Abstract
Studies of genetically engineered flies and mice have revealed the role that orthologs of the human LIM homeobox LHX4 have in the control of motor-neuron–identity assignment and in pituitary development. Remarkably, these mouse strains, which bear a targeted modification of Lhx4 in the heterozygous state, are asymptomatic, whereas homozygous animals die shortly after birth. Nevertheless, we have isolated the human LHX4 gene, as well as the corresponding cDNA sequence, to test whether it could be involved in developmental defects of the human pituitary region. LHX4, which encodes a protein 99% identical to its murine counterpart, consists of six coding exons and spans >45 kb of the q25 region of chromosome 1. We report a family with an LHX4 germline splice-site mutation that results in a disease phenotype characterized by short stature and by pituitary and hindbrain (i.e., cerebellar) defects in combination with abnormalities of the sella turcica of the central skull base. This intronic mutation, which segregates in a dominant and fully penetrant manner over three generations, abolishes normal LHX4 splicing and activates two exonic cryptic splice sites, thereby predicting two different proteins deleted in their homeodomain sequence. These findings, which elucidate the molecular basis of a complex Mendelian disorder, reveal the fundamental pleiotropic role played by a single factor that tightly coordinates brain development and skull shaping during head morphogenesis.
Introduction
The pituitary gland, an endocrine gland that occupies the center of the head, plays vital roles in controlling functions as various as postnatal growth, reproduction, and stress response. The mature gland, in humans, is composed of two lobes: the posterior hypophysis and the anterior hypophysis. The anterior hypophysis contains a heterogeneous population of highly specialized cells producing six hormones: growth hormone (GH), luteinizing hormone (LH), follicle-stimulating hormone (FSH), thyroid-stimulating hormone (TSH), prolactin (PRL), and adrenocorticotropin (ACTH) (Ikeda et al. 1988; Japon et al. 1994). The posterior hypophysis originates from the brain neuroectoderm, whereas the anterior hypophysis originates from Rathke's pouch, an invagination of the oral ectoderm that derives from the most anterior part of the neural ridge. At the end of embryonic development, the two lobes of the pituitary gland are grouped together within the sella turcica, a depression of the upper surface of the central body of the sphenoid bone (Couly and Le Douarin 1988; Eagleson and Harris 1990; Osumi-Yamashita et al. 1994; Dubois et al. 1997). Although the factors that regulate the coordinated formation of the pituitary gland and surrounding structures are still unknown, one might suggest that the proper development of this region could be affected by alterations in the recently identified pituitary-specific transcriptional cascades (Watkins-Chow and Camper 1998).
Several developmental defects of the pituitary region have indeed been reported in humans. These disorders are characterized by an extreme variability in expression of the disease phenotype. Patients present with clinical features revealing a deficit of one or several pituitary hormones, the latter condition being referred to as “combined pituitary-hormone deficiency.” These conditions, which are clinically and genetically heterogeneous, have been linked with rare abnormalities of genes encoding transcription factors necessary for pituitary development (Pfaffle et al. 1992; Radovick et al. 1992; Tatsumi et al. 1992; Dattani et al. 1998; Wu et al. 1998; Netchine et al. 2000; Lamolet et al. 2001). One of these genes, which has been identified in mice, is Lhx4 (MIM 602146)—a LIM-homeobox gene—which has been shown to regulate, together with the closely related Lhx3, proliferation and differentiation of pituitary lineages (Sheng et al. 1997). Although genetically engineered mice bearing a targeted modification of Lhx4 in the heterozygous state are asymptomatic—and although homozygous animals, which have an abnormal pituitary phenotype, die shortly after birth—we sought out the human ortholog of this gene and evaluated its role in the pathogenesis of developmental defects of the pituitary region.
Patients and Methods
Patients
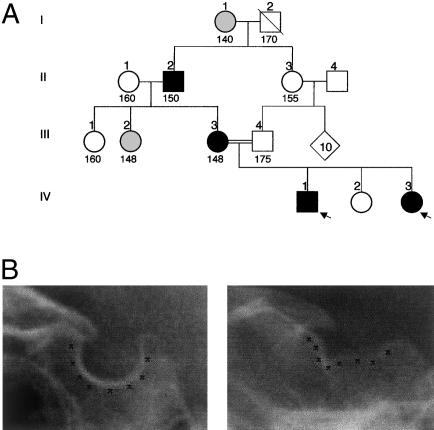
The probands presented at Robert Debré Hospital in Paris, with combined deficiency of three anterior pituitary hormones (i.e., GH, TSH, and ACTH). Levels of gonadotrophins (i.e., LH and FSH) were not investigated because of the early age of the probands. Radiographs and magnetic-resonance imaging of the head showed a small sella turcica, a persistent craniopharyngeal canal, a hypoplastic anterior hypophysis associated with a deformation, into a pointed configuration, of the cerebellar tonsils, and an ectopic posterior hypophysis; the posterior hypophysis was detected as a bright spot near the optic chiasma, in patient IV1, and in the middle of the pituitary stalk, in patient IV3 (see below). The probands’ mother (III3) is 148 cm tall. She has a small sella turcica and a hypoplastic anterior hypophysis associated with a deformation, into a pointed configuration, of the cerebellar tonsils; the posterior hypophysis is at its normal anatomical position. III3 has two sisters, who are 160 cm and 140 cm tall. The probands’ maternal grandfather (II2) is 150 cm tall and has a small sella turcica. The probands’ maternal great-grandmother is 140 cm tall.
Cloning and Sequencing of the Human LHX4 cDNA and Genomic DNA
To isolate the human ortholog of Lhx4, we searched for homology between the Lhx4 cDNA sequence and the sequences in the human expressed-sequence tag (EST) database, by use of BLAST. These alignments indicated the existence of two human ESTs that may correspond to the human LHX4 transcript. In silico mapping of these ESTs in GeneMap'99 led to the identification of a YAC (802b5; Centre d'Étude du Polymorphisme Humain) containing LHX4. On the basis of both the mouse Lhx4 cDNA sequence (GenBank accession number AF135415) and the human EST sequences, we designed exonic primers that permitted us to amplify a fragment spanning exons 4–6 of the human LHX4 gene. Thereafter, by combining in silico (BLAST) and in vitro methods, we designed sense and antisense intronic and exonic primers that permitted us to amplify exons 1–3. We isolated the human LHX4 cDNA by reverse-transcriptase PCR (RT-PCR) amplifications performed with adult-human pituitary poly(A+) RNA (Clontech) as a template, by use of two seminested couples of exonic primers designed on the basis of the human LHX4 sequence. All sequences were determined, for both strands.
Mapping of Human LHX4
The chromosomal localization of human LHX4 was determined by FISH of human-chromosome spreads with the biotin-labeled YAC clone 802b5.
Mutation Analysis
For each individual, we isolated genomic DNA from peripheral-blood samples, according to standard techniques. First, we tested the implication of the LHX3 gene in the disease phenotype of patient IV1, by LHX3-specific primers (Netchine et al. 2000); all LHX3 exons and intronic boundaries displayed a normal sequence (data not shown). Then, we amplified the LHX4 coding exons (exons 1–3 and a fragment spanning exons 4–6) and their flanking intronic sequences, by intronic and exonic primers, and sequenced the corresponding PCR products, on both strands.
Constructions
A human LHX4 cDNA fragment spanning exons 1–4 was fused to a genomic fragment spanning exons 4–6 and originating from the normal LHX4 allele. This minigene was cloned into the pTracer-CMV expression vector (Invitrogen), according to standard procedures, by use of T4 DNA Ligase (Biolabs). This product was subsequently subjected to site-directed mutagenesis by the QuickChange kit (Stratagene), with oligonucleotides designed to introduce the G→C substitution that was found in the patients.
Transfections
Chinese hamster ovary (CHO) cells were obtained from the American Type Culture Collection and were grown in Iscove medium (Life Technologies) containing 10% fetal-calf serum (Life Technologies). Transfections were performed using the Lipofectamine-Plus method (Life Technologies) in OptiMEM medium, according to the manufacturer's standard protocol. Approximately 48 h after transfection of either the wild-type or the mutant minigenes, total RNA was isolated. RT-PCR was performed using several sets of primers located on the LHX4 coding sequence, a procedure allowing us to study the effect that the mutation identified in the patients had on the processing of LHX4 primary transcripts.
Results
Cloning and Mapping of LHX4
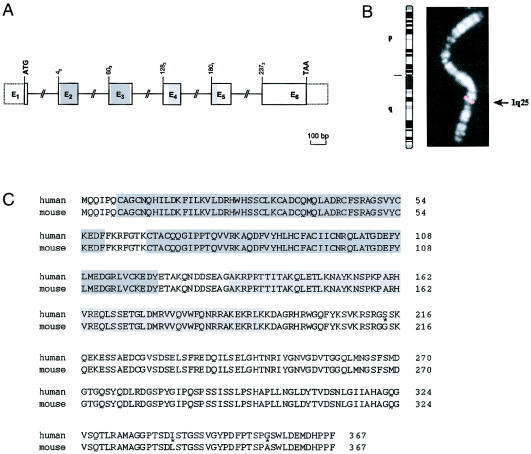
We isolated the human ortholog of mouse Lhx4 by combining in silico and in vitro methods. The human gene, which spans >45 kb, is composed of six translated exons, as deduced by the alignment of the genomic LHX4 sequence and the corresponding cDNA sequence, the latter of which was obtained using human-pituitary cDNA as a template. Sequences from exons 2 and 3 encode the first and second LIM domains, respectively, whereas sequences from exons 4 and 5 encode the homeodomain (fig. 1A). The human gene is located on the long arm of chromosome 1 (fig. 1B), in a region (q25) syntenic to the mouse chromosome 1 region that was shown to contain Lhx4 (Yamashita et al. 1997). Comparison of murine and human LHX4 amino acid sequences, as deduced from their cDNA sequences, showed that LHX4 has been extremely conserved (i.e., 99% identical) between these two species (fig. 1C)—a result demonstrating that this sequence is indeed the human ortholog of mouse Lhx4.
Figure 1.
Genomic structure, chromosomal localization, and predicted amino acid sequence of human LHX4. A, Intron-exon organization of human LHX4. The six exons (E1–E6) encoding LHX4 are indicated by boxes with either solid or dashed borders, depicting translated or untranslated sequences, respectively. Sequences encoding the two LIM domains and the homeodomain are shaded dark gray and light gray, respectively. The locations of the ATG initiation codon and of the TAA stop codon are shown. The number of the first codon in each exon is indicated; exons beginning with the first, second, or third base of a codon are indicated by the subscripts “1,” “2,” or “3,” respectively. Exons are drawn to scale. B, Chromosomal mapping of human LHX4 by FISH. The fine localization of LHX4 to 1q25 is indicated by the black arrow (right). An ideogram of chromosome 1 is given (left). C, Comparison of deduced amino acid sequences of human and mouse LHX4. Different residues are indicated by an asterisk. The two LIM domains and the homeodomain are shaded dark gray and light gray, respectively.
Identification of an LHX4 Splice Mutation
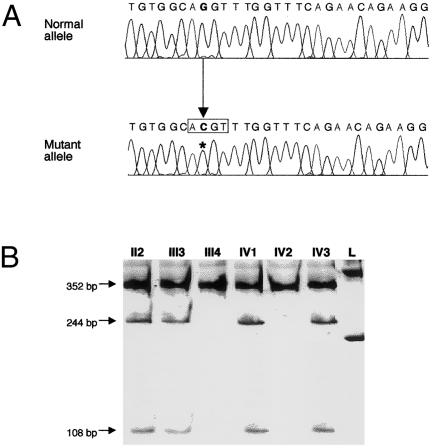
The phenotypic abnormalities of the probands (patients IV1 and IV3, as shown in the pedigree presented in fig. 2A; also see table 1) included short stature due to GH deficiency (as well as deficits of other anterior pituitary hormones associated with a hypoplastic anterior hypophysis), poorly developed sella turcica (fig. 2B), and deformation, into a pointed configuration, of the cerebellar tonsils (which is characteristic of the tonsillar herniation, outside the skull, that defines the Chiari malformation [MIM 118420]). DNA samples from six members of this family were screened for mutations within the LHX4 gene. This study revealed that patients II2, III3, IV1, and IV3 displayed a heterozygous intronic point mutation (G→C transversion) involving the invariant dinucleotide (AG) of the splice-acceptor site preceding exon 5, whereas individuals III4 and IV2, who both exhibited a normal phenotype, bore the intact AG sequence. This nucleotide substitution results in the creation of a recognition site for MaeII, thereby allowing the distinction, by RFLP analysis, between normal and mutant alleles (fig. 3A and B).
Figure 2.
Pedigree and phenotypic features of kindred presenting with brain and skull developmental abnormalities. A, Pedigree of family. Black symbols denote affected individuals, gray symbols denote individuals with uncertain phenotypic status, and white symbols denote unaffected individuals; circles denote females, and squares denote males; a slash through a symbol denotes a deceased individual. Although individuals I1 and III2 exhibit short stature—as in the patients’ phenotype—their phenotypic status remains uncertain, because no further clinical and biological information was available. Heights (in cm) are indicated under the corresponding symbols. Probands are indicated by arrows. B, Radiographs of sella turcica region. The shapes of sella turcica from a normal individual (left) and from patient IV3 (right) are highlighted by black Xs.
Table 1.
Phenotypic Features of the Two Probands[Note]
| Patient | Peak GHa(μg/liter) | FreeThyroxineb(pmol/liter) | Basal PRLc(μg/liter) | BasalCortisold(μg/liter) |
| IV1 | .8 | 3.7 | 8 | 80 |
| IV3 | 3.3 | 9.8 | 5 | 50 |
Note.— Both probands have a hypoplastic anterior hypophysis, an ectopic posterior hypophysis, a poorly developed sella turcica, and deformation of the cerebellar tonsils.
After arginine-insulin stimulation (normal value >10 μg/liter).
Normal value is >10 pmol/liter.
Normal baseline range is 3–15 μg/liter.
Normal value is >100 μg/liter.
Figure 3.
Identification of LHX4 molecular defect. A, Nucleotide sequences of normal (top) and mutant (bottom) LHX4 alleles, as determined after cloning of the LHX4 PCR products into TOPO-XL cloning vectors (Invitrogen). The G→C substitution (boldface; position indicated by asterisk) identified in the mutant allele creates a recognition sequence for the MaeII restriction enzyme (box). B, Segregation analysis of splice-acceptor–site mutation in the family. The presence of the mutation in the genomic DNA of six family members was assessed by means of MaeII digestion of PCR products generated by primers bracketing the splice site. This assay confirmed that all affected members tested (lanes II2, III3, IV1, and IV3) have one mutant (244 bp and 108 bp) and one normal (352 bp) allele, whereas the unaffected individuals (III4 and IV2) have only normal (352 bp) alleles. The size marker (lane L) is the “Smart-ladder” (Eurogentec).
One Mutant LHX4 Allele Predicting Two Mutant LHX4 Proteins
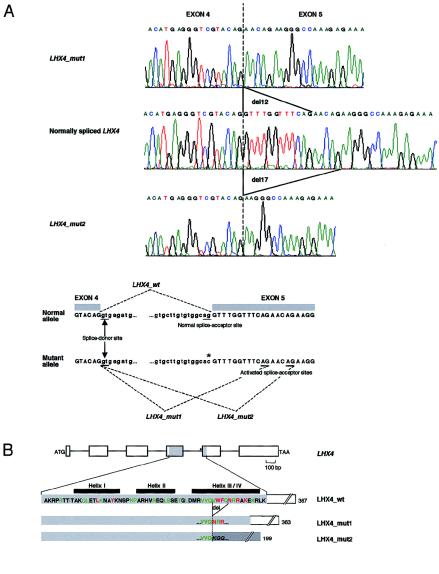
To determine the consequences that this splice-acceptor-site mutation has on the processing of pituitary LHX4 transcripts, CHO cells were transfected with normal or mutant LHX4 allelic constructs. Because of the large size of the gene, a human LHX4 cDNA fragment spanning exons 1–4 was fused to a genomic fragment spanning exons 4–6 and corresponding to either the normal allele or the mutant allele. RT-PCR amplification of LHX4 transcripts isolated from cells transfected with the wild-type minigene yielded a 1,264-bp amplicon consistent with the normal splicing of the intron located between exons 4 and 5. In contrast, a similar assay, performed using the mutant construct, generated two fragments, of 1,252 bp and 1,247 bp (17 and 3 clones, respectively, of the 20 clones tested). Sequence analysis of these latter products revealed that they resulted from the use of two cryptic splice-acceptor sites located within exon 5 (fig. 4A); at the protein level, the use of the first splice-acceptor site would lead to the in-frame deletion of four highly conserved amino acids in the third helix (i.e., helix III/IV) of the homeodomain (i.e., deletion of VWFQ at positions 47–50 in the homeodomain); the use of the second splice-acceptor site would alter the reading frame at position 47 of the homeodomain, leading to a premature termination codon within exon 5 (fig. 4B). The nature and the location of this mutation therefore are consistent with a drastic alteration of the function of LHX4.
Figure 4.
Consequences of LHX4 intronic mutation, at RNA level and protein level. A, Effect that mutation has on splicing of LHX4 primary transcripts. Top, Sequence electropherograms of normal and mutant LHX4 transcripts. The mutant allele generates two populations of transcripts—one with a deletion of 12 nucleotides (LHX4_mut1) and another with a deletion of 17 nucleotides (LHX4_mut2), both located within the coding sequence of exon 5. Bottom, Schematic representation of in vitro splicing of normal and mutant LHX4. The location of the G→C substitution is indicated by an asterisk. The invariant splice-donor site and the three splice-acceptor sites (i.e., the site normally used, as well as the two cryptic splice sites that are activated in the mutated allele) are underlined. B, Evolutionary conservation of amino acid residues involved in LHX4-homeodomain deletions generated by mutant allele. Top, Schematic presentation of genomic structure of LHX4. Exons are drawn to scale and are depicted by rectangles. (Introns are not drawn to scale.) Sequences from exons 4 and 5 encode the homeodomain (shaded). The splice-acceptor–site mutation (indicated by an asterisk) is located upstream from exon 5. Bottom, Normal and mutant LHX4-homeodomain amino acid sequences. The amino acid sequence of normal LHX4 (LHX4_wt) is shown against a light-gray background. Helices are denoted by black boxes. In the consensus sequence, the seven positions occupied by amino acids that are invariant in >95% of the homeodomain proteins thus far identified are indicated by red letters. Highly conserved residues (i.e., at the most, three different amino acids at these locations in all homeodomain proteins thus far identified) are indicated by green letters (Burglin, T. R.). The mutant allele generates two populations of transcripts (encoding mutant LHX4 proteins)—one maintaining the normal reading frame and another changing the reading frame and resulting in the creation of a premature termination codon within exon 5. The four amino acids, which are deleted when either of the two cryptic splice-acceptor sites is used, are indicated on the normal LHX4-homeodomain sequence (del). The LHX4-homeodomain products (LHX4_mut1 and LHX4_mut2) resulting from the mutation are drawn to scale, in proportion to the length of the normal homeodomain. Part of the nonsense sequence, which results from the frameshift introduced by the activation of the splice-acceptor site located 17 nucleotides downstream of the normal one, is shown in italic against a dark-gray background. Numbers indicate the total length of predicted amino acid sequences.
Discussion
We have described the molecular basis of a complex Mendelian disorder characterized by short stature and pituitary and cerebellar defects in combination with abnormalities of the central skull base. We have shown that this condition is due to a germline mutation within the LIM-homeobox transcription factor LHX4, a result revealing previously unsuspected functions of this gene.
In this study, we have provided evidence that a G→C substitution in the intron preceding exon 5 of LHX4 is indeed the defect responsible for autosomal dominant syndromic short stature, on the basis of the following data: (a) Within this large family, over three generations, there is a perfect cosegregation of this mutation with the disease phenotype. (b) This mutation involves the last invariant nucleotide of a splice-acceptor site, the functional importance of which, in the proper splicing of primary transcripts, is well documented (Shapiro and Senapathy 1987; Burset et al. 2001). (c) As shown by means of in vitro–expression studies, this mutation abolishes normal LHX4 splicing and activates two cryptic splice-acceptor sites located within exon 5. (d) If translated, the two aforementioned abnormal transcripts would generate LHX4 proteins with mutations in their helix III/IV of the homeodomain—one transcript would generate an in-frame deletion of four highly conserved residues of the 60-amino-acid-long homeodomain, and the other would generate a truncated protein in which the homeodomain is severely affected; helix III/IV is the region of the homeodomain-containing transcription factors that is in contact with the major groove of the DNA (Gehring et al. 1994). This helix therefore is essential for the formation of homeodomain-specific DNA-target complexes. Altogether, these data strongly suggest that the LHX4 intronic variation identified herein is the disease-causing mutation, which most probably results in the inability of LHX4 to control the expression of as-yet-unknown target genes.
Remarkably, in mice, the targeted disruption of Lhx4 is asymptomatic in the heterozygous state. In contrast, the LHX4 mutation identified in this kindred is responsible for a complex disease phenotype in the heterozygous state, thereby raising the question of what the mechanism is by which such a defect impairs LHX4-dependent transcriptional cascades. Also, we cannot exclude the possibility that this mutation may be lethal in the homozygous state, since that is the case in animals carrying the Lhx4−/− genotype (Li et al. 1994; Sheng et al. 1997).
It may seem somewhat surprising that the probands, who were born of a consanguineous union, carry the defect at the LHX4 locus in the heterozygous state; however, several lines of evidence indicate that consanguinity is irrelevant to the occurrence of the disease condition. First, the pathological trait is transmitted in a dominant manner, affecting only the maternal side of the kindred. Second, the mutant allele cosegregates perfectly, over at least three generations, with the complex disease phenotype (i.e., all individuals who bear one copy of the mutant allele exhibit short stature and the defect in the sella turcica). In addition, variability in clinical expression of the disease phenotype—which segregates in a fully penetrant manner—has been clearly documented in this kindred. This phenomenon, which is a common feature of dominantly inherited genetic disorders, may reflect the existence of epigenetic events and/or modifying factors (Peltonen and McKusick 2001). In the family presented herein, such variability in clinical expression of the disease phenotype is indeed observed in more than one aspect of the phenotype—although the probands have a deficit of several anterior pituitary hormones, the other patients present with an isolated GH deficiency revealed by short stature. In addition, the two probands display an ectopia of the posterior hypophysis, but this phenotypic feature—the expression of which varies between these two patients—is not observed in all affected individuals.
Finally, the disease phenotype includes the pointed cerebellar tonsils observed in patients with Chiari malformation (Meadows et al. 2000), a heterogeneous condition of unknown etiology. We therefore assume that LHX4-dependent transcriptional cascades could be implicated in the pathogenesis of a developmental anomaly that, together with the abnormal location of the posterior hypophysis, somehow is reminiscent of the cell-pathfinding defect in Lhx4−/− mice described elsewhere (Sharma et al. 1998). It is also worth noting that, in addition to the pituitary and hindbrain defects, the patients display a poorly developed sella turcica; this strongly suggests that an LHX4-induced signaling pathway therefore is required for proper shaping of the sphenoid bone underneath the pituitary gland. Overall, these data, which elucidate the molecular basis of a complex developmental disorder, reveal unsuspected functions of LHX4, a homeodomain transcription factor that hence emerges as a morphogenetic organizer of cephalic development in humans.
Acknowledgments
We are grateful to the family for participating in this study. We wish to thank Dr. Catherine Garel for evaluation of radiographs and magnetic-resonance imaging of the head. This work was supported by Assistance Publique/Hôpitaux de Paris grant CRC96085 and by a grant from the Association Française contre les Myopathies. K.M. and J.P. are the recipients of fellowships from the Ministère de l’Education Nationale, de la Recherche et de la Technologie, and from the Institut National de la Santé et de la Recherche Médicale, respectively.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Burglin, T. R., http://www.biosci.ki.se/groups/tbu/homeo/consensus.gif (for information relevant to homeobox proteins and their classification and evolution)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for mouse Lhx4 cDNA sequence [accession number AF135415] and for human LHX4 cDNA sequence [accession number AF282899])
- GeneMap'99, http://www.ncbi.nlm.nih.gov/genemap99/ (for in silico mapping of human ESTs)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Chiari type I malformation [MIM 118420] and Lhx4 [MIM 602146])
References
- Burset M, Seledtsov IA, Solovyev VV (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res 29:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couly G, Le Douarin NM (1988) The fate map of the cephalic neural primordium at the presomitic to the 3-somite stage in the avian embryo. Development 103:101–113 [DOI] [PubMed] [Google Scholar]
- Dattani MT, Martinez-Barbera JP, Thomas PQ, Brickman JM, Gupta R, Martensson IL, Toresson H, Fox M, Wales JK, Hindmarsh PC, Krauss S, Beddington RS, Robinson IC (1998) Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nat Genet 19:125–133 [DOI] [PubMed] [Google Scholar]
- Dubois PM, El Amraoui A, Héritier AG (1997) Development and differentiation of pituitary cells. Microsc Res Tech 39:98–113 [DOI] [PubMed] [Google Scholar]
- Eagleson GW, Harris WA (1990) Mapping of the presumptive brain regions in the neural plate of Xenopus laevis. J Neurobiol 21:427–440 [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Affolter M, Burglin T (1994) Homeodomain proteins. Annu Rev Biochem 63:487–526 [DOI] [PubMed] [Google Scholar]
- Ikeda H, Suzuki J, Sasano N, Niizuma H (1988) The development and morphogenesis of the human pituitary gland. Anat Embryol 178:327–336 [DOI] [PubMed] [Google Scholar]
- Japon MA, Rubinstein M, Low MJ (1994) In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem 42:1117–1125 [DOI] [PubMed] [Google Scholar]
- Lamolet B, Pulichino A, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J (2001) A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell 104:849–859 [DOI] [PubMed] [Google Scholar]
- Li H, Witte DP, Branford WW, Aronow BJ, Weinstein M, Kaur S, Wert S, Singh G, Schreiner CM, Whitsett JA (1994) Gsh-4 encodes a LIM-type homeodomain, is expressed in the developing central nervous system and is required for early postnatal survival. EMBO J 13:2876–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J, Kraut M, Guarnieri M, Haroun RI, Carson BS (2000) Asymptomatic Chiari type I malformations identified on magnetic resonance imaging. J Neurosurg 92:920–926 [DOI] [PubMed] [Google Scholar]
- Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, Marcos E, Duriez B, Cacheux V, von Moers A, Goossens M, Gruters A, Amselem S (2000) Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet 25:182–186 [DOI] [PubMed] [Google Scholar]
- Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K (1994) The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of mouse embryos. Dev Biol 164:409–419 [DOI] [PubMed] [Google Scholar]
- Peltonen L, McKusick VA (2001) Genomics and medicine: dissecting human disease in the postgenomic era. Science 291:1224–1229 [DOI] [PubMed] [Google Scholar]
- Pfaffle RW, DiMattia GE, Parks JS, Brown MR, Wit JM, Jansen M, Van der Nat H, Van den Brande JL, Rosenfeld MG, Ingraham HA (1992) Mutation of the POU-specific domain of Pit-1 and hypopituitarism without pituitary hypoplasia. Science 257:1118–1121 [DOI] [PubMed] [Google Scholar]
- Radovick S, Nations M, Du Y, Berg LA, Weintraub BD, Wondisford FE (1992) A mutation in the POU-homeodomain of Pit-1 responsible for combined pituitary hormone deficiency. Science 257:1115–1118 [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL (1998) LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell 95:817–828 [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Moriyama K, Yamashita T, Li H, Potter SS, Mahon KA, Westphal H (1997) Multistep control of pituitary organogenesis. Science 278:1809–1812 [DOI] [PubMed] [Google Scholar]
- Tatsumi K, Miyai K, Notomi T, Kaibe K, Amino N, Mizuno Y, Kohno H (1992) Cretinism with combined hormone deficiency caused by a mutation in the PIT1 gene. Nat Genet 1:56–58 [DOI] [PubMed] [Google Scholar]
- Watkins-Chow DE, Camper SA (1998) How many homeobox genes does it take to make a pituitary gland? Trends Genet 14:284–290 [DOI] [PubMed] [Google Scholar]
- Wu W, Cogan JD, Pfaffle RW, Dasen JS, Frisch H, O'Connell SM, Flynn SE, Brown MR, Mullis PE, Parks JS, Phillips JA, Rosenfeld MG (1998) Mutations in PROP1 cause familial combined pituitary hormone deficiency. Nat Genet 18:147–149 [DOI] [PubMed] [Google Scholar]
- Yamashita T, Moriyama K, Sheng HZ, Westphal H (1997) Lhx4, a LIM homeobox gene. Genomics 44:144–146 [DOI] [PubMed] [Google Scholar]