Abstract
Mutations in the COL3A1 gene that encodes the chains of type III procollagen result in the vascular form of Ehlers-Danlos syndrome (EDS), EDS type IV, if they alter the sequence in the triple-helical domain. Although other fibrillar collagen–gene mutations that lead to allele instability or failure to incorporate proα-chains into trimers—and that thus reduce the amount of mature molecules produced—result in clinically apparent phenotypes, no such mutations have been identified in COL3A1. Furthermore, mice heterozygous for Col3a1 “null” alleles have no identified phenotype. We have now found three frameshift mutations (1832delAA, 413delC, and 555delT) that lead to premature termination codons (PTCs) in exons 27, 6, and 9, respectively, and to allele-product instability. The mRNA from each mutant allele was transcribed efficiently but rapidly degraded, presumably by the mechanisms of nonsense-mediated decay. In a fourth patient, we identified a point mutation, in the final exon, that resulted in a PTC (4294C→T [Arg1432Ter]). In this last instance, the mRNA was stable but led to synthesis of a truncated protein that was not incorporated into mature type III procollagen molecules. In all probands, the presenting feature was vascular aneurysm or rupture. Thus, in contrast to mutations in genes that encode the dominant protein of a tissue (e.g., COL1A1 and COL2A1), in which “null” mutations result in phenotypes milder than those caused by mutations that alter protein sequence, the phenotypes produced by these mutations in COL3A1 overlap with those of the vascular form of EDS. This suggests that the major effect of many of these dominant mutations in the “minor” collagen genes may be expressed through protein deficiency rather than through incorporation of structurally altered molecules into fibrils.
Introduction
Ehlers-Danlos syndrome (EDS) type IV (EDS type IV [MIM 130050]), the vascular or ecchymotic type (Beighton et al. 1998), is a dominantly inherited disorder that results from mutations in the COL3A1 gene, which encodes the constituent chains of type III procollagen (Pope et al. 1977; Byers and Schwarze, in press). Many affected individuals have fragile, thin, or translucent skin through which the venous pattern is readily visible. They generally bruise easily and often have unexplained ecchymoses in well-protected areas. Some have a “characteristic” facies with tight skin, thin nose, and a “stare” (Pope et al. 1980, 1988). Large-joint mobility is generally normal, with hypermobility limited to the small joints of the hands. The major complications are arterial, bowel, and uterine rupture. Because of these dramatic complications, life expectancy is shortened to a mean of <50 years (Pepin et al. 2000).
The causative gene, COL3A1 (accession numbers 193023938–193062278 in the Human Genome Project Working Draft at UCSC), located at 2q31-q32, contains 52 exons (of which exons 4 and 5 are fused to create a single exon) distributed over 44 kb (Chu and Prockop 1993). New mutations are common, and approximately one-half of identified individuals with the condition have no apparent family history of the disorder. More than 200 mutations in the COL3A1 gene have been identified (Kuivaniemi et al. 1997; Pepin et al. 2000) (see the “Mutations in COL3A1” database), all of which lead to synthesis of an abnormal type III procollagen protein. Approximately two-thirds of the mutations are single-nucleotide substitutions that result in substitutions for glycine residues in the triple-helical domain of the proα1(III) chain. Most of the rest are splice-site mutations that result in exon skipping, although some have more-complex outcomes (Kuivaniemi et al. 1990; Schwarze et al. 1997), and a small number are larger genomic deletions (Kuivaniemi et al. 1997). No correlations have been discerned between the nature or location of the mutation and the type or frequency of major complications (Pepin et al. 2000).
To date, no COL3A1 mutations that result in loss of transcript stability—and, thus, in half-normal amounts of type III procollagen—have been reported. One hypothesis to explain the failure to identify this class of mutations is that the phenotype caused by COL3A1 haploinsufficiency differs, with respect to its severity or its symptomatic range, from the usual presentation of EDS type IV and has been excluded from analysis. Heterozygous mice with a Col3a1 null allele generated by targeted gene inactivation were phenotypically normal (Liu et al. 1997), although late onset of signs would have been missed if they had occurred after the follow-up period of 18 mo. The notion that milder or late-onset vascular involvement may be a candidate phenotype for the COL3A1 null-allele genotype was weakened by the data from two large studies of individuals with cerebral aneurysms (Kuivaniemi et al. 1993) and abdominal aortic aneurysms (Tromp et al. 1993), inasmuch as the majority of affected individuals had evidence of expression of both COL3A1 alleles.
We have now identified mutations in four individuals that resulted in effective COL3A1 haploinsufficiency. Each of the individuals had a clinical course compatible with EDS type IV, with significant morbidity due to vascular complications. There were two molecular mechanisms. In three of the individuals, frameshift mutations resulted in premature termination codons (PTCs) and nonsense-mediated decay (NMD) of mRNA; in the fourth individual, a nonsense mutation, in the terminal exon, which resulted in a PTC, was associated with both stability of the mRNA and a shortened protein that failed to participate in trimer formation and that thus reduced the amount of the mature molecule produced.
Subjects, Material, and Methods
Clinical Summary
Thirty-nine individuals whose cells appeared to make only type III procollagen molecules of normal mobility were selected, to determine whether mRNA from both COL3A1 alleles was present. On the basis of clinical evaluation, EDS type IV was the most likely diagnosis in 26 individuals and (because of familial aneurysms or spontaneous pneumothorax) was a reasonable differential diagnosis in 6 individuals. Five additional individuals who had signs consistent with classic EDS (i.e., EDS types I/II) were studied to rule out EDS type IV. Two additional individuals had been diagnosed with EDS type VIII. In the first group, cells from 3 (individuals P1–P3) of 26 individuals expressed stable COL3A1 mRNA from only one allele and were selected for further studies; also in the first group, in cells from 5 additional individuals, which had stable COL3A1 mRNA expression from both alleles, the region of mRNA encoding the C-propeptide was sequenced directly, and one abnormality (in individual P4) was identified.
Individual 1 (P1) was a 31-year-old female with characteristic physical findings: a “pinched” nose, thin lips, lobeless ears, scant subcutaneous tissue with easily visible veins, multiple ecchymoses, numerous wide scars from surgical procedures, and hypermobility of the finger joints. Easy bruisability since childhood was reported. At age 21 years, she had undergone colectomy because of “polyposis coli,” and the bowel tissue was noted to be paper thin. Several laparotomies for the repair of enterocutaneous fistulae were complicated by bowel anastomoses and wound dehiscence. At age 29 years, rupture of a common-hepatic-artery aneurysm resulted in a large retroperitoneal hematoma that required surgical intervention, and the blood vessels were described as very fragile. Postoperative angiography showed a renal aneurysm and a superior-mesenteric-artery dissection. At age 30 years, a sciatic-nerve palsy developed because of a spontaneous gluteal hematoma. At age 31 years, she experienced a subarachnoid hemorrhage. Cerebral angiography revealed multiple aneurysms and marked tortuosity of all cervical arteries. During surgery, the ophthalmic artery, after very minor manipulation, tore off the carotid artery, which resulted in a massive cerebral infarct from which the patient died. The patient’s mother and sister had a similar facial appearance, very thin skin, easy bruisability, and several vascular events, including vertebral- and internal-carotid-artery dissections resulting in brain-stem and hemispheric infarcts.
Individual 2 (P2) is a 46-year-old female who had a history of easy bruising with minor or no trauma and some hypermobility of the small joints of her hands. Her skin was not translucent but showed markedly decreased mechanical strength when a punch biopsy was performed. At age 44 years, she presented to the emergency room with an atraumatic hemoperitoneum and hemorrhagic shock. Exploratory laparotomy revealed multiple bleeding vessels; bleeding from the spleen, from the pancreatic tail bed, and from lacerations of the liver; and a torn mesocolon. During surgery, tissue friability was not noted to be abnormal, but the postoperative course was complicated by signs of continued bleeding that required re-exploration. Postoperative angiography showed numerous aneurysms of multiple large- and medium-size abdominal arteries and a distal abdominal aortic aneurysm with diffuse dilatation of the common iliac arteries and of the external and internal iliac arteries. Smaller wounds usually healed well, with wide scars, whereas the surgical wounds caused chronic problems, with herniations of viscera, which have required repeated hospitalization. This individual's family history is remarkable only for easy bruisability, by report, in her mother and for facial features in one of her daughters that are reminiscent of EDS type IV.
Individual 3 (P3) is a 48-year-old male with soft, velvety, and translucent skin and some atrophic scars on his lower extremities. He had no major illness until age 47 years, when he had sudden onset of headache followed by acute left-flank pain. Abdominal computed tomography (CT) revealed multiple infarcts in the left kidney, and subsequent angiography showed dissection of the left renal artery, with clot formation in the false lumen. Ultrasonography of the carotid arteries revealed no abnormalities. A few days later, this individual had another acute episode of abdominal pain. Angiography revealed multiple dissections involving the splenic and hepatic branches of the celiac artery and both renal arteries, as well as occlusion of the gastroduodenal artery. He became critically ill, with signs of cardiovascular instability, adrenal insufficiency, and acute respiratory failure. Because of developing symptoms of abdominal-compartment syndrome, he underwent exploratory laparotomy with temporary closure and mesh placement. He had good wound healing after the procedure, but skin grafting was required for complete closure of the abdominal wall. Bilateral abdominal tissue expanders were placed, with a gradual increase in the injected volume. A few months after this procedure, a rupture of a subcutaneous vessel occurred. Except for a large superficial hematoma, an abdominal CT did not disclose other abnormalities. Surgery for abdominal-wall closure is planned. There are no other family members with clinical signs consistent with EDS type IV.
Individual 4 (P4) is a 46-year-old black female with a lifelong history of easy bruising. At age 45 years, she was evaluated for anemia and heavy menses. Pelvic ultrasonography and abdominal CT revealed an aneurysm of the left common iliac artery. Three months later, she had sudden onset of severe abdominal pain, at which time CT and angiography revealed a new infrarenal abdominal aortic aneurysm and a dissection extending from the infrarenal aorta to the left common iliac artery. She underwent surgery, with ligation of the infrarenal abdominal aorta combined with an axillobifemoral bypass and fenestration of the iliac dissection. The postoperative course was complicated by dehiscence of the axillary anastomosis, which required surgical revision, from which she recovered without further complications. The proband’s parents died in their 60s, the father from cancer and the mother from unknown cause during her sleep, so that the diagnosis of EDS type IV in the mother cannot be excluded. Of the proband’s three sisters (who were ages 54, 52, and 48 years), the oldest was evaluated for EDS type IV, because of easy bruising, soft and thin skin, hypermobile joints, inguinal hernias, aneurysmal dilatation of the superior mesenteric artery, and repeated colorectal surgeries due to frequent episodes of abdominal pain, bowel obstruction, and fistulae in the absence of inflammatory bowel disease. In the two younger sisters, no vascular complications or spontaneous bowel perforation have been reported. The proband’s only child (a 15-year-old male) is healthy.
Cell Culture and Analysis of Collagenous Proteins
Dermal fibroblasts were obtained from explants of skin-biopsy specimens from the four individuals, after appropriate consent had been obtained. Growth and maintenance of those fibroblasts and of control cell strains, radiolabeling of collagenous proteins, and analysis of proα chains and α chains by SDS-PAGE were performed as described elsewhere (Bonadio et al. 1985).
Preparation of cDNA and Genomic DNA, and Analysis of Expressed Polymorphisms in COL3A1
Total cellular RNA and genomic DNA were extracted from cultured dermal fibroblasts from P1–P4 and of controls, by RNeasy Mini Kit (Qiagen) and QIAamp DNA Mini Kit (Qiagen), respectively. In addition, nuclear and cytoplasmic RNA were prepared from cultured fibroblasts from P1, P3, and a control cell strain, which were incubated in the absence or presence of cycloheximide (CHX [100μg/ml]) for 6 h before the cell harvest, by guanidinium thiocyanate-phenol-chloroform extraction (Chomczynski and Sacchi 1987), as described elsewhere (Schwarze et al. 1999). Complementary DNA was synthesized by priming with random hexamers in the presence of Superscript II reverse transcriptase (Gibco BRL) in total RNA or in nuclear/cytoplasmic RNA after treatment with DNase I (Boehringer-Mannheim) for 20 min at 37°C with the incubation buffer recommended by the manufacturer. To check for residual genomic DNA in the RNA preparation, a control PCR was performed without prior reverse transcription.
Three single-nucleotide polymorphisms, in exons 27, 31, and 33, and an “AG” insertion/deletion polymorphism in the 3′UTR of COL3A1, with a combined heterozygosity of ∼.80, were analyzed in genomic DNA and cDNA (cDNA reference sequence is GenBank accession number NM_000090). Fragments were amplified, cleaved with the appropriate restriction enzyme, and separated by PAGE. The polymorphic variants used were G/A at nucleotide 1851 (with reference to the A of the initiating methionine, which is considered to be nucleotide 1 in the cDNA sequence) in exon 27 (Tromp et al. 1993), G/A at nucleotide 2092 in exon 31 (Zafarullah et al. 1990), and C/T at nucleotide 2244 in exon 33 (Tromp et al. 1991). 1851G/A was assayed as a DdeI site; 2092G/A as a SpeI site, by use of a mismatch primer (table 1), and also as an AluI site for P1; and 2244C/T as a HaeIII site in genomic DNA and as a Fnu4HI site in cDNA. The amplification primers are listed in table 1. Unless otherwise specified, all amplifications consisted of 35 PCR cycles.
Table 1.
Restriction Enzymes Employed to Distinguish between Alleles—and Primers Used to Amplify Genomic DNA and cDNA around Polymorphic Sites—in COL3A1 Exons 27, 31, and 33
| Polymorphic Site | Amplification Primers(5′→3′) | AnnealingTemperature(°C) |
| DdeI 1851G/A (exon 27): | ||
| Genomic DNA | GCTCCTGGTAAGAATGGAGAACG (exon 26) | 58 |
| GGGTCCTGTGTCTCCTTTGTC (exon 28) | ||
| cDNA | TCCTGGAAAGAATGGTGAAACTG (exon 27) | 60 |
| CTTGCCTCCTGGAGCTCCAGGTG (exon 30) | ||
| SpeI 2092G/A (exon 31): | ||
| Genomic DNA | AAGTATACAAATTTCTAGATTG (intron 30) | 46 |
| CTCCTTCGGGACCAGGGGGACTAG (exon 31)a | ||
| cDNA | GTGATGCCGGTGCACCTGGAGCT (exon 30) | 60 |
| CTCCTTCGGGACCAGGGGGACTAG (exon 31)a | ||
| AluI 2092G/A (exon 31): | ||
| Genomic DNA | AAGTATACAAATTTCTAGATTG (intron 30) | 46 |
| ATAAATGATCAGAAGGAAATCA (intron 31)b | ||
| cDNA | GTGATGCCGGTGCACCTGGAGCT (exon 30) | 58 |
| GCAAGCTTTCCCTGGGACACCAT (exon 33)b | ||
| 2244C/T (exon 33): | ||
| Genomic DNA (HaeIII) | CAACACTCCTGGAAAGTAATCG (intron 32) | 56 |
| AGTGCAGGACTGTCCCATATG (intron 33) | ||
| cDNA (Fnu4HI) | AGGAATTCCTGGAGAAAGAGGAG (exon 32) | 60 |
| TTCACCTTTGACACCTTGGGGA (exon 38) |
Underscored is the mismatched nucleotide introduced to create a SpeI site.
Antisense primer end-labeled with γ[32P]-ATP (3,000 Ci/mmol; Amersham) by T4 polynucleotide kinase (New England BioLabs) and added to the last cycle of a PCR that consisted of 26 cycles to prevent heteroduplex formation and an artificial bias of cut:uncut product ratio.
Heterozygosity screening for an AG insertion/deletion polymorphism in the 3′UTR of COL3A1 (U. Schwarze, unpublished data) was performed by amplification of a 568–570-bp fragment, by use of primers 5′-TCTTTGAATCCTAGCCCATCTG-3′ and 5′-TGTGACAAAAGCAGCCCCATAA-3′ (10 s at 95°C, 20 s at 55°C, and 35 s at 72°C, for 35 cycles) and separation of the products both on 6% acrylamide and on MDE (BMA) gels. The nucleotide sequence of the 3′UTR was derived from GenBank (accession number NM_000090). 5302A was either deleted or replaced by AG.
Identification of Mutations That Led to Unstable mRNA in P1 and P2
Depending on which allele was determined to be stable on the basis of the analysis of 2092G/A in exon 31, the primers listed in table 2 were used to amplify the COL3A1 coding sequence in two large fragments, through 15 cycles. A 10-μl aliquot was digested with the appropriate restriction enzyme (table 2), under conditions recommended by the manufacturer (New England BioLabs), and a 2-μl aliquot was reamplified through 35 cycles with the same primer pairs that had been used for the initial amplification. The second amplification preferentially amplified the allele that had not been cut (i.e., the unstable allele), and the products were separated on 6% polyacrylamide gels and were excised from the gel. The gel slices were submerged in dH2O overnight at room temperature, and the supernatant was used to amplify, in 15 overlapping fragments, the COL3A1 coding sequence. The sequences of the primers (see the Appendix) were derived from the previously published cDNA sequence for COL3A1 (Ala-Kokko et al. 1989; Benson-Chanda et al. 1989). The PCR fragments were separated on 6% polyacrylamide gels. If a heteroduplex was generated, this fragment was sequenced selectively. If all fragments were indistinguishable from the control, then all fragments were sequenced with the amplification sense primer. Sequence was determined either by the dideoxy-chain–termination method (Sanger et al. 1977) using T7 polymerase (Sequenase version 2.0; US Biochemicals) after the fragment of interest was directly cloned into the PCR II vector, according to instructions provided in the TA cloning kit (Invitrogen), or by an ABI PRISM BigDye Terminator cycle sequencing reaction and ABI PRISM 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems). Identified mutations were confirmed in genomic DNA by amplification of the region of interest, followed by direct sequencing.
Table 2.
Primers Used to Amplify COL3A1 Coding Sequence in Two Large Fragments
| Stable Allele2092G/A (exon 31) | Amplification Primers(5′→3′) | RestrictionEnzymea |
| G allele | CTGAATTCCAGGGAACAACTTGATG (exon 1) | BsrBI |
| CTCCTTCGGGACCAGGGGGACCCG (exon 31)b | ||
| GGCCCCAGGACTTAGAGGTGCA (exon 31)b | BsgI | |
| TGATCCATGTATGCAATGCTAT (exon 51) | ||
| A allele | CTGAATTCCAGGGAACAACTTGATG (exon 1) | SpeI |
| CTCCTTCGGGACCAGGGGGACTAG (exon 31)b | ||
| GGGCCCCAGGACTTAGAGTTCGA (exon 31)b | BstBI | |
| AGGTAGTCTCACAGCCTTGCGTGT (exon 52) |
Allowed selective digestion of stable allele.
Underscored is (are) the mismatched nucleotide(s) introduced to create, for the restriction enzyme shown, a recognition sequence on the stable allele.
Identification of the Mutation That Led to Unstable mRNA in P3
Complementary DNA synthesized from nuclear RNA from P3 and from a control was used to amplify, in 15 overlapping fragments, the COL3A1 coding sequence (see above), and the products were separated on 6% polyacrylamide gels. Heteroduplex bands in P3 were excised from the gel and were submerged in dH2O overnight at room temperature, and the supernatant was used for reamplification by the same primer pair. The mutation was confirmed in genomic DNA by direct sequencing.
Mutation Identification in P4
The region of COL3A1 cDNA that encodes the carboxyl-terminus was amplified by primers in exon 45 (5′-TCCTGCTGGTTCCCGAGGTG-3′) and exon 52 (5′-GTTGACAAGATTAGAACAAGAGGAACA-3′) and were directly sequenced with the amplification primers. The mutation was confirmed in genomic DNA by the restriction endonuclease SspI.
Western Blot Analysis of Type III Procollagen
Proα chains from the medium and from the cell layer of cultured fibroblasts from P1 and from a control cell strain were separated by SDS-PAGE (7%) under reducing conditions, were transferred to nitrocellulose membrane, and were immunodetected by LF-72 antiserum (a polyclonal antibody that binds the amino-terminal telopeptide of proα1(III), which was kindly provided by Larry Fisher of the National Institute of Dental Research) (Fisher et al. 1995) and ECL reagents (Amersham).
Pulse-Chase Analysis of Procollagen Assembly
Fibroblasts from P4 and from a control cell strain were preincubated in serum-free medium and ascorbic acid for 4 h and then were pulse-labeled with 200 μCi of tritiated proline (85–130 Ci/mmol; Amersham) for 80 min. For chase, cells were rinsed with serum-free medium supplemented with 10 mM proline and were incubated in the same medium for 0, 20, 40, 60, or 80 min. Proteins from the medium and proteins from the cell layer were harvested separately by ethanol precipitation and were analyzed by SDS-PAGE under nonreducing conditions.
Results
Analysis of Type III Collagen Production by Patients’ Cultured Fibroblasts
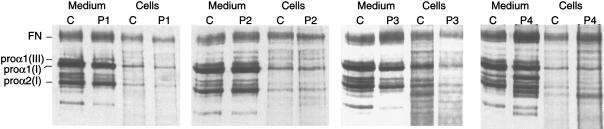
Radiolabeled procollagens from the medium and from the fibroblast cell layer were analyzed by SDS-PAGE, first after addition of a reducing agent (fig. 1) and then under nonreducing conditions after partial digestion, with pepsin, to remove the precursor-specific peptides at both ends of the triple-helical domain (not shown). In all cell strains (i.e., P1–P4), there was no overmodification or intracellular retention of proα1(III) chains and, thus, no evidence of a structural defect (fig. 1). Only in P1 did the amount of structurally normal type III procollagen secreted into the medium appear to be substantially diminished relative to the amount of type I procollagen and compared to that in the control cell strain (fig. 1).
Figure 1.
SDS-PAGE of radiolabeled procollagens from medium and fibroblasts cell layer, analyzed under reducing conditions, from a control cell line (lanes C) and cell strains from four individuals with EDS type IV (P1–P4). In cells from P1 the diminution in the amount of proα1(III) is evident, but it is not clear in the other three individuals. FN = fibronectin.
A Single Stable COL3A1 Allele in Cells from Three Patients with EDS type IV
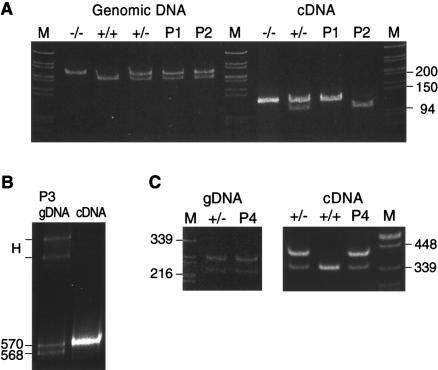
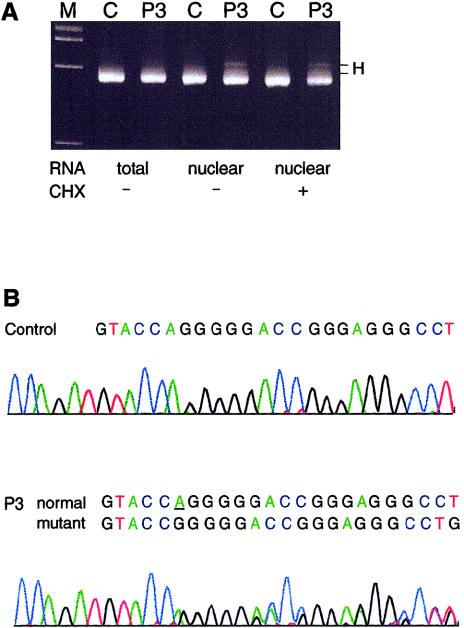
Although the evidence of reduced synthesis of type III procollagen was limited to a single cell strain, the clinical profile of all probands was sufficient to warrant further investigation to determine whether both COL3A1 alleles were effectively expressed. Samples of genomic DNA were analyzed for heterozygosity of COL3A1 markers. P1 and P2 were heterozygous for 2092G/A in exon 31 (fig. 2A). When cDNA amplified from exon 30 to exon 31 was digested with SpeI, in the sample from P1, only the G allele, SpeI(−), was detectable. In the cDNA sample from P2, the abundant species was the A allele, SpeI(+), although there was a very small amount of the G allele present (fig. 2A). P3 was homozygous, in genomic DNA, for the markers in exons 27, 31, and 33 but was heterozygous for the AG insertion/deletion in the 3′UTR. When cDNA was amplified and analyzed by PAGE, the heteroduplex that indicated the presence of both alleles in genomic DNA was greatly diminished (not shown). Separation of genomic and cDNA amplification products on MDE gel showed that only transcripts derived from the AG-insertion allele were stable (fig. 2B). P4 was heterozygous, in genomic DNA, for 2244C/T in exon 33, and both alleles were detectable in cDNA, in amounts comparable to those in the heterozygous control individual (fig. 2C).
Figure 2.
Demonstration of loss of expression of one COL3A1 allele in three of four individuals with EDS type IV. A, Heterozygosity for the exon 31 (2092G/A) polymorphism assayed as a SpeI site in genomic DNA (left) from P1 and P2. The cDNA from P1 contained only the SpeI(−) allele. In the cDNA from P2, only the SpeI (+) allele was stable. B, Amplification products for the AG insertion/deletion in the 3′UTR, separated on MDE gel. In genomic DNA from P3 (left lane) both alleles were detectable in equal amounts, whereas in cDNA (right lane) only the AG-insertion allele was present, which resulted in the disappearance of the heteroduplex (H). C, Heterozygosity for the exon 33 (2244C/T) polymorphism assayed as a HaeIII site in genomic DNA (left) from P4. In cDNA, after digestion with Fnu4HI, both alleles were present in P4, comparable to the heterozygous control.
Identification of Mutations That Led to Unstable mRNA
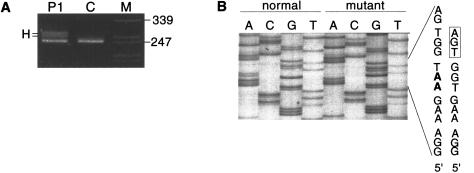
After enrichment for the unstable allele (see the Subjects, Material, and Methods section), analysis of cDNA from P1 revealed a heteroduplex in the amplification product that extended from exon 22 to exon 28 (not shown). Excision of the heteroduplex bands and reamplification with the same primer set, followed by sequencing, identified a 2-bp deletion in exon 27 (1831–1832delAA) and, in the same exon, a PTC (i.e., TGA) 3 nucleotides downstream of the deletion site. The mutation was confirmed by amplification of exon 27 in genomic DNA (fig. 3A) and sequence determination after separation of the normal and mutant allele by the TA cloning procedure (fig. 3B).
Figure 3.
Amplification of COL3A1 exon 27 in genomic DNA from P1 and from a control individual. A, Apparent heteroduplex (H) in P1, owing to deletion of 2 bp in one COL3A1 allele. B, Nucleotide sequence in the normal (left four lanes) and mutant (right four lanes) alleles from P1. The two deleted nucleotides (AA) are in boldface in the letter code for the normal sequence. The consequence of the 2-bp deletion in P1 is a PTC (boxed triplet) immediately downstream of the deletion site.
Because the same approach to analysis of cDNA from P2 failed to demonstrate a heteroduplex, all 51 exons were sequenced after amplification, in 15 overlapping fragments, of the COL3A1 cDNA. The amplification product that spanned exons 2–8 showed superimposition of two sequences, because of a 1-bp deletion located within six consecutive C’s in fusion exon 4/5 (413delC), affecting the 12th of 13 Gly-Xaa-Yaa repeats that encode the minor triple helix of the N-propeptide. Consequent to the C deletion, a PTC (i.e., TAG) was encountered 77 nucleotides downstream in the N-telopeptide–encoding portion of exon 6. The mutation was confirmed by amplification of exon 4/5 in genomic DNA and direct sequencing (fig. 4).
Figure 4.
COL3A1 exon 4/5 genomic sequence around the 1-bp deletion (underlined) in P2.
In P3, amplification, in 15 overlapping fragments, of COL3A1 cDNA, derived from nuclear RNA, revealed a heteroduplex in the fragment that extended from exon 6 to exon 13 (fig. 5A). Exposure of cultured fibroblasts to cycloheximide (100 μg/ml) for 6 h before isolation of nuclear RNA did not enhance the appearance of the heteroduplex (fig. 5A). Excision of the heteroduplex bands, reamplification with the same primer set, and direct sequencing identified a 1-bp deletion in exon 7 (555delT) and a PTC (i.e., TAG) 80 nucleotides downstream in exon 9. The mutation was confirmed by amplification of exon 7 in genomic DNA and direct sequencing (fig. 5B).
Figure 5.
A, Amplification of COL3A1exons 6–13 in cDNA of a control individual and in P3. Complementary DNA was derived from either total cellular RNA, nuclear RNA, or nuclear RNA, after treatment with CHX (100 μg/ml). When nuclear RNA was used, in P3 a heteroduplex (H) became visible that was barely detectable in total RNA and that was not enhanced by treatment with CHX. B, Exon 7 genomic sequence around the 1-bp deletion (underlined) in P3, obtained by an antisense sequencing primer located in intron 7.
Comparison of RNA Stability in pre-mRNA and in Nuclear and Cytoplasmic RNA in P1
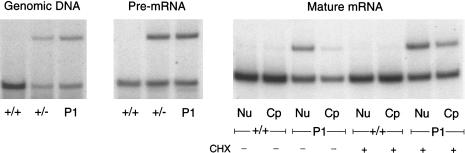
Exon 31 was amplified in genomic DNA and cDNA derived from pre-mRNA (by use of the nuclear-RNA preparation), with primers in introns 30 and 31 (table 1), and in cDNA derived from nuclear and cytoplasmic RNA, with primers in exons 30 and 33 (table 1). The respective antisense primer was end-labeled with γ[32P]-ATP (3,000 Ci/mmol; Amersham) by T4 polynucleotide kinase (New England BioLabs) and was added to the last cycle of a PCR, which consisted of 26 cycles, to prevent heteroduplex formation and an artificial bias of the cut:uncut product ratio. All amplification products were digested with AluI. The AluI(−):AluI (+) allele ratio in pre-mRNA from P1 was identical to that seen both in genomic DNA from P1 and in pre-mRNA and genomic DNA from the heterozygous control (fig. 6). In nuclear RNA, the AluI(−) allele (2092A) appeared to be markedly reduced, and it was barely detectable in cytoplasmic RNA (fig. 6). The addition of cycloheximide (100 μg/ml) to cultured fibroblasts for 6 h prior to RNA isolation resulted in partial stabilization of the unstable AluI(−) allele in both the nuclear and cytoplasmic compartments (fig. 6).
Figure 6.
Comparison of RNA stability in pre-mRNA and in nuclear and cytoplasmic RNA from P1. The two COL3A1 alleles were distinguishable after restriction digestion with AluI, owing to heterozygosity for 2092G/A. The AluI(−):AluI (+) allele ratio in pre-mRNA from P1 was similar to that seen in the heterozygous control. In nuclear (lanes Nu) RNA the AluI(−) allele was markedly reduced, and in cytoplasmic (lanes Cp) RNA it was barely detectable. The treatment of cells with CHX prior to RNA isolation resulted in increased appearance of the AluI(−) allele in both cellular compartments.
Western Blot Analysis of Secreted Type III Procollagen in P1
The location of the PTC in transcripts from the mutant allele in P1 predicted truncated proα1(III) chains of ∼67 kD, yet the transcript instability demonstrated earlier made it unlikely that those shortened chains were synthesized and secreted into the medium. Immunodetection of type III procollagen by an antibody against the N-telopeptide yielded a signal only for full-length proα1(III) chains, in both medium and cell layer and, in addition, for partially converted chains, pNα1(III), from which the C propeptide had been removed, in the medium (not shown).
Identification of a Nonsense Mutation in P4 That Did Not Affect mRNA Stability
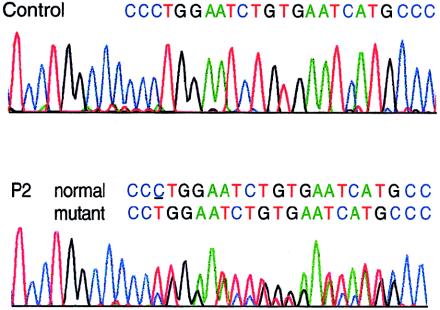
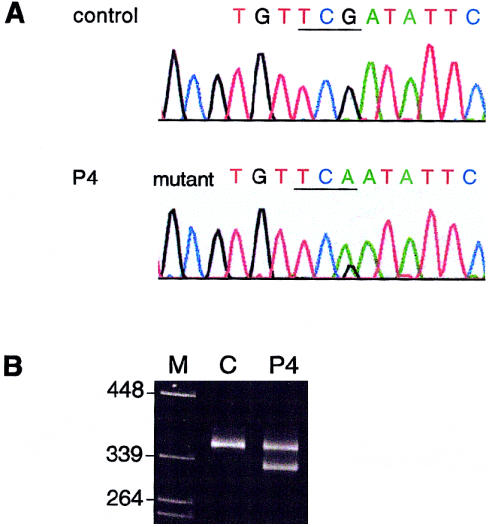
In P4, although mRNA from both COL3A1 alleles was found to be stable, the most carboxyl-terminal exons were sequenced in cDNA, to exclude the possibility of a PTC that did not affect mRNA stability but that resulted in truncated proα1(III) chains that either impaired or were excluded from proper chain trimerization. Direct sequencing of the cDNA fragment that comprised exons 45–52 identified, in exon 52, a nonsense mutation (4294C→T [R1432X]) that predicted truncation by 35 amino acid residues of proα1(III) chains derived from the mutant allele (fig. 7A). The truncation led to elimination of the most-carboxyl-terminal of eight cysteine residues in the C propeptide (C1464), a residue that forms an intrachain disulfide bond with the cysteine at position 1302, a process that precedes chain association into trimers. The mutation was confirmed in genomic DNA by the restriction endonuclease SspI, a recognition site for which was created by the nucleotide substitution (fig. 7B). Restriction digestion demonstrated the presence of the mutation also in the proband’s three sisters but its absence from the proband’s son.
Figure 7.
Heterozygosity for a nonsense mutation identified in P4, in exon 52 of COL3A1. A, Sequence around the site of the mutation obtained by an antisense sequencing primer. The substitution of T for C resulted in the conversion of a codon for arginine (CGA) to a PTC (TGA). B, Restriction digestion, with SspI, of genomic-amplification products from a control individual and from P4. The mutation shown in panel A created a recognition sequence for SspI.
Pulse-Chase Analysis of Procollagen Assembly in P4
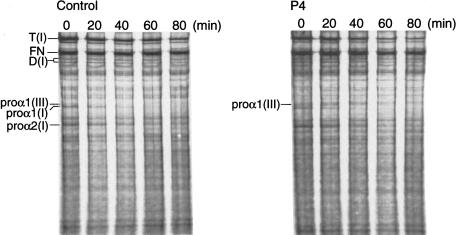
Because intrachain disulfide-bond formation in the C propeptide precedes chain trimerization during procollagen assembly, the effect that the absence of the most-carboxyl-terminal cysteine residue in shortened proα1(III) chains in P4 had on incorporation into trimers was examined by labeling of proteins with [3H]proline for 80 min and chase, for ⩽80 min, with excess unlabeled proline (fig. 8). In the cell layer of control fibroblasts, proα1(III)-chain monomers disappeared after 20–40 min (fig. 8). In contrast, in P4, defective proα1(III) chains persisted as monomers, with increasingly delayed electrophoretic mobility, for >80 min and were not rapidly degraded. To address the possibility that small amounts of defective proα1(III) chains were participating in trimer formation and were being secreted, proteins harvested from the medium layer during pulse-chase were separated by SDS-PAGE under reducing conditions, were transferred to nitrocellulose membrane, and were immunodetected by LF-72 antiserum. Both in the control and in P4, proα1(III) chains of normal length were detected, and there was no evidence of shortened chains in P4 (not shown).
Figure 8.
Pulse-chase analysis of procollagen assembly. Cells from a control and from P4 were pulse-labeled, and the cell-layer proteins were harvested either immediately or after differing periods of chase and were separated by SDS-PAGE under nonreducing conditions. In control cells, proα1(III) chain monomers were detectable only at times 0 min and 20 min. In P4, proα1(III) chain monomers with increasingly delayed electrophoretic mobility were detectable throughout the entire chase period, whereas the proα1(I) chains that migrated directly below the proα1(III) chains had completely disappeared at 60 min, in both the control and P4, owing to incorporation into dimers and trimers of type I procollagen, which are denoted by “D(I)” and “T(I),” respectively]. FN = fibronectin.
Discussion
Because mice heterozygous for a nonexpressed Col3a1 allele were reported to have no vascular phenotype (Liu et al. 1997), it was unexpected that people with either similar mutations or mutations that resulted in secretion of only half the normal amount of type III procollagen would have arterial fragility. It is striking that, in the present study, the individuals in whom we identified COL3A1 “null” mutations had a clinical course similar to that seen in individuals with EDS type IV (Pepin et al. 2000), all of whom had mutations that affected the structure of type III procollagen. All individuals in the present study had significant morbidity due to vascular rupture, and in at least one family those complications occurred during the 3d decade of life. There were no instances of bowel rupture, although the total number of identified individuals was small and bowel rupture accounts for only ∼20% of complication in individuals with mutations in COL3A1 that alter the structure of the protein product (Pepin et al. 2000). The findings in our study suggest that a 50% reduction of the amount of structurally normal type III procollagen is as deleterious to the vascular system as are other types of mutations. Because the number of individuals identified is still small, we are uncertain whether individuals heterozygous for COL3A1 null alleles are more likely to have a longer time to symptomatic arterial events. The length of the follow-up period in mice heterozygous for Col3a1 null mutations was ∼18 mo, so late onset of signs of arterial involvement might have been missed. Alternatively, mouse models might not replicate human disease phenotypes in all aspects.
Ultrastructural analyses of tissues derived from homozygous Col3a1−/− mutant mice, which have a phenotype that resembles human EDS type IV, have shown that the tissues most affected were those which have the greatest amount of type III collagen and are most severely affected in EDS type IV (Liu et al. 1997). No studies of the heterozygous mice have been described. In the Col3a1−/− mice, collagen fibrils were either absent from or severely reduced in number in the tunica media of the aorta. The diameter of collagen fibrils in the adventitia of the aorta was highly variable, and the number of fibrils was reduced to approximately one-third that in the normal aorta. The collagen fibrils in the skin were disorganized and highly variable in diameter; and collagen fibrils were either absent from or markedly reduced in number in the submucosa and serosa of the intestine. The collagen fibrils in all the tissues studied consist primarily of type I collagen, so it is likely that type III collagen plays an important role in the early organization of the collagenous network that provides tensile strength to these tissues.
Mutation analysis in individuals in whom the diagnosis of EDS type IV was made on clinical grounds has been very productive when cultured fibroblasts synthesized a cohort of abnormal type III procollagen molecules. In all instances identified thus far, the underlying mutations occur in one COL3A1 allele and result in either substitutions for single-glycine residues in the triple-helical domain of proα1(III) chains, small in-frame deletions, or splice-site mutations (Kuivaniemi et al. 1997; Pepin et al. 2000; Byers and Schwarze, in press) (see the “Mutations in COL3A1” database). In each instance, the effect of an abnormal allele product is amplified, since type III procollagen is a homotrimer. As a consequence, an abnormal product from one allele is expected to result in seven of eight molecules being abnormal. In contrast, the mutations described here appear simply to reduce the amount of type III collagen in a tissue, by virtue of either loss of the mRNA products of one allele or inability of the protein product to assemble into trimers. COL3A1 mutations that result in either loss of stable transcript or in functional loss of the protein products of one COL3A1 allele of type III procollagen appear to be either rare or difficult to identify, since we are unaware of additional examples of these types of mutations among the >200 mutations that have been compiled in the COL3A1 mutation database.
Heterozygosity for mutations that lead to loss of the products of one allele have been identified in several other fibrillar collagen genes, including COL1A1 (Willing et al. 1990, 1994, Willing et al. 1996; Stover et al. 1993; Redford-Badwal et al. 1996; Korkko et al. 1998; Ward et al. 2001), COL2A1 (Kuivaniemi et al. 1997; Freddi et al. 2000), and COL5A1 (Schwarze et al. 2000; Wenstrup et al. 2000). This class of mutations in the COL1A1 and COL2A1 genes leads to reduced production of their allele products and to the mildest of the phenotypes that result from mutations in those genes: osteogenesis imperfecta type I, in the case of the COL1A1 gene, and Stickler syndrome, in the case of the COL2A1 gene. In both instances, the genes encode the major protein products in the tissues in which they are expressed (type I procollagen/collagen and type II procollagen/collagen), the mutations reduce the amount of each mature protein to half-normal levels, and the proteins made are normal. In contrast, mutations in COL5A1 that alter either the amount produced or chain structure of type V collagen molecules, a minor product in most tissues, appear to have similar phenotypes—the EDS type I/II clinical picture. This effect is similar to that seen with the different types of mutation in the COL3A1 gene and thus may be characteristic of genes that do not encode the major structural product of the tissue.
Given the model of COL1A1 and COL2A1 genes in which haploinsufficiency mutations produce a milder variant of the more severe phenotype, it had been expected that such would be the case for mutations in the COL3A1 gene. Candidate phenotypes included isolated aneurysmal disease, but studies of individuals with either cerebral (Kuivaniemi et al. 1993) or abdominal aortic (Tromp et al. 1993) aneurysms failed to identify COL3A1 mutations, and the majority of affected individuals had evidence of expression of both alleles. Because the study was done by direct sequence analysis of cDNAs, nonexpressed alleles or unstable mRNA products would not have been detected in the minority of individuals who were homozygous (or hemizygous) at known polymorphic sites.
It has been difficult to correlate either the nature of the mutation, the type of substitution for glycine in the triple helix, the location of exon skipping, or the presence of insertions or deletions with variations in the phenotype that result from mutations in the COL3A1 gene. Pope et al. (1996) have suggested that individuals with mutations that alter sequences near the carboxyl-terminal end of the triple helix are more likely to have acrogeria. We noted a paucity of bowel rupture among individuals who had substitutions for glycine in the C-terminal quarter of the triple helix; but their ages were younger, and we could not determine whether the difference was significant (Pepin et al. 2000). The factors that might be considered to be variables in a “severity” scale include age at first major event (e.g., arterial rupture or bowel rupture) and the types of events seen in a family (e.g., arterial alone, bowel rupture alone, or both). However, when the ascertainment criteria include one or more of these features, it is not surprising that “genotype-phenotype” correlations are difficult to identify.
Both the role of type III collagen in tissues and the mechanisms by which mutations in the COL3A1 gene produce disease are still largely unknown. Type III collagen is expressed in multiple tissues and usually colocalizes with type I collagen in the same fibril (Kadler et al. 1996). It is especially prevalent in tissues that have multidirectional extensibility, such as the intestine, large vessels, and the uterine wall, in which type III collagen may constitute 35%–45% of the total collagen (Seyer and Kang 1996). In general, the type III:type I collagen ratio is inversely proportional to the diameter of the collagen fibrils, so that highly compliant tissues have both a higher percentage of type III collagen and thinner fibrils (Lapiere et al. 1977; Romanic et al. 1991). With reduced synthesis of type III collagen, an increase in the proportion of type I collagen is expected to result in altered physical properties of tissues—including reduced tissue distensibility, which may lead to vascular and hollow-organ rupture.
In the cells that we have described here, there are two mechanisms by which the production of type III procollagen is reduced. In P1–P3 frameshifts result in PTCs in exons 27, 6, and 9, whereas in P4 a nucleotide substitution results in a new termination codon in exon 52, the last exon of the gene. In the first three cell strains, the mRNA from the mutant allele is unstable, although, as demonstrated by the equal amounts of precursor mRNA in the nucleus of cells from P1, both alleles appear to be transcribed with the same efficiency. The mRNA from the mutant allele was stabilized by the translation inhibitor cycloheximide in cells from P1, although the nuclear precursor RNA in cells from P3 was less responsive to treatment with the drug. These findings are compatible with degradation of the abnormal mRNA by the mechanisms of NMD (Maquat and Carmichael 2001). This cellular strategy identifies PTCs that are separated from the constitutive termination codon by one or more introns, possibly by “marking” domains just upstream from previous exon-intron boundaries with proteins that associate with mRNA (Le Hir et al. 2000; Lykke-Andersen et al. 2000). Translation then leads to displacement of these “marks,” but, if translation is terminated prior to the constitutive termination site, these proteins remain bound, recruit exonucleases to the mRNA, and lead to its degradation (Lykke-Andersen et al. 2000; Lykke-Andersen 2001).
In the cells from P4, a C→T transition converts a codon for arginine to a PTC in exon 52, the ultimate exon of the COL3A1 gene. As predicted by the model, the mRNA from cells from P4 is stable, transported to the cytoplasm, and appears to encode a polypeptide chain that is truncated by 35 amino acids. This protein is translated but, although reasonably stable, is not incorporated into mature type III procollagen molecules, because it cannot fold into a productive conformation. The terminal 35 residues include a cysteine that is vital to the formation of one of the intrachain disulfide bonds that stabilizes a chain prior to trimer formation. In this instance, diminished production of type III procollagen reflects the failure of chain association rather than mRNA instability.
It has proved difficult to detect reduced production of type III procollagen by analysis of the proteins produced by cultured cells. As a result, analysis of the products of the two COL3A1 alleles has been a particularly valuable adjunct in the search for this type of mutation, but even that has failed to identify the mutation that did not result in mRNA instability. Direct genomic analysis is thus likely to be a more efficient mechanism for identification of these cells strains, now that the clinical phenotype has been recognized.
Acknowledgments
This work was supported, in part, by National Institutes of Health grant AR21557.
Appendix
Table A1.
Primer Sets for Amplification of COL3A1 Coding Sequence
| Primer Set | Exona | Primer Sequence(5′→3′)b | Wild-Type Sequence(5′→3′) |
| 1 | 1F | CTGAATTCCAGGGAACAACTTGATG | CTGAAGGGCAGGGAACAACTTGATG |
| 3R | TGGATCTCCCTTGGGGCCTTG | ||
| 2 | 2F | GTCAGTCCTATGCGGATAGAGA | |
| 8R | CAGCTTGCCCAGGTTCACCAGGGGAT | CAGCTTGCCCAGGTTCACCAGGGGGT | |
| 3 | 6F | AAGAATTCTCCCCAGTATGATTCA | AACTATTCTCCCCAGTATGATTCA |
| 13R | TTGTCGACTGGAAGACCATTTTCA | TTTTCGCCTGGAAGACCATTTTCA | |
| 4 | 12F | ACGAATTCGAGAAAAGGGTGAAAC | ACGAAATGGAGAAAAGGGTGAAAC |
| 19R | CCATTAGCACCGGCTGGTCC | ||
| 5 | 17F | TCAGGGACACGCTGGTGCTCAA | |
| 23R | GGACCTCCAGGGACGCCATC | ||
| 6 | 22F | GGACCTGCTGGACCAAATGGC | |
| 28R | GGGTCCTGTGTCTCCTTTGTC | ||
| 7 | 26F | GCTCCTGGTAAGAATGGAGAACG | |
| 31R | CTCCTTCGGGACCAGGGGGACTAG | CTCCTTCGGGACCAGGGGGACCAG | |
| 8 | 31F | GGGCCCCAGGACTTAGAGTTCGA | GGGCCCCAGGACTTAGAGGTGGA |
| 33R | GCAAGCTTTCCCTGGGACACCAT | GCCATCTTTCCCTGGGACACCAT | |
| 9 | 32F | AGGAATTCCTGGAGAAAGAGGAG | AGGAATGCCTGGAGAAAGAGGAG |
| 38R | TTCACCTTTGACACCTTGGGGA | ||
| 10 | 37F | TGAAGGAGGCCCTCCTGGAGTT | |
| 41R | AGGACCTGGCATGCCTGGTGGTC | ||
| 11 | 40F | TCCCAGCGGTTCTCCAGGCA | |
| 43R | GACCATCTGATCCAGGGTTTC | ||
| 12 | 42F | ACCAGGAGCTAACGGTCTCAG | |
| 47R | TCGTCGACTGCCGATTGCACC | TCCTGGACTGCCGATTGCACC | |
| 13 | 45F | TCCTGCTGGTTCCCGAGGTG | |
| 49R | GAGAACCATCAGGACTAATGAG | ||
| 14 | 48F | CAGGGCCTCGAGGTAACAGAGGT | |
| 50R | TTGTCGACAACATTCAAAGGATTG | TTCCGTGGAACATTCAAAGGATTG | |
| 15 | 49F | CTCATTAGTCCTGATGGTTCTC | |
| 51R | TGATCCATGTATGCAATGCTAT |
The numeral denotes the exon; F = forward; R = reverse.
Mismatched nucleotides, to introduce restriction-endonuclease sites for directed cloning, are underlined.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for cDNA reference sequence [accession number NM_000090])
- Human Genome Project Working Draft at UCSC, http://genome.ucsc.edu/ (for COL3A1 gene sequence, chromosome 2 [accession numbers 193023938–193062278])
- Mutations in COL3A1,http://www.le.ac.uk/genetics/collagen/col3a1.html
- Online Mendelian Inheritance in Man (OMIM), http://www3.ncbi.nlm.nih.gov/Omim/ (for EDS type IV [MIM 130050])
References
- Ala-Kokko L, Kontusaari S, Baldwin CT, Kuivaniemi H, Prockop DJ (1989) Structure of cDNA clones coding for the entire prepro alpha 1 (III) chain of human type III procollagen: differences in protein structure from type I procollagen and conservation of codon preferences. Biochem J 260:509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton P, De Paepe A, Steinmann B, Tsipouras P, Wenstrup RJ (1998) Ehlers-Danlos syndromes: revised nosology, Villefranche, 1997: Ehlers-Danlos National Foundation (USA) and Ehlers-Danlos Support Group (UK). Am J Med Genet 77:31–37 [DOI] [PubMed] [Google Scholar]
- Benson-Chanda V, Su MW, Weil D, Chu ML, Ramirez F (1989) Cloning and analysis of the 5′ portion of the human type-III procollagen gene (COL3A1). Gene 78:255–265 [DOI] [PubMed] [Google Scholar]
- Bonadio J, Holbrook KA, Gelinas RE, Jacob J, Byers PH (1985) Altered triple helical structure of type I procollagen in lethal perinatal osteogenesis imperfecta. J Biol Chem 260:1734–1742 [PubMed] [Google Scholar]
- Byers PH, Schwarze U. Ehlers-Danlos syndrome. In: Rimoin DL, Connor JM, Pyeritz RE (eds) Emery and Rimoin's principles and practice of medical genetics. Harcourt, London (in press) [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed] [Google Scholar]
- Chu M-L, Prockop DJ (1993) Collagen: gene structure. In: Royce PM, Steinmann B (eds) Connective tissue and its heritable disorders. Wiley-Liss, New York, pp 149–165 [Google Scholar]
- Fisher LW, Stubbs JT, Young MF (1995) Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop Scand Suppl 266:61–65 [PubMed] [Google Scholar]
- Freddi S, Savarirayan R, Bateman JF (2000) Molecular diagnosis of Stickler syndrome: a COL2A1 stop codon mutation screening strategy that is not compromised by mutant mRNA instability. Am J Med Genet 90:398–406 [PubMed] [Google Scholar]
- Kadler KE, Holmes DF, Trotter JA, Chapman JA (1996) Collagen fibril formation. Biochem J 316:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkko J, Annunen S, Pihlajamaa T, Prockop DJ, Ala-Kokko L (1998) Conformation sensitive gel electrophoresis for simple and accurate detection of mutations: comparison with denaturing gradient gel electrophoresis and nucleotide sequencing. Proc Natl Acad Sci USA 95:1681–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivaniemi H, Kontusaari S, Tromp G, Zhao MJ, Sabol C, Prockop DJ (1990) Identical G+1 to A mutations in three different introns of the type III procollagen gene (COL3A1) produce different patterns of RNA splicing in three variants of Ehlers-Danlos syndrome. IV. An explanation for exon skipping some mutations and not others. J Biol Chem 265:12067–12074 [PubMed] [Google Scholar]
- Kuivaniemi H, Prockop DJ, Wu Y, Madhatheri SL, Kleinert C, Earley JJ, Jokinen A, Stolle C, Majamaa K, Myllylä VV, Norrgård Ö, Schievink WI, Mokri B, Fukawa O, ter Berg JWM, De Paepe A, Lozano AM, Leblanc R, Ryynänen M, Baxter BT, Shikata H, Ferrell RE, Tromp G (1993) Exclusion of mutations in the gene for type III collagen (COL3A1) as a common cause of intracranial aneurysms or cervical artery dissections: results from sequence analysis of the coding sequences of type III collagen from 55 unrelated patients. Neurology 43:2652–2658 [DOI] [PubMed] [Google Scholar]
- Kuivaniemi H, Tromp G, Prockop DJ (1997) Mutations in fibrillar collagens (types I, II, III, and XI), fibril-associated collagen (type IX), and network-forming collagen (type X) cause a spectrum of diseases of bone, cartilage, and blood vessels. Hum Mutat 9:300–315 [DOI] [PubMed] [Google Scholar]
- Lapiere CM, Nusgens B, Pierard GE (1977) Interaction between collagen type I and type III in conditioning bundles organization. Connect Tissue Res 5:21–29 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19:6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wu H, Byrne M, Krane S, Jaenisch R (1997) Type III collagen is crucial for collagen I fibrillogenesis and for normal cardiovascular development. Proc Natl Acad Sci USA 94:1852–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J (2001) mRNA quality control: marking the message for life or death. Curr Biol 11:R88–R91 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121–1131 [DOI] [PubMed] [Google Scholar]
- Maquat LE, Carmichael GG (2001) Quality control of mRNA function. Cell 104:173–176 [DOI] [PubMed] [Google Scholar]
- Pepin M, Schwarze U, Superti-Furga A, Byers PH (2000) Clinical and genetic features of Ehlers-Danlos syndrome type IV, the vascular type. N Engl J Med 342:673–680 [DOI] [PubMed] [Google Scholar]
- Pope FM, Martin GR, McKusick VA (1977) Inheritance of Ehlers-Danlos type IV syndrome. J Med Genet 14:200–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope FM, Narcisi P, Nicholls AC, Germaine D, Pals G, Richards AJ (1996) COL3A1 mutations cause variable clinical phenotypes including acrogeria and vascular rupture. Br J Dermatol 135:163–181 [PubMed] [Google Scholar]
- Pope FM, Narcisi P, Nicholls AC, Liberman M, Oorthuys JW (1988) Clinical presentations of Ehlers Danlos syndrome type IV. Arch Dis Child 63:1016–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope FM, Nicholls AC, Jones PM, Wells RS, Lawrence D (1980) EDS IV (acrogeria): new autosomal dominant and recessive types. J R Soc Med 73:180–186 [PMC free article] [PubMed] [Google Scholar]
- Redford-Badwal DA, Stover ML, Valli M, McKinstry MB, Rowe DW (1996) Nuclear retention of COL1A1 messenger RNA identifies null alleles causing mild osteogenesis imperfecta. J Clin Invest 97:1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanic AM, Adachi E, Kadler KE, Hojima Y, Prockop DJ (1991) Copolymerization of pNcollagen III and collagen I: pNcollagen III decreases the rate of incorporation of collagen I into fibrils, the amount of collagen I incorporated, and the diameter of the fibrils formed. J Biol Chem 266:12703–12709 [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze U, Atkinson M, Hoffman GG, Greenspan DS, Byers PH (2000) Null alleles of the COL5A1 gene of type V collagen are a cause of the classical forms of Ehlers-Danlos syndrome (types I and II). Am J Hum Genet 66:1757–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze U, Goldstein JA, Byers PH (1997) Splicing defects in the COL3A1 gene: marked preference for 5′ (donor) splice-site mutations in patients with exon-skipping mutations and Ehlers-Danlos syndrome type IV. Am J Hum Genet 61:1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze U, Starman BJ, Byers PH (1999) Redefinition of exon 7 in the COL1A1 gene of type I collagen by an intron 8 splice-donor-site mutation in a form of osteogenesis imperfecta: influence of intron splice order on outcome of splice-site mutation. Am J Hum Genet 65:336–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyer JM, Kang AH (1996) Connective tissues of the subendothelium. In: Loscalzo J, Creager MA, Dzau VJ (eds) Vascular medicine: a textbook of vascular biology and diseases. Little, Brown, Boston, pp 39–67 [Google Scholar]
- Stover ML, Primorac D, Liu SC, McKinstry MB, Rowe DW (1993) Defective splicing of mRNA from one COL1A1 allele of type I collagen in nondeforming (type I) osteogenesis imperfecta. J Clin Invest 92:1994–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromp G, Kleinert C, Kuivaniemi H, Prockop DJ (1991) C to T polymorphism in exon 33 of the COL3A1 gene. Nucleic Acids Res 19:681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromp G, Wu Y, Prockop DJ, Madhatheri SL, Kleinert C, Earley JJ, Zhuang J, Norrgård O, Darling RC, Abbott WM, Cole CW, Jaakkola P, Ryynänen M, Pearce WH, Yao JST, Majamaa K, Smullens SN, Gatalica Z, Ferrell RE, Jimenez SA, Jackson CE, Michels VV, Kaye M, Kuivaniemi H (1993) Sequencing of cDNA from 50 unrelated patients reveals that mutations in the triple-helical domain of type III procollagen are an infrequent cause of aortic aneurysms. J Clin Invest 91:2539–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LM, Lalic L, Roughley PJ, Glorieux FH (2001) Thirty-three novel COL1A1 and COL1A2 mutations in patients with osteogenesis imperfecta types I–IV. Hum Mutat 17:434 [DOI] [PubMed] [Google Scholar]
- Wenstrup RJ, Florer JB, Willing MC, Giunta C, Steinmann B, Young F, Susic M, Cole WG (2000) COL5A1 haploinsufficiency is a common molecular mechanism underlying the classical form of EDS. Am J Hum Genet 66:1766–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing MC, Cohn DH, Byers PH (1990) Frameshift mutation near the 3′ end of the COL1A1 gene of type I collagen predicts an elongated Pro alpha 1(I) chain and results in osteogenesis imperfecta type I. J Clin Invest 85:282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing MC, Deschenes SP, Scott DA, Byers PH, Slayton RL, Pitts SH, Arikat H, Roberts EJ (1994) Osteogenesis imperfecta type I: molecular heterogeneity for COL1A1 null alleles of type I collagen. Am J Hum Genet 55:638–647 [PMC free article] [PubMed] [Google Scholar]
- Willing MC, Deschenes SP, Slayton RL, Roberts EJ (1996) Premature chain termination is a unifying mechanism for COL1A1 null alleles in osteogenesis imperfecta type I cell strains. Am J Hum Genet 59:799–809 [PMC free article] [PubMed] [Google Scholar]
- Zafarullah K, Kleinert C, Tromp G, Kuivaniemi H, Kontusaari S, Wu YL, Ganguly A, Prockop DJ (1990) G to A polymorphism in exon 31 of the COL3A1 gene. Nucleic Acids Res 18:6180 [DOI] [PMC free article] [PubMed] [Google Scholar]