and lymphoblasts, which have a lower expected expression of enzyme activity, as compared with organs like liver, kidney, and pancreas, which have the highest activity levels in the body (Van Pilsum et al. 1972).
The DGGE method, in comparison with other mutation screening methods, detects almost 100% of mutations, requires no label, and separates mutant molecules from wild-type molecules for analysis (Cotton 2000). In the DGGE method developed for mutation screening of AGAT deficiency, increased electrophoretic resolution was achieved by heteroduplex formation. As a result, the zygosity of the patient sample could rapidly be determined before final confirmation by sequencing (Guldberg and Güttler 1993)
In both patients, almost complete restoration of extremely low pretreatment cerebral creatine levels by means of oral creatine supplementation was paralleled by a favorable clinical response, as shown by significant improvement of highly abnormal developmental scores (Bianchi et al. 2000). This observation suggests that AGAT deficiency is a treatable inborn error of creatine synthesis and that early recognition and treatment of this disease might effectively prevent neurological sequelae. Determination of guanidinoacetate in body fluids, the enzyme assay and DGGE mutation screening method developed for the biochemical and molecular characterization of the first two index patients, will provide valuable tools for the noninvasive diagnosis of this new disorder.
Acknowledgments
This work has been supported by Austrian National Bank Scientific Trust grants ONB 8676 (to C.B.I.) and ONB 7115 (to C.S.) and by Italian Ministry of Health grants RC 1/2000 (to G.C.) and RC 4/2000 (to M.T.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for AGAT genomic DNA [accession number 1432573] and AGAT mRNA [accession number S68805])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AGAT [MIM 602360], GAMT [MIM 601240], and X-linked creatine transporter [MIM 300036])
References
- Bianchi MC, Tosetti M, Fornai F, Alessandrì MG, Cipriani P, De Vito G, Canapicchi R (2000) Reversible brain creatine deficiency in two sisters with normal blood creatine level. Ann Neurol 47:511–513 [PubMed] [Google Scholar]
- Bodamer OA, Bloesch SH, Stöckler-Ipsiroglu S, O'Brien WE (2001) Analysis of guanidinoacetate and creatine by isotope dilution electrospray tandem mass-spectrometry. Clin Chim Acta 308:173–178 [DOI] [PubMed] [Google Scholar]
- Cecil KM, Salomons G, Ball WS, Wong B, Chuck G, Verhoeven NM, Jakobs C, DeGrauw TJ (2001) Irreversible brain creatine deficiency with elevated serum and urine creatine: a creatine transporter defect? Ann Neurol 49:401–404 [DOI] [PubMed] [Google Scholar]
- Cotton RG (2000) Methods in clinical molecular genetics. Eur J Pediatr 159:S179–S182 [DOI] [PubMed] [Google Scholar]
- Cotton RGH, Scriver CR (1998) Proof of “disease causing” mutation. Hum Mutat 12:1–3 [DOI] [PubMed] [Google Scholar]
- Ganesan V, Johnson A, Connelly A, Eckhardt S, Surtees RAH (1997) Guanidinoacetate methyltransferase deficiency: new clinical features. Pediatr Neurol 17:155–157 [DOI] [PubMed] [Google Scholar]
- Greber-Platzer S, Guldberg P, Scheibenreiter S, Item C, Schuller E, Patel N, Strobl W (1997) Molecular heterogeneity of classical and duarte galactosemia: mutation analysis by denaturing gradient gel electrophoresis. Hum Mutat 10:49–57 [DOI] [PubMed] [Google Scholar]
- Guimbal C and Kilimann MW (1993) A Na+-dependent creatine transporter in rabbit brain, muscle, heart, and kidney. J Biol Chem 268:8418–8421 [PubMed] [Google Scholar]
- Guldberg P, Güttler F (1993) A simple method for identification of point mutations using denaturing gradient gel electrophoresis. Nucleic Acids Research 21:2261–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humm A, Fritsche E, Mann K, Gohl M, Huber R (1997) Recombinant expression and isolation of human l-arginine:glycine amidinotransferase and identification of its active-site cysteine residue. Biochem J 322:771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunneman DH and Hanefeld F (1997) GC-MS determination of guanidinoacetatein urine and plasma. J Inher Metab Dis 20:450–452 [DOI] [PubMed] [Google Scholar]
- Ilas J, Mühl A, Stöckler-Ipsiroglu S (2000) Guanidinoacetate methyltransferase (GAMT) deficiency: non invasive enzymatic diagnosis of a newly recognized inborn error of metabolism. Clin Chim Acta 290:179–188 [DOI] [PubMed] [Google Scholar]
- Leuzzi V, Bianchi MC, Tosetti M, Carducci C, Cerquiglini C, Cioni G, Antonozzi I (2000) Brain creatine depletion: guanidinoacetate methyltransferase deficiency (improving with creatine supplementation). Neurology 55:1407–1409 [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Upadhyaya M (1999) Evaluation of the protein truncation test and mutation detection in NF1 gene: mutational analysis of 15 known and 40 unknown mutations. Hum Genet 105:327–332 [DOI] [PubMed] [Google Scholar]
- Salomons GJ, van Dooren SJM, Verhoeven NM, Cecil KM, Ball WS, DeGrauw TJ, Jakobs C (2001) X-linked creatine transporter (SLC6A8 gene) defect: a new creatine deficiency syndrome. Am J Hum Genet 68:1497–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Hess T, Wevers R, Mayatepek E, Bachert P, Marescau B, Knopp MV, De Deyn PP, Bremer HJ, Rating D (1997) Creatine deficiency syndrome caused by guanidinoacetate methyltransferase deficiency: diagnostic tools or a new inborn error of metabolism. J Pediatr 131:626–631 [DOI] [PubMed] [Google Scholar]
- Sora I, Richman J, Santoro G, Wei G, Wang Y, Vanderah T, Horwath R, Nguyen M, Waite S, Roeske WR, Yamamüra HI (1994) The cloning and expression of a human creatine transporter. Biochem Biophys Res Commun 204:419–427 [DOI] [PubMed] [Google Scholar]
- Stöckler S, Hanefeld F, Frahm J (1996a) Creatine replacement therapy in guanidinoacetate methyltransferase deficiency, a novel inborn error of metabolism. Lancet 348:789–790 [DOI] [PubMed] [Google Scholar]
- Stöckler S, Holzbach U, Hanefeld F, Marquardt I, Helms G, Requart M, Hänicke W, Frahm J (1994) Creatine deficiency in the brain: A new treatable inborn error of metabolism. Pediatr Res 36:409–413 [DOI] [PubMed] [Google Scholar]
- Stöckler S, Isbrandt D, Hanefeld F, Schmidt B, von Figura K (1996b) Guanidinoacetate methyltransferase deficiency: the first inborn error of creatine metabolism in man. Am J Hum Genet 58:914–922 [PMC free article] [PubMed] [Google Scholar]
- Stöckler S, Marescau B, DeDeyn PP, Trijbels JMF, Hanefeld F (1997) Guanidino compounds in guanidinoacetate methyltransferase deficiency, a new inborn error of creatine synthesis. Metabolism 46:1189–1193 [DOI] [PubMed] [Google Scholar]
- Stöckler-Ipsiroglu S (1997) Creatine deficiency syndromes: A new perspective on metabolic disorders and a diagnostic challenge. J Pediatr 131:510–511 [PubMed] [Google Scholar]
- Struys EA, Jansen EEW, Ten Brink HJ, Verhoeven NM. van der Knapp MS, Jacobs C (1998) An accurate stable isotope dilution gas chromatograpic-mass spectrometric approach to the diagnosis of guanidinoacetate methyltransferase deficiency. J Pharm Biomed Anal 18:659–665 [DOI] [PubMed] [Google Scholar]
- Walker JB (1979) Creatine: biosynthesis, regulation, and function. Adv Enzymol 50:177–242 [DOI] [PubMed] [Google Scholar]
- van der Knaap, M.S, Verhoeven NM, Maaswinkel-Mooij P, Powels PJW, Onkenhout W, Peeters EA, Stöckler-Ipsiroglu S, Jakobs C (2000) Mental retardation and behavioural problems as presenting signs in a creatine synthesis defect. Ann Neurol 47:540–543 [DOI] [PubMed] [Google Scholar]
- Van Pilsum JF, Stephens GC, Taylor D (1972) Distribution of creatine, guanidinoacetate and the enzymes for their biosynthesis in the animal kingdom. Implications for phylogeny. Biochem J 126:325–345 [PubMed] [Google Scholar]
- von Figura K, Hanefeld F, Isbrandt D, Stöckler-Ipsiroglu S (2000) Guanidinoacetate methyltransferase deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular basis of inherited disease. McGraw Hill, New York, pp 1897–1908 [Google Scholar]