Abstract
X inactivation is the mammalian method for X-chromosome dosage compensation, but some features of this developmental process vary among mammals. Such species variations provide insights into the essential components of the pathway. Tsix encodes a transcript antisense to the murine Xist transcript and is expressed in the mouse embryo only during the initial stages of X inactivation; it has been shown to play a role in imprinted X inactivation in the mouse placenta. We have identified its counterpart within the human X inactivation center (XIC). Human TSIX produces a >30-kb transcript that is expressed only in cells of fetal origin; it is expressed from human XIC transgenes in mouse embryonic stem cells and from human embryoid-body–derived cells, but not from human adult somatic cells. Differences in the structure of human and murine genes indicate that human TSIX was truncated during evolution. These differences could explain the fact that X inactivation is not imprinted in human placenta, and they raise questions about the role of TSIX in random X inactivation.
Introduction
X inactivation is the mammalian method for equalization of the dosage of X-linked genes in males and females; this is accomplished by down-regulating the transcriptional output of X chromosomes in females, so that only one X is active in diploid somatic cells of both sexes (Lyon 1962). This transcriptional silencing is initiated in female embryonic blastocysts and can be induced in murine embryonic stem (ES) cells. Inactivation is manifested by underacetylated histones, methylated CpG islands, and delayed DNA replication, as reviewed by Heard et al. (1997). The initial events responsible for a single active X in mammalian cells are largely unknown, but some players have been identified. The key control region on the X chromosome, from which inactivation is initiated and signals are spread, has been mapped to Xq13.2 and is referred to as the “X inactivation center” (XIC in humans and Xic in mouse). X chromosomes can be programmed to inactivate because of the X inactive specific transcript gene (XIST in human and Xist in mouse [MIM 314670]) located within this region. The expression of XIST starts a cascade of events that results in silencing of the chromosome from which it is transcribed.
Prior to the time that an inactive X is seen in female embryos, XIST is expressed at low levels from all X chromosomes, even from the single X in male embryos (Ray et al. 1997). Therefore, it seems likely that an early step in this developmental pathway is one that ensures that the XIST locus on one X chromosome is turned off in cells of both sexes; in this way, one and only one X chromosome will be functional. The XIST allele that is repressed in female cells is usually chosen randomly; all other X chromosomes in the cell are inactivated by default, via propagation of the XIST-induced inactivating signal(s) along the chromosome. The choice is nonrandom in mouse placental tissues, where the Xist locus on the maternal X is always repressed; hence, the maternal X is always active. The XIST RNA is untranslated and remains in the nucleus, associated with the inactive X (Clemson et al. 1996). The stabilization and accumulation of this RNA around the inactive X is thought to propagate the inactivating signal along the chromosome (Panning et al. 1997).
Clearly, XIST is not the only player in this process; if it were, there would be no active X chromosome, since all X chromosomes would be silenced. Something else is needed to repress the XIST locus on the active X in both sexes. The X controlling element (Xce), a locus downstream of murine Xist, affects the randomness of inactivation in a manner not yet known (Simmler et al. 1993). The orthologous locus has not been identified on the human X chromosome. In addition, Lee et al. (1999) discovered Tsix (MIM 300181), the gene encoding an RNA that is, in part, antisense to the murine Xist transcript. Tsix was initially described as a 40-kb transcript starting downstream of Xist and overlapping the entire Xist locus. More recently, Sado et al. (2001) have found a processed Tsix transcript in murine ES cells. Like Xist, Tsix has no open reading frames (ORFs), and its RNA is found in the nucleus. However, unlike Xist, the Tsix RNA is associated with the future active X. There is some evidence that the Tsix transcript may be an antagonist of Xist. Studies of mice carrying a Tsix deletion show that Tsix is required for repression of the maternal Xist allele in placental tissues where the paternal X chromosome is preferentially silenced. Lee (2000) and Sado et al. (2001) have proposed that Tsix is the maternally expressed factor that protects the maternal X from imprinted X inactivation in extra-embryonic tissues. The role of Tsix in random inactivation remains to be elucidated (this will be discussed below).
We recently showed that it was feasible to study the human XIC region through use of human transgenes introduced into murine ES cells, as well as through chimeric mice derived from them. Six tandem copies of one transgene with 480 kb of the human XIC region, including the XIST locus, inserted into mouse chromosome 11 was able to induce random X inactivation in male chimeric mice, resulting in inactivation of the murine X chromosome in some XY cells carrying the transgene (Migeon et al. 1999). The other, a single-copy transgene with <300 kb of the human XIC region carried on mouse chromosome 6, was recognized as an XIC in male ES cells but could not induce inactivation in mice (Migeon et al. 2001), presumably because of the low copy number (Heard et al. 1999). Unlike XIST/Xist, which is expressed in all cells with an inactive X at any age, murine Tsix is expressed only transiently, prior to and during the time that X inactivation is being established. Murine ES cells recapitulate the stages of X inactivation seen in the mouse embryo proper (Lee and Lu 1999) and are an accepted model for studies of this developmental process. Therefore, to determine whether there was a human gene orthologous to Tsix, we looked for other human transcripts in the mouse ES cells carrying the human XIC transgene. As a result, we have identified a transcript that is antisense to XIST and overlaps the XIST locus at its 3′ end. This transcript is also present in human-embryo–derived stem cells but is absent from adult somatic cells. On the basis of our observations, we suggest that human TSIX is not involved in genomic imprinting and may not be an antagonist of human XIST expression.
Material and Methods
Transgenes and Cell Lines
Mouse ES cells carrying the ES-10 and ES-5 transgenes were derived from mouse J1 ES cells and have been reported elsewhere (Migeon et al. 1999, 2001). ES-10 contains six copies of the full transgene, and ES-5 contains a single copy of a truncated one. In addition, we studied XY somatic cells from chimeric mouse embryos 949.1 and 850.3, which carry the ES-10 and ES-5 transgenes on autosomes and are hereafter referred to as “ESch10” and “ESch5,” respectively. The murine ES cell transfectants were kept undifferentiated on feeders in medium with leukemia inhibitory factor (LIF). For differentiation, they were passaged twice without feeders on gelatinized dishes in the presence of LIF and then were cultured in suspension without LIF, for aggregation into embryoid bodies (EBs).
The human embryonic cells used for analysis were four genetically distinct cell lines originating from primordial germ cells at 5–11 wk postfertilization (pf). These fibroblast cultures were established from human EBs by J. Gearhart and are referred to as human “embryoid-body–derived” (EBD) cells. They are no longer totipotent but do transcribe some genes that are expressed early in development (Shamblott et al. 2001). Cell lines LV (5 wk pf) and SD (11 wk pf) were 46, XX, whereas SL (6 wk pf) and LU2 (7 wk pf) were 46, XY. Our use of the human EBD cells was approved by the university committee concerned with patents and by Geron, Inc., according to the licensing agreement. They were maintained in EGM2MV medium on collagen plates, as described by Shamblott et al. (2001).
RNA Analysis
Primers
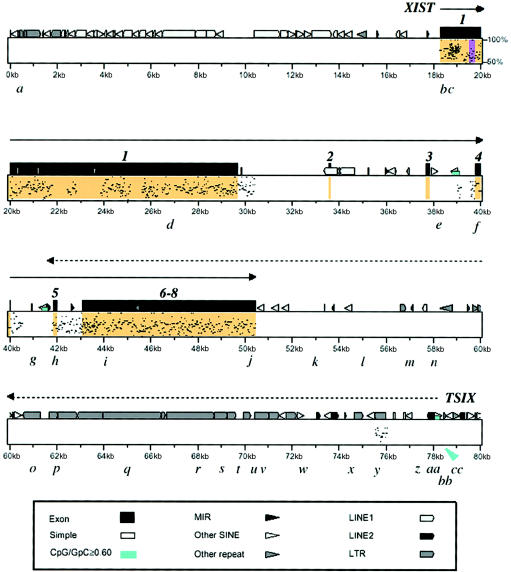
XIST is oriented in the opposite direction of the mouse gene, so that the 3′ XIST is closest to the centromere. Both the ES-10 and ES-5 transgenes include the ∼50-kb human XIST locus and ∼70 kb centromeric (proximal) to the 3′ end of XIST. They differ in the amount of sequence telomeric (distal) to the 5′ end of XIST (360 kb for ES-10 and <180 kb for ES-5). Sequence data for both transgenes are provided in PAC 92E23 (GenBank), which has been sequenced in two parts, found under GenBank accession numbers U80459 and U80460. The locations of primers used for RNA analysis are shown in figure 1 and table 1 and are based on the U80460 sequence. Specific primer sequences will be provided on request.
Figure 1.
PipMaker percent identity plot of the human XIC region in ES-10, showing the location of a novel transcript and homology to the comparable region in the murine genome. Nucleotides 0–80000 of the U80460 sequence (0–80 kb) are shown. Plotting of XIST exons (orange) is based on all known transcribed sequences (Brown et al. 1992; Hong et al. 2000). The black dot pattern shows the percent homology (50%–100%) with the comparable mouse Xic region. The XIST transcript is shown as a solid line with an arrow. The TSIX transcript (dotted line with arrow) is antisense to XIST and is initiated from four clustered transcription start sites, the most 5′ of which shown (see fig. 3). The purple vertical bar in XIST exon 1 represents the single CpG island identified by the Grail program (version 1.3). The small blue boxes indicate a CpG:GpC frequency of ⩾.6; the one near the TSIX start site is marked by a blue arrow. The locations of RT-PCR primers (a–cc) are shown in lowercase letters. The lack of homology outside of the XIST region has also been documented in a PipMaker plot by Nesterova et al. (2001, fig. 5e). MIR = mammalianwide interspersed repeats; SINE = short interspersed elements.
Table 1.
RT-PCR Analysis of Human XIST / TSIX Transcripts[Note]
|
Mouse ES Cell Transgenes |
Human EBD Cells (Female) |
||||
| Primer(Position)a | Human Somatic Cell (Female) | ES-10 | ES-5 | SD | LV |
| a (.3 kb) | − | − | − | − | − |
| b (XIST-pro) | −/− | +/− | −/ − | −/− | −/− |
| c (5’ XIST) | +/− | +/− | +/− | ND | +/− |
| d (XIST-ex1) | +/− | +/− | +/− | +/− | +/− |
| e (XIST-int3) | − | − | − | − | − |
| f (XIST-ex4) | +/− | +/− | +/− | +/− | +/ |
| g (XIST-int4) | ND/− | ND/− | ND/− | ND/− | ND/− |
| h (XIST-ex5) | +/− | +/+ | +/+ | +/+ | +/+ |
| i (XIST-ex6) | +/− | +/+ | +/+ | +/+ | +/+ |
| j (3’ XIST) | +/− | +/+ | +/+ | +/+ | +/+ |
| k (53 kb) | − | + | + | + | + |
| l (55 kb) | − | + | + | ND | ND |
| m (57 kb) | −/− | −/+ | −/+ | + | + |
| n (58 kb) | − | + | + | + | + |
| o (61 kb) | − | + | + | ND | ND |
| p (62 kb) | − | + | + | ND | ND |
| q (65 kb) | − | + | + | + | + |
| r (68 kb) | − | + | + | ND | ND |
| s (69 kb) | − | + | + | ND | ND |
| t (69.8 kb) | − | + | + | + | + |
| u (70.4 kb) | − | + | ND | ND | ND |
| v (70.8 kb) | − | + | + | + | + |
| w (72.6 kb) | − | + | + | + | + |
| x (74.5 kb) | − | + | ND | − | − |
| y (75.5 kb) | − | + | + | − | − |
| z (77.4 kb) | −/− | −/+ | −/+ | − | − |
| aa (77.8 kb) | − | + | + | − | − |
| bb (78.2 kb) | − | − | − | − | − |
| cc (79 kb) | − | − | − | ND | ND |
Note.— Controls (human male and mouse J1 ES cells) were negative for each primer set. + = product present; − = product absent. A single symbol indicates a result of random priming of cDNA, and two symbols (i.e., +/+, −/−, or +/−) indicate sense/antisense (XIST/TSIX), the results of strand-specific priming. ND = not done.
Primers are shown in figure 1. Values shown in kilobases indicate the position in U8046. Pro = promotor, ex = exon, int = intron.
Reverse-Transcriptase PCR
RNA was obtained from ES cells before and after differentiation into EBs. Total RNA was isolated with TRIZOL (Life Technologies) and treated with DNaseI (at 37°C for 1 h). Poly A+ RNA was isolated using the Nucleotrap mRNA separation kit (Clontech). To screen for transcripts, reverse-transcriptase PCR (RT-PCR) was performed (1 μg RNA, 35 cycles; Migeon et al. 1999). The cDNA was generated using random priming for sequences outside the XIST transcription unit, whereas strand-specific priming was used for the portion of TSIX that overlaps XIST.
For strand-specific RT-PCR, first-strand cDNA was synthesized using either sense or antisense primers, as described by Lee et al. (1999), with minor modifications. RNA (0.5–1 μg) with strand-specific primer (3 pmol) was incubated at 70°C for 5 min and then was equilibrated to 50°C. The cDNA was synthesized with Superscript II reverse transcriptase (200 U, Life Technologies) at 50°C for 1 h. The RT was heat-inactivated at 80°C for 45 min. After treatment with RNaseH (Life Technologies), first-strand cDNA was amplified with Taq polymerase for 35 cycles, using paired sense and antisense primers. Controls containing RNA and strand-specific primer but no reverse transcriptase were processed in parallel.
5′ Rapid Amplification of cDNA Ends (RACE)
Total RNA (10 μg) was treated with RNase-free DNaseI (Ambion). 5′ RACE (was performed using the GeneRacer kit (Invitrogen), according to manufacturer instructions. First-strand cDNA was primed with a gene-specific primer (U80460 nucleotides (nt) 77426–77443), was extended with Superscript II at 42°C for 50 min and was treated with RNaseH. 5′ ends were amplified using the GeneRacer 5′ primer and a gene-specific primer (U80460 nt 77565–77588). Nested PCR was performed with the GeneRacer 5′ nested primer and two gene-specific primers (U80460 nt 77578–77603 and U80460 nt 77678–77704). PCR products were either gel purified or were cloned into the pCR4-TOPO vector (Invitrogen) and were sequenced using [33P]-labeled chain terminators (Thermo Sequenase Cycle Sequencing Kit, USB).
Sequence Analysis
Once identified, the TSIX sequence from U80460 (GenBank) was analyzed for CpG islands, using the Grail program (version 1.3; Oak Ridge National Laboratory Web site). Potential TATA boxes were identified using the HCtata program on the WebGene server. Only strong consensus sequences were considered. We assembled the mouse Xic sequence (GenBank accession numbers AJ010350, M97167, L04961, U41394, and X999460) through use of Sequencher software and compared it to the human XIC sequence (U80460). Both strands were searched for homology by the Advanced PipMaker (Schwartz et al. 2000; PipMaker Web site). The human sequence was masked using the RepeatMasker program (RepeatMasker Web site). Percent identity plot (PIP) and dot plot graphics from the Advanced PipMaker were modified in Adobe PhotoShop, for clarity of presentation.
Results
Search for Transcripts from the Human XIC Transgene in Mouse ES Cells
Both human XIC transgenes include nt 1–119,000 of the U80460 sequence; the XIST transcription unit extends from nt 18317 to 50425 (Brown et al. 1992; Hong et al. 2000). Initially, we looked for novel transcripts, using RT-PCR primers at 10-kb increments, starting from U80460 nt 1–119000 (70 kb proximal and 18 kb distal to XIST). We also used primers at the distal end of U80459 (50 kb upstream of the 5′ end of XIST), a region also present in both ES-10 and ES-5 cells.
Figure 1 is a PipMaker PIP (Schwartz et al. 2000) showing the U80460 portion of ES-5 and ES-10 transgenes that includes XIST and the only other transcript identified in the region. RT-PCR analysis using cDNA obtained by random priming showed that the novel transcript originates ∼27 kb from the 3′ end of XIST. Table 1 shows that it extended as far as XIST exon 5; RT-PCR results were positive with primer set h but negative with primer set g, 600 bp upstream in XIST intron 4. The primers that gave negative results in the RNA analysis were positive in the DNA analysis of the same cells. This transcript was not seen in any of the control cells (human male and female or parental J1 ES cells).
Novel Transcript Is Antisense to XIST
Strand-specific priming of the cDNA showed that this transcript was present in poly A+ RNA. This transcript was antisense to XIST, since it was amplified from the cDNA primed by the forward primers (relative to XIST) and not from that primed by the reverse primers (fig. 2). Strand-specific priming also showed that this antisense transcript overlapped the XIST transcript, extending from 3′ XIST through exon 5 (table 1 and fig. 1). Because this novel transcript is transcribed from the opposite strand as human XIST and overlaps XIST, we believe that it is the counterpart of murine Tsix; therefore, we will subsequently refer to this gene as “TSIX.”
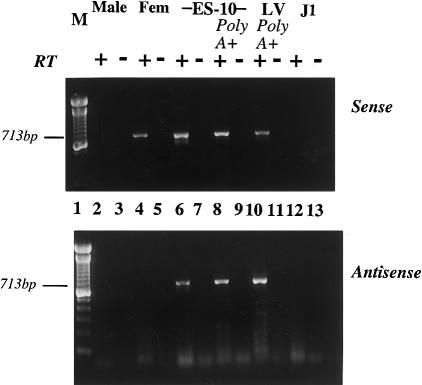
Figure 2.
Strand-specific RT-PCR showing polyadenylation and orientation of transcripts. Top, Sense (XIST) transcript (cDNA synthesized using reverse primer). Bottom, Antisense (TSIX) transcript (cDNA synthesized using forward primer). RNA samples were total RNA, unless otherwise indicated. Lane 1, 100-bp marker; lanes 2 and 3, male; lanes 4 and 5, female; lanes 6–9, ES-10; lanes 10 and 11, human EBD LV cells; lanes 12 and 13, J1, mouse ES cell control. Samples were assayed using primer set g in the presence (+) or absence (−) of reverse transcriptase (RT). Note that ES-10 and LV cells have both sense and antisense transcripts with XIST exon 6 primers.
Despite numerous attempts, we have obtained little evidence of a processed TSIX transcript. Using a large number of closely spaced primers, which in the TSIX unique region were 0.1–3 kb apart, with an average interval of 1.15 kb (fig. 1), we found a contiguous transcript. We did identify a small intron (763 bp) at the start of the transcript in ES-10 (fig. 3; discussed below). In addition, no small transcripts were detected on northern blots.
Figure 3.
Identification of TSIX transcription start sites in ES-10 cells. Transcription start sites (arrows) were determined by 5′ RACE using ES-10 as template. The TSIX transcript is initiated from four clustered transcription start sites (bases 77657, 77865, 78267, and 78860 in the U80460 sequence). The location of repetitive elements are indicated as follows: black vertical lines denote Alu elements, gray boxes denote LTRs, diagonal lines denote LINE elements, and horizontal lines denote MER1 elements.
TSIX Has No ORF
The coding potential of the TSIX transcript was investigated in silico. The sequence from nt 30000 to nt 80000 of U80460 (complementary strand) was analyzed using ORFGene (Rogozin et al. 1996; ORFGene 2 Web site) and GeneBuilder (Milanesi et al. 1999; GeneBuilder Web site) software. ORFGene did not identify any ORFs in this interval. GeneBuilder predicted a series of exons across the interval, which were inconsistent with the results of RT-PCR analysis (table 1 and data not shown). The predicted ORF lacked an initiation codon and showed no homology to any protein in the SWISSPROT database.
TSIX Has Little Homology to Tsix
Figure 1, which is based on the GenBank sequence, shows the homology between the human and mouse DNA sequence in the XIST-TSIX region. There are many repeats within this region. Virtually all the significant homology (>50%) lies within the XIST locus. The only >50% homology in the region of the TSIX transcript is minimal and is limited to a single region at ∼76 kb, near the start of the TSIX transcript in ES-10; this is most likely the region of human homology to mouse Tsix observed by Lee et al. (1999, fig. 6).
Identification of TSIX Transcription Start
5′ RACE was used to define the 5′ boundary of the TSIX transcript in ES-10 cells (fig. 3). Four transcription start sites were identified, at nt 77657, 77865, 78267, and 78860 of U80460. The first three start sites are located within repetitive elements MER58B, AluY, and L2, respectively. Transcription from positions 78860 and 77865 may be directed by TATA box sequences within the repeats. An additional upstream exon was identified in the 78860 transcript. The alternate first exon is 23 bases in length, followed by an intron of 763 bases—perhaps the remnant of the small intron between murine Tsix exons 2 and 3 (Sado et al. 2001). Splice sites conform to consensus sequences (Shapiro and Senapathy 1987).
Evidence that Human TSIX is Truncated Relative to Tsix
On the basis of the results of 5′ RACE studies and homology searches, the start sites in the human gene are at what may be analogous to Tsix exons 2 and 3 in the mouse (Sado et al. 2001) (see region with homology in fig. 1). Unlike murine Tsix, which has a CpG island at its 5′ end (Lee and Lu 1999), the human TSIX gene lacks a proper CpG island (fig. 1). A search of the U80460 sequence by the Grail program (version 1.3) revealed only a single CpG island—the one that overlaps the 5′ end of XIST (nt 19545–19756; see figs. 1 and 4). Only a small CpG-rich region remains near the TSIX start site (fig. 1, arrowed blue box).
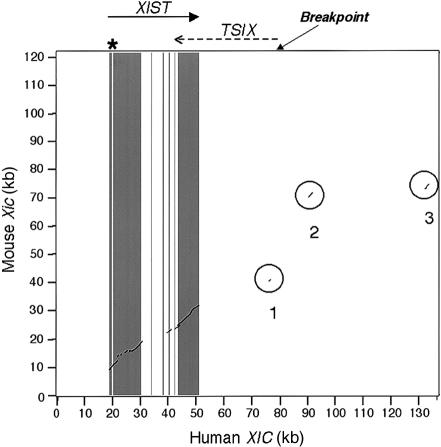
Figure 4.
Dot plot showing the evolutionary breakpoint within the human XIC. The region shown is an extension of that in figure 1 and includes the region from 0–136 kb of U80460. The dark gray bars represent the XIST exons; the light bar shows the CpG island in XIST exon 1 (asterisk). The effective evolutionary breakpoint is shown as occurring at the start of the TSIX transcript; the actual breakpoint is difficult to determine. The areas circled and numbered 1–3 show the only conserved regions outside of the XIST gene. Region 1 is homologous to murine Tsix, and regions 2 and 3 are homologous to the murine testis gene (Tsx) exons 5 and 4, respectively, which are separated in the human genome by ∼40 kb. This figure, a PipMaker dot plot comparing human and mouse XIC sequences, is similar to the traditional dot plot of this region, shown by Lee et al. (1999, fig. 6); however, we now know that regions 2 and 3 are homologous to TSX and not TSIX.
To insure that the absence of a CpG island from the 5′ end of TSIX is not an artifact, the U80460 sequence was compared to that obtained from a whole-genome shotgun assembly of genomic DNA from five unrelated individuals (Venter et al. 2001; Celera Web site). The sequences obtained from the two data sets are colinear over the entire length of U80460, exhibiting 99.96% identity, with the largest discrepancy being a 10-bp insertion/deletion within a pentanucleotide repeat sequence.
The lack of the expected CpG island at the start of TSIX suggested an evolutionary breakpoint. Evidence of this comes from the analysis of a neighboring gene. The X-linked testes specific gene (Tsx), which resides within 7 kb of murine Tsix (Sado et al. 2001), is expressed in rodent testes but has no counterpart expressed in human testes (Simmler et al. 1996). In fact, there is no human ortholog, since only homologous fragments of mouse Tsx are present in the human genome. Human TSX sequences are scattered in the human X chromosome: exon 1 is in Xq23 and exon 6 is in Xq21. Remnants of exon 5 and exon 4 are present within U80460, but these fragments are 13–20 kb and >50 kb from the 5′ end of TSIX, respectively (areas 2 and 3, respectively, of fig. 4).
TSIX is Transcribed in Human Embryonic Cells
Because transcripts expressed from our human transgene could have been artifacts of transfection, we were encouraged by the preliminary evidence of a low-level antisense transcript originating “downstream of XIST exon 8 and extending only into the 3′ end of XIST” in the NT2 human embryonal carcinoma line derived from a testicular teratocarcinoma (Chow et al. 2000). To find the TSIX transcript that was not present in adult human cells, we chose to examine fetal cells derived from human EBs (Shamblott et al. 2001). To determine the X inactivation status of these cells, we assayed them for XIST transcripts. The two female human EBD cell lines (SD and LV) expressed XIST, but the male human EBD lines (SL and LU2) did not. Since XIST is expressed in male blastocysts prior to the time of X inactivation (Ray et al. 1997), SL and LU2 have passed the developmental stage when XIST is expressed. Therefore, both male and female EBD cell lines were at a stage subsequent to the onset of X inactivation.
Since X inactivation had already occurred in these cells, we were surprised to find the TSIX transcript in the two female EBD lines. In both of them, the transcript was ∼3.8–5.6 kb shorter than that found in ES–10 cells. The start site was difficult to determine, because of the density of repeated sequences in this region (fig. 1), but transcription of TSIX most likely originates between nt 73254 and nt 75031 of U80460. The repeated sequences also made it difficult to design primers for 5′ RACE. Since primers within the unique sequence failed to generate RACE products, it is difficult to determine the TSIX transcription start site in the human EBD cells more precisely. On the other hand, our analysis shows that the 3′ end of the TSIX transcript was identical to that in ES-5 and ES-10 cells, since it extended only as far as XIST exon 5 (table 1).
TSIX is Transcribed in Mouse Embryonic Fibroblasts Carrying the Human Transgene
We examined ESch5 and ESch10 fibroblasts originating from 13.5-d and 18-d embryos chimeric for the ES-5 or ES-10 transgenes. Both cell lines expressed not only XIST, but also TSIX . Assayed with TSIX-specific primer sets l, n, o, t, w, aa, bb, and cc (see fig. 1), these fibroblasts had RT-PCR products with all primer sets except cc, indicating the same expression pattern as the ES transfectants from which they were derived (cf. ES-5 and ES-10; table 1).
Characteristics of TSIX Expression in Embryonic Cells
According to observations of Lee et al. (1999) and Sado et al. (2001), murine Tsix is not present in eight-cell embryos, is expressed in blastocysts from both X chromosomes before X inactivation, and is expressed at the onset of inactivation only from the allele on the active X. Tsix expression disappears from ES cells during the 2–9 d following their differentiation into EBs. In chimeras, the level of expression is greater in the placenta than in the fetus proper and is absent from both by 12–15 d of gestational age. The expression of human TSIX seems to persist longer, at least in a mouse environment, since it is found in ES-10 cells long after their differentiation into EBs (at least to the 19th passage after differentiation) and in the fetal somatic cells derived from 18-d and 13.5-d ES-10 and ES-5 chimeric fetuses, at a time when murine Tsix transcripts are minimal or no longer expressed.
Another difference has to do with lack of evidence that human TSIX influences XIST expression. In the female embryo-derived cells, TSIX transcripts were coexpressed with those from XIST, so that human TSIX expression did not repress the expression of XIST. Since the male embryo-derived cells did not express TSIX, the transcript is not associated with active X chromosomes in males. Coexpression was also seen in the ES-10 and ES-5 EBs and their chimeric mouse derivatives. In this case, both human transcripts had to come from the transgene, and, hence, from the same chromosome.
Discussion
Comparing Human TSIX with Murine Tsix
The results of our analysis show that human TSIX, like its murine counterpart, is expressed only in embryo-derived cells, initiates downstream of the 3′ end of XIST, and produces an untranslated RNA. This RNA is transcribed from the opposite strand as XIST and is, in part, antisense to XIST. However, the human TSIX gene differs from its murine counterpart in that it is truncated at the 5′ end, does not cover the XIST promoter, and does not have the CpG island that is essential for function of murine Tsix.
The mouse Tsix gene was originally described as a 40-kb RNA without introns, originating 15 kb downstream of Xist and transcribed across the Xist locus. More-recent studies indicate that it is at least partially processed, giving a 2.7-kb transcript originating from the major promoter located near Tsix exon 2. Sado et al. (2001) also found a 4.3-kb transcript originating from the minor promoter, located ∼28 kb downstream of Xist, suggesting that Tsix starts 13 kb further downstream than originally reported. In addition, these investigators show that exon 2 is associated with a CpG island, which is known to be differentially methylated on active and inactive chromosomes (Prissette et al. 2001). Although the human TSIX transcripts from ES-10 and ES-5 cells also begin ∼27 kb downstream of XIST, they have a much shorter overlap with XIST, ending in exon 5; they do not overlap the 5′ end of the gene (table 1). The only transcript found in ES-10 in the region of the XIST promoter was shown by strand-specific RT-PCR to be attributable to XIST and not TSIX (table 1 and data not shown). Therefore, the TSIX transcript in ES-5 and ES-10 cells is ∼35 kb in length. The TSIX transcript from human EBD cells has the same limited overlap but is 3.8–5.6 kb shorter; this difference may reflect the influence of the mouse environment on expression of the human transgenes.
Analysis of TSIX Identifies XIC Breakpoint
All of our evidence suggests that the 5′ end of the human TSIX locus may have been decapitated by an evolutionary breakpoint. The existence of an XIC in Xq13 was originally postulated on the basis of the conserved order of genes on murine and human X chromosomes. However, subsequent studies have revealed many microrearrangements in this region, including an inversion, which affects the transcriptional orientation of the XIST locus (Brockdorff et al. 1992). Although the exact site of the breakpoint cannot be determined, it is clear that there was an evolutionary change in the human TSIX region that resulted in loss of the human counterpart of the mouse testes-specific gene (Tsx) distal to murine Tsix. This breakage event shattered the TSX gene, spreading exons throughout the long arm of the X chromosome in the ancestor who gave rise to the human population. The extraordinary distance between TSX exons 4 and 5 has also been observed by Nesterova et al. (2001, fig. 5e). This breakpoint at ∼27 kb from human XIST probably defines one boundary of the essential XIC in both species (indicated by an arrow in fig. 4).
An important question is whether this breakpoint influences the function of TSIX or the process of X inactivation. Some insights come from deletions in the murine Tsix gene, induced by Sado et al. (2001). Disrupting Tsix exon 1 (located ∼28 kb from murine Xist)—but not the CpG island (∼14 kb from Xist)—neither affected the second promoter nor interfered with Tsix function. However, the deletion in the Tsix exon 2 region, including part of the CpG-rich region, blocked Tsix transcripts running across the Xist gene, which these investigators suggest is responsible for the loss of Tsix function. In addition, the targeted (3.5-kb) deletion by Lee (2000) of this CpG island also disrupted imprinted X inactivation. The evolutionary breakpoint in human TSIX not only eliminated exon 1 and most likely part of exon 2, but also resulted in the loss of most of the CpG-rich region near the start of TSIX, either directly or through evolutionary drift. The lack of significant homology over the TSIX region (figs. 1 and 4) and the large number of repetitive sequences—mostly long tandem repeats (LTRs) (figs. 1 and 3) not seen in the murine Tsix region (data not shown)—suggest that significant drifting has occurred. In any case, although the exon 1 deletion may not interfere with TSIX function, it seems likely that elimination of the CpG island would interfere with parental imprinting.
Significance of Species Differences with Respect to X Inactivation
There are many ways to compensate for sex differences in X gene dosage; compensatory mechanisms are directed at transcription and range from up-regulating the transcriptional output of the single X in males to down-regulating that of the X chromosomes in females. However, only mammals compensate by X inactivation. Since both marsupials and placental mammals use X inactivation, the basic mechanisms should be the same for all mammals. Therefore, features of X inactivation that differ among mammals are merely variations on the basic theme (Migeon 1990). These species variations tell us which details of the X inactivation scheme are not essential for establishing a single active X in mammalian cells. Such variable features include time of onset, choice of active X (either random or maternal), and stability of inactivation once established. Variations affecting stability result from differences in the DNA methylation status of CpG islands on the inactive X chromosome (Migeon 1990). Most likely, differences in the time of onset are responsible, at least in part, for the paternal (imprinted) inactivation in marsupials and in placental tissues of rodents and for the well-documented lack of imprinted X inactivation in human placental tissues (Migeon and Do 1978, 1979; Migeon et al. 1985, 1986; Mohandas et al. 1989; van den Hurk et al. 1997). Other variable features include the XIST locus itself, which has undergone inversion event(s) during mammalian evolution and whose DNA sequence is so poorly conserved among mammals that the marsupial gene has not yet been identified. Such species differences in the details of X inactivation reflect variations that do not interfere with the basic process responsible for the single active X.
What do our studies reveal about the role of Tsix/TSIX in imprinted X inactivation? On one hand, they support the notion that murine Tsix plays a role in imprinted X inactivation in mice, perhaps as a factor to block Xist expression on the maternal X chromosome. Studies of knockouts of the mouse Tsix locus clearly show that it has a role in paternal X inactivation in the placenta; Tsix deletions carried on the maternal X lead to silencing of the maternal as well as the paternal X chromosome in many cells, resulting in placental abnormalities and fetal death (Lee 2000; Sado et al. 2001). On the other hand, along with the lack of paternal X inactivation in human placental tissues, the evolutionary changes we observe suggest that human TSIX does not share this function. The structural features of the murine Tsix locus seem to be suited to its imprinting function. Like many imprinted genes, Tsix is initiated near a CpG island that is differentially methylated; it produces an antisense transcript, which, whether processed or not, overlaps the promoter of its target gene and is monoallelically expressed from the opposite chromosome as Xist. The nature of the interaction between the antisense transcript and its target is not yet known, and the possibility that the abbreviated human TSIX transcript interacts with XIST at the critical moment in development cannot be excluded. However, the fact that the human gene is a truncated version of the murine one has implications for its function. Because the human transcript does not cover the XIST promoter, the means by which it interacts with XIST cannot involve direct physical contact with the promoter. In addition, since TSIX lacks the differentially methylated CpG island, which, when deleted in the mouse gene, disrupts imprinted X inactivation, it probably cannot imprint XIST expression. Further, we obtained no evidence that human TSIX inhibits XIST expression; the fact that TSIX and XIST are transcribed simultaneously from the same transgene in ES-10 and ES-5 cells suggests that it does not.
Jacob (1977) has suggested that Mother Nature is more tinkerer than engineer and that she uses materials at hand to modify her work. Tsix in mice—and, perhaps, a comparable RNA in marsupials—may have been used to reinforce the parental imprinting needed because of the earlier onset of X inactivation in these species. In any case, our observations could explain why X inactivation is not imprinted in human placental tissues.
The role of Tsix in the basic scheme of random (nonimprinted) X inactivation is less clear. Tsix knockouts do skew the patterns of X inactivation in the embryo proper, since the chromosome bearing the deletion is exclusively the inactive one. However, the effect of a knockout of Tsix function on random (nonimprinted) X inactivation must be distinguished from its effect on imprinted inactivation in the placenta. In addition, a nonspecific effect of the deletion on Xist function, unrelated to antisense function, cannot be excluded. In fact, Morey et al. (2001) studied mice carrying a 65-kb deletion that removed all of Tsix except that which overlaps Xist and which was associated with exclusive inactivation of the deleted X. They showed that restoration of Tsix function represses Xist RNA accumulation in cis but does not restore random inactivation. These investigators suggest that although Tsix represses the initiation of X inactivation in cis, it is insufficient for normal random choice. Conceivably, human TSIX is an evolutionary relic of the murine gene, which continues to be expressed in fetal tissues, in much the same way that XIST continues to be expressed long after the critical time in development—because its expression does no harm.
In addition, it is unlikely that any cis-acting X-linked gene could, by itself, bring about the initial choice of the single active X. The existence of a trans-acting factor to block XIST expression was postulated by Lyon (1996) and others before the discovery of Tsix. Evidence from triploid cells (Jacobs et al. 1979; Migeon et al. 1979) suggests that the factor(s) needed to induce the silencing of XIST on a single X chromosome in each cell most likely operates in trans and is autosomal in origin. In the presence of a trans-acting factor to repress XIST expression on the active X and DNA methylation to lock in the silence of the locus, Tsix would not be essential to the inactivation process but would merely reinforce the repression of maternal Xist in the species in which it is needed.
Acknowledgments
We are grateful to Dr. John Gearhart for the human EBD cells. We thank Kimesha Hammett and Joyce Axelman, for cell cultures; Tonya Watkins, for assistance with PCR studies; Roxanne Ashworth, for her contributions in designing primers; and Cheron Jones, for DNA sequencing. We gratefully acknowledge Drs. Mohamed Khalifa, Mita Mukherjee, Kirby Smith, and Ethylin W. Jabs, for helpful suggestions about the manuscript. This work was supported by NIH grants HD05465 (to B.R.M.) and AR44702 (to I.M.). J.A.D. is a student in the predoctoral program in human genetics, supported by National Institutes of Health training grant GM07814.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Celera, http://public.celera.com/index.cfm (for publicly available human genome sequence)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human XIC reference sequence [accession number U80460] and mouse XIC reference sequences [accession numbers AJ010350, M97167, L04961, U41394, X999460])
- GeneBuilder, http://125.itba.mi.cnr.it/~webgene/genebuilder.html (for gene-structure prediction)
- HCtata, http://125.itba.mi.cnr.it/~webgene/wwwHC_tata.html (for identification of promoter elements)
- Oak Ridge National Laboratory Gene Recognition and Assembly Internet Link, http://compbio.ornl.gov/Grail-1.3/ (Grail, version 1.3, for CpG island identification)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for XIST [MIM 314670] and Tsix [MIM 300181])
- ORFGene 2, http://125.itba.mi.cnr.it/~webgene/wwworfgene2.html (for identification of ORFs)
- PipMaker, http://bio.cse.psu.edu/pipmaker/ (for homology analysis)
- RepeatMasker, http://ftp.genome.washington.edu/RM/RepeatMasker.html
References
- Brockdorff N, Ashworth A, Kay G, McCabe V, Norris DP, Cooper P, Swift S, Rastan S (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71:515–526 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71:527–542 [DOI] [PubMed] [Google Scholar]
- Chow JC, Clemson CM, Lawrence JB, Brown CJ (2000) XIST expression in human embryonal carcinoma lines: identification of novel sense and antisense transcripts. Am J Hum Genet Suppl 67:44 [Google Scholar]
- Clemson CM, McNeil JA, Willard H, Lawrence JB (1996) RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/ chromosome structure. J Cell Biol 132:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P (1997) X-chromosome inactivation in mammals. In: Campbell A, Anderson W, Jones EW (eds) Annual review of genetics. Vol 31. Annual Reviews Inc, Palo Alto, California, pp 572–610 [DOI] [PubMed] [Google Scholar]
- Heard E, Mongelard F, Arnaud D, Avner P (1999) Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol 19:3156–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y-K, Ontiveros SD, Strauss WM (2000) A revision of the human XIST gene organization and structural comparison with mouse Xist. Mamm Genome 11:220–224 [DOI] [PubMed] [Google Scholar]
- Jacob F (1977) Evolution and tinkering. Science 196:1161–1166 [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Masuyama AM, Buchanan IM, Wilson C (1979) Late replicating X chromosomes in human triploidy. Am J Hum Genet 31:446–457 [PMC free article] [PubMed] [Google Scholar]
- Lee J (2000) Disruption of imprinted X inactivation by parent-of-origin-effects at Tsix. Cell 103:17–27 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D (1999) Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21:400–404 [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N (1999) Targeted mutagenesis of Tsix leads to nonrandom inactivation. Cell 99:47–57 [DOI] [PubMed] [Google Scholar]
- Lyon MF (1962) Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet 14:135–148 [PMC free article] [PubMed] [Google Scholar]
- ——— (1996) Pinpointing the centre. Nature 379:116–117 [DOI] [PubMed] [Google Scholar]
- Migeon BR (1990) Insights into X chromosome inactivation from studies of species variation, DNA methylation and replication, and vice versa. Genetical Res (Cambridge) 56:91–98 [DOI] [PubMed] [Google Scholar]
- Migeon BR, Do TT (1978) In search of nonrandom X inactivation: studies of the placenta from newborns heterozygous for glucose-6-phosphate dehydrogenase. In: Russell LB (ed) Genetic mosaics and chimeras in mammals. Plenum Publishing Corporation, New York, pp 379–391 [DOI] [PubMed] [Google Scholar]
- ——— (1979) In search of non-random X inactivation: studies of fetal membranes heterozygous for glucose-6-phosphate dehydrogenase. Am J Hum Genet 31:581–585 [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Kazi E, Haisley-Royster C, Hu J, Reeves RH, Call L, Lawler A, Moore CS, Morrison H, Jeppesen P (1999) Human X inactivation center induces random X inactivation in male transgenic mice. Genomics 59:113–121 [DOI] [PubMed] [Google Scholar]
- Migeon BR, Schmidt M, Axelman J, Ruta-Cullen C (1986) Complete reactivation of X chromosomes from human chorionic villi with a switch to early DNA replication. Proc Natl Acad Sci USA 83:2182–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon B, Sprenkle JA, Do TT (1979) Stability of the “two active X” phenotype in triploid cells. Cell 18:637–641 [DOI] [PubMed] [Google Scholar]
- Migeon BR, Winter H, Kazi E, Chowdhury AK, Hughes A, Haisley-Royster C, Morrison H, Jeppesen P (2001) Low-copy-number human transgene is recognized as an X inactivation center in mouse ES cells, but fails to induce cis-inactivation in chimeric mice. Genomics 71:156–162 [DOI] [PubMed] [Google Scholar]
- Migeon BR, Wolf SF, Axelman J, Kaslow DC, Schmidt M (1985) Incomplete X dosage compensation in chorionic villi of human placenta. Proc Natl Acad Sci USA 82:3390–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanesi L, D'Angelo D, Rogozin I (1999) GeneBuilder: interactive in silico prediction of gene structure. Bioinformatics 15:612–621 [DOI] [PubMed] [Google Scholar]
- Mohandas TK, Passage MB, Williams JW, Sparkes RS, Yen PH, Shapiro LJ (1989) X-chromosome inactivation in cultured cells from human chorionic villi. Somat Cell Mol Genet 15:131–136 [DOI] [PubMed] [Google Scholar]
- Morey C, Arnaud D, Avner P, Clerc P (2001) Tsix-mediated repression of Xist accumulation is not sufficient for normal random X inactivation. Hum Mol Genet 10:1403–1411 [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Slobodyanyuk SY, Elizaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM (2001) Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res 11:833–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panning B, Dausman J, Jaenisch R (1997) X chromosome inactivation is mediated by Xist RNA stabilization. Cell 90:907–916 [DOI] [PubMed] [Google Scholar]
- Prissette M, El-Maarri O, Arnaud D, Walter J, Avner P (2001) Methylation profiles of DXpas34 during the onset of X-inactivation. Hum Mol Genet 10:31–38 [DOI] [PubMed] [Google Scholar]
- Ray PF, Winston RML, Handyside AH (1997) XIST expression from the maternal X chromosome in human male preimplantation embryos at the blastocyst stage. Hum Mol Genet 6:1323–1327 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Milanesi L, Kolchanov NA (1996) Gene structure prediction using information on homologous protein sequence. Comput Appl Biosci 12:161–170 [DOI] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E (2001) Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128:1275–1286 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Kelly A, Smit A, Reimer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamblott MJ, Axelman J, Littlefield JW, Blumenthal PD, Huggins GR, Cui Y, Cheng L, Gearhart JD (2001) Human embryonic germ cell derivatives express a broad range of developmentally distinct markers and proliferate extensively in vitro. Proc Natl Acad Sci USA 98:113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmler M, Cattanach BM, Rasberry C, Rougeulle C, Avner P (1993) Mapping the murine Xce locus with (CA)n repeats. Mamm Genome 4:523–530 [DOI] [PubMed] [Google Scholar]
- Simmler M-C, Cunningham DB, Clerc P, Vermat T, Caudron B, Cruaud C, Pawlac A, Szpirer C, Weissenbach J, Claverie JM, Avner P (1996) A 94 kb genomic sequence 3′ to the murine Xist gene reveals an AT rich region containing a new testis specific gene Tsx. Hum Mol Genet 5:1713–1726 [DOI] [PubMed] [Google Scholar]
- van den Hurk JA, Hendriks W, van de Pol DJ, Oerlemans F, Jaissle G, Ruther K, Kohler K, Hartmann J, Zrenner E, van Bokhoven H, Wieringa B, Ropers HH, Cremers FP (1997) Mouse choroideremia gene mutation causes photoreceptor cell degeneration and is not transmitted through the female germline. Hum Mol Genet 6:851–858 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]