Abstract
A total of 61 strains of Staphylococcus aureus and 177 coagulase-negative staphylococcal strains were isolated from the blood of patients with bloodstream infections and from the skin of both children under cancer treatment and human immunodeficiency virus-positive patients. The MIC analyses revealed that 118 isolates (50%) were resistant to quaternary ammonium compound-based disinfectant benzalkonium chloride (BC). The frequencies of resistance to a range of antibiotics were significantly higher among BC-resistant staphylococci than among BC-sensitive staphylococci. Of 78 BC-resistant staphylococcal isolates, plasmid DNA from 65 (83%), 2 (3%), 43 (55%), and 15 (19%) isolates hybridized to qacA or -B (qacA/B), qacC, blaZ, and tetK probes, respectively. The qacA/B and blaZ probes hybridized to the same plasmid in 19 (24%) staphylococcal strains. The plasmids harboring both qacA/B and blaZ genes varied from approximately 20 to 40 kb. The Staphylococcus epidermidis Fol62 isolate, harboring multiresistance plasmid pMS62, contained qacA/B and blaZ together with tetK. Molecular and genetic studies indicated different structural arrangements of blaZ and qacA/B, including variable intergenic distances and transcriptional directions of the two genes on the same plasmid within the strains. The different organizations may be due to the presence of various genetic elements involved in cointegration, recombination, and rearrangements. These results indicate that qac resistance genes are common and that linkage between resistance to disinfectants and penicillin resistance occurs frequently in clinical isolates in Norway. Moreover, the higher frequency of antibiotic resistance among BC-resistant strains indicates that the presence of either resistance determinant selects for the other during antimicrobial therapy and disinfection in hospitals.
Staphylococci, including Staphylococcus aureus and the coagulase-negative staphylococci (CNS), and enterococci account for approximately one-third of all bloodstream infections and up to 50% of nosocomial bloodstream infections. In the past, CNS were considered to be very rarely involved in disease, but since the 1980s there has been increasing evidence that CNS can act as opportunistic pathogens and can be a frequent and important cause of disease. In recent years there has been a dramatic increase in the prevalence of multiple-drug-resistant strains. Worldwide emergence of antimicrobial resistance in staphylococci is probably a serious and increasing problem, especially in hospitals (16).
Disinfectants based on quaternary ammonium compounds (QACs), e.g., benzalkonium chloride (BC), cetylpyridinium chloride, cetrimide, proceine, and detizor, are frequently used in hospitals to disinfect and to prevent the spread of pathogens. It has been suggested that the widespread use of QACs may impose a selective pressure and contribute to the emergence of disinfectant-resistant microorganisms in these environments (22, 26). Known qac resistance genes reported in clinical staphylococci, qacA, qacB, and qacC/smr, are generally plasmid borne and are widely distributed in the environment (12, 14, 15, 18).
Large consumption of antibiotics by both humans and animals has resulted in the development and spread of a large number of antibiotic resistance determinants among bacterial populations, thus creating critical public health problems. Concerns have arisen regarding the potential emergence of cross-resistance and coresistance between widely used disinfectants and antibiotics (20, 22, 26). Resistance genes on transferable genetic elements such as plasmids and transposons may lead to the epidemic spread of resistance between species. On some of the published plasmids (pST6, pSK4, and pSK41) and transposons (Tn552 and Tn4002), qac resistance determinants are located together with antibiotic resistance genes encoding resistance to gentamicin (GEN), trimethoprim (TMP), penicillin, kanamycin (KAN), and tobramycin (3, 12, 14, 27).
Staphylococcal β-lactamase structural gene blaZ and two closely linked genes, blaI and blaR, that control its expression have been identified on several transposons, e.g., Tn552 (24, 25), and large plasmids, e.g., pST6 (27), as well as on the chromosome (23). Staphylococcal β-lactamase transposon Tn552 is closely related to staphylococcal transposons Tn4002, Tn3852, and Tn4201 and to Tn4201-like elements. Staphylococcal transposons contains terminal inverted repeats (TIR) at their ends that serve as recognition sequences for transposase in their role of fusing the ends of the transposon with the recipient DNA. The three genes (blaI, blaR, and blaZ) involved in β-lactamase production constitute the right half of transposon Tn552 (Tn552-like bla gene module). The left half of Tn552 consists of genes p271, encoding a potential ATP-binding protein, p480, encoding a transposase, binL, encoding a resolvase, and a resolution site, resL. Partial and complete genetic structural arrangements of Tn552 in staphylococci from different geographic locations have been described (3, 4, 21, 27). Some staphylococci harbor only the right or left half of Tn552. Staphylococcal insertion sequence IS257, associated with Tn552 and IS256, has been shown to be an active mobile genetic element (5, 6, 13, 17, 27).
There is limited knowledge concerning the frequency of QAC resistance and the genetic linkage between qac-mediated resistance and antibiotic resistance genes. In this study, we investigated the presence and genetic linkage of known qac resistance determinants and antibiotic resistance genes, along with genetic mobile elements, in staphylococci isolated from clinical environments.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
A total of 61 isolates of S. aureus and 177 CNS isolates were included in this study (Table 1). The staphylococci were isolated from patients in four different hospitals. S. aureus (n = 61) and CNS (n = 60) were isolated from patients with bloodstream infections at two hospitals (Molde County Hospital and Norwegian Radium Hospital) during 1991 to 1992 and 1995 to 1996 (10). The CNS isolates (n = 56) from the skin of human immunodeficiency virus (HIV)-positive patients were from Ullevål Hospital, while the remaining 61 CNS isolates were obtained at two hospitals (Ullevål Hospital and Rikshospitalet University Hospital) from the skin of children under cancer treatment receiving cytostatica and undergoing multiple-antibiotic cures. In all cases, repeat isolates from the same patient were excluded. This strain collection represents staphylococci from different types of hospitals and patients subject to various degrees of antimicrobial treatment. Reference strains were Staphylococcus haemolyticus NVH97A (resistant to BC and penicillin) and S. aureus RN4220, S. haemolyticus DSM20623, and S. aureus ATCC 25923 (sensitive to BC and penicillin). All staphylococcus strains were cultured in Mueller-Hinton (MH) broth or on MH agar plates (Difco Laboratories, Detroit, Mich.) at 37°C. Isolates were preserved in MH broth with 15% glycerol at −80°C.
TABLE 1.
Screening for phenotypic susceptibility to BC and PEN among clinical staphylococci
| Source | Bacterium | Na | No. of strains with indicated susceptibilityb to:
|
|||
|---|---|---|---|---|---|---|
| BC
|
PEN
|
|||||
| R | S | R | S | |||
| Skin isolatec | CNS | 61 | 30 | 31 | 22 | 39 |
| Skin isolated | CNS | 56 | 17 | 39 | 30 | 26 |
| Bloodstream infection | S. aureus | 61 | 37 | 24 | 41 | 20 |
| CNS | 60 | 34 | 26 | 50 | 10 | |
| Total | 238 | 118 | 120 | 143 | 95 | |
N, number of strains studied.
Susceptibility was tested by the microdilution method. R, resistant; S, sensitive.
Isolates from children with cancer.
Isolates from HIV-positive outpatients.
Antimicrobial agents.
BC was from the Norwegian Medical Depot (Oslo, Norway). Ampicillin (AMP), penicillin G (PEN), methicillin (MET), tetracycline (TET), erythromycin (ERY), KAN, GEN, chloramphenicol, TMP, and ethidium bromide (EBR) were purchased from Sigma Chemical Co. (St. Louis, Mo.), and chlorhexidine (CHX) was from Nycomed (Oslo, Norway). Amphoteric disinfectant Tego 103G was purchased from Otto Olsen AS (Lillestrøm, Norway). Neo-Sensitabs disks (Rosco, Medkjemi A/S, Taastrup, Denmark) containing antimicrobial agents were used for susceptibility testing.
Susceptibility tests.
MICs of antimicrobial agents and dyes were determined by the microdilution method (28) and by E test (10) using MH medium. The lowest concentration of an antimicrobial agent and dye that totally inhibited bacterial growth after 24 h of incubation at 37°C was considered the MIC. Antibiotic susceptibility patterns were also examined by the disk diffusion method (Neo-Sensitabs user's guide, Rosco). The diameters of the inhibition zones around antimicrobial disks were measured in millimeters after 2 days of incubation at 37°C and compared to standard MIC breakpoints recommended by “Susceptibility testing standardization groups” (Neo-Sensitabs user's guide, Rosco). The results were interpreted as recommended by the Norwegian AFA Group (Neo-Sensitabs user’s guide). Susceptibility tests for the isolates resistant to BC and PEN were repeated two times on different days.
MIC testing with disinfectants BC and Tego 103G was carried out by using 1-μg/ml intervals from 0 to 10 μg/ml. For CHX the interval was 0.5 μg/ml from 0 to 4 μg/ml. MIC tests with PEN, AMP, and EBR were performed by using twofold dilutions starting from 320 μg/ml.
DNA isolation and analysis.
Cells were lysed with lysostaphin (Sigma-Aldrich, St. Louis, Mo.) at a concentration of 40 μg/ml and incubated at 37°C for 30 min. Total DNA and plasmid DNA were isolated by Easy-DNA (Invitrogen, Carlsbad, Calif.) and QIAprep spin (Qiagen GmbH, Hilden, Germany) kits, respectively. After agarose gel electrophoresis, the DNA was stained with EBR and visualized under UV light.
Plasmid curing.
Strains were grown in increasing sublethal concentrations of novobiocin in MH broth as described by Heir et al. (7).
Southern blotting and hybridization.
Plasmid DNA and total DNA were transferred from agarose gel to a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) along with positive and negative control DNA by vacuum blotting, according to the manufacturer's instructions (Pharmacia, Uppsala, Sweden). PCR products (see below) specific for genes qacA and qacB (common probe), qacC, qacG, qacH, blaZ, blaR, blaI, ermC, cat, tetK, aacA-aphD, and dfrA and insertion sequences IS257 and IS256 were used as probes. The PCR products were purified with a Qiagen PCR product purification kit and sequenced on an ABI PRISM 377 (Applied Biosystems, Perkin-Elmer Cetus Corp., Norwalk, Conn.) before being used as probes. The labeling of probes and hybridization were done with an AlkPhos direct gene image kit in accordance with the manufacturer's instructions (Amersham Pharmacia Biotech). Membrane stripping was done as recommended by the manufacturer.
PCR amplification.
The primers used for PCR amplifications were designed from previously known sequences (Table 2). All the designed primers were first tested for their specificity on known positive and negative control strains. To obtain probes for DNA hybridization, standard PCR was carried out for qacA or -B (qacA/B), qacC, qacG, qacH, blaZ, blaR, blaI, ermC, cat, tetK, aacA-aphD, dfrA, IS257, and IS256. Each PCR mixture contained 50 ng of plasmid DNA, 2 μM (each) primer, 200 μM (each) deoxynucleoside triphosphate, 2.5 μM MgCl2, 1× reaction buffer (Promega Corp., Madison, Wis.), and 2.5 U of Taq DNA polymerase (Promega Corp.) in a total volume of 50 μl. Reaction mixtures were subjected to 30 cycles of amplification. The conditions for each cycle were denaturation for 1 min at 95°C, annealing for 1 min at between 50 and 55°C depending on the primer set, and primer extension for 2 min at 72°C. Finally, reaction mixtures were incubated at 72°C for 10 min. The PCR products were separated by electrophoresis in a 1% agarose gel, stained with EBR, and visualized under UV light.
TABLE 2.
Primers used in PCR and XL-PCR amplification
| Primer | Sequence (5′-3′)a | Annealing positionb or source | Accession no. or reference |
|---|---|---|---|
| qacA/B R | TGGCCCTTTCTTTAGGGTTT | 1278-1259 | X56628 |
| qacA/B F | ATTCCATTGAGTGCCTTTGC | 1061-108 | X56628 |
| qacA/B | GCTGCATTTATGACAATGTTTG | 1693-1713 | X56628 |
| qac II | AATCCCACCTACTAAAGCAG | 2321-2302 | X56628 |
| qac IIR | CTGCTTTAGTAGGTGGGATT | 2302-2321 | X56628 |
| qac IV | TTTAAATGGCGAATGGTGT | 249-267 | X56628 |
| qacC F | GGCTTTTCAAAATTTATACCATCCT | 621-645 | Z37964 |
| qacC R | ATGCGATGTTCCGAAAATGT | 870-850 | Z37964 |
| qacG F | TAACTTACGCAACATGGGCA | 170-190 | Y16944 |
| qacG R | TCAATGGCTTTCTCCAAATAC | 325-303 | Y16944 |
| qacH F | CAAGTTGGGCAGGTTTAGGA | 180-200 | Y16945 |
| qacH R | TGTGATGATCCGAATGTGTTT | 321-300 | Y16945 |
| blaI F | ATGTCTCGCAATTCTTCAA | 3190-3208 | X52734 |
| blaI R | CTATGGCTGAATGGGAT | 3520-3503 | X52734 |
| blaI R2 | CAAAGAAATTGAAGAATTGCGA | 3195-3216 | X52734 |
| blaR F | CATCTGATAAATGTGTAGC | 3612-3630 | X52734 |
| blaR R | GGTATCTAACTCTTCTTGC | 5177-5159 | X52734 |
| blaZ 1F | TACAACTGTAATATCGGAGGG | 5372-5392 | X52734 |
| blaZ 2F | GAGGCTTCAATGACATATAGTG | 5741-5762 | X52734 |
| blaZ R | CAATAGGTTCAGATTGGCCC | 6149-6129 | X52734 |
| blaZ 3F | CACCTGCTGCTTTCGGTAAGAC | 5916-5937 | X52734 |
| blaZ 4F | GTTGATAAGTGAAACCG | 6201-6218 | X52734 |
| tetK F | CTACTCCTGGAATTACAA | 138-154 | S67449 |
| tetK R | TACTATACACTCCAGAAG | 1291-1273 | S67449 |
| dfrA F | CACTTGTAATGGCACGGAAA | 119-139 | AF045472 |
| dfrA R | CTGGTCAATCATTGCTTCGT | 335-315 | AF045472 |
| ermC F | ATCTTTGAAATCGGCTCAGG | 2639-2620 | J01755 |
| ermC R | CAAACCCGTATTCCACGATT | 2345-2364 | J01755 |
| cat F | ATGGTTCGGGGAAATTGTTT | 1627-1647 | J01754 |
| cat R | TCCTGCATGATAACCATCACA | 1853-1832 | J01754 |
| aacA-aphD F | CAGAGCCTTGGGAAGATGAA | 150-170 | M18086 |
| aacA-aphD R | TTGCCTTAACATTTGTGGCA | 564-544 | M18086 |
| mecA F | GTGGAATTGGCCAATACAGG | 478-497 | X52594 |
| mecA R | TGAGTTCTGCAGTACCGGAT | 1797-1816 | X52594 |
| IS256 F | CAGAACAGCTGGATCCTATGG | 523-543 | M18086 |
| IS256 R | GTCGACTTTTAGCCTCACGCG | 970-990 | M18086 |
| IS257 F | TTGGGTTCAAGAATATGCCC | 202-222 | U40386 |
| IS257 R | CTTCGTTGAAGGTGCCTGAT | 473-453 | U40386 |
| Bio-qacA/Bc | Biotin-CTGCATTTATGACAATGTTTG | 1692-1713 | X56628 |
| Bio-blaZ 2Fc | Biotin-GAGGCTTCAATGACATATAGTG | 5740-5762 | X52734 |
| Bio-FPc | Biotin-CAGTTCAAGCTTGTCCAGGAATTC | This study | CRe |
| Degen-FP1d | CAGTTCAAGCTTGTCCAGGAATTCNNNNNNNGGCCT | This study | CR |
| Degen-FP2d | CAGTTCAAGCTTGTCCAGGAATTCNNNNNNNGCGCT | This study | CR |
| Degen-FP3d | CAGTTCAAGCTTGTCCAGGAATTCNNNNNNNGCCCT | This study | CR |
| Degen-FP4d | CAGTTCAAGCTTGTCCAGGAATTCNNNNNNNGCGGT | This study | CR |
Underlined sequences indicate differences among degenerate primers.
Annealing position in published sequence.
Biotinylated primer.
Degenerate primer.
CR, Sørensen et al., Dynalogue customer report 3:2-3, 1999.
XL-PCR.
The primers used for extralong PCR (XL-PCR) amplification are included in Table 2. The XL-PCR was carried out with a GeneAmp XL-PCR kit (Applied Biosystems) in accordance with the manufacturer's instructions with slight modifications. The GeneAmp XL-PCR kit (Perkin-Elmer Cetus Corp.) was used for amplification of the DNA regions between the β-lactamase genes and disinfectant resistance gene qacA/B. The primer combinations used were qac IV and blaI R2 and qac IV and blaZ 2F (see Fig. 2; Table 2). The DNA region between qacA/B and staphylococcal insertion element IS257 was amplified by using primers qac IV and IS257 F (see Fig. 2). Each PCR mixture contained 10 ng of plasmid DNA, 2 μM (each) PCR primer, 200 μM (each) deoxynucleoside triphosphate, 1 mM MgCl2, and 1.5 U of rTh DNA polymerase-XL in a total volume of 50 μl. The cycle conditions were preliminary denaturation for 2 min at 93°C, followed by 25 cycles of denaturation at 93°C for 1 min, annealing for 45 s at between 47 and 50°C depending on the primer set, and primer extension for 7 min at 68°C. Finally, reaction mixtures were incubated at 68°C for 5 to 12 min depending on the primer set. The PCR products were examined by electrophoresis in a 0.7 or 1.0% agarose gel, stained with EBR, and visualized under UV light.
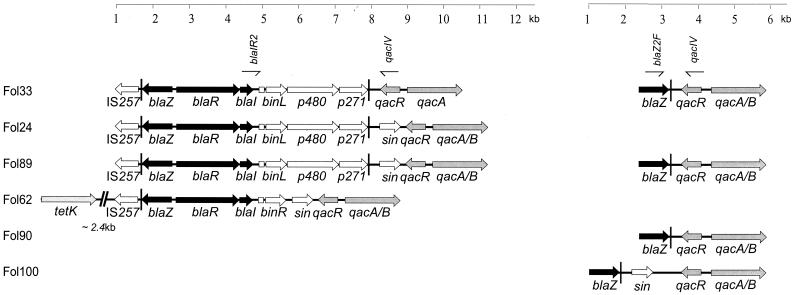
FIG. 2.
Genetic organization of qac and β-lactam resistance genes in staphylococcal isolates Fol24, Fol33, Fol62, Fol89, Fol90, and Fol100. Genes involved in qac and β-lactam resistance are gray and black, respectively. qacA/B encodes QAC resistance; qacR encodes a putative repressor of qacA/B; sin is a putative staphylococcal recombinase gene; p271 encodes a potential ATP-binding protein; p480 encodes a transposase; binR and binL are resolvase-encoding genes; blaZ is a β-lactamase structural gene; blaR and blaI are β-lactamase regulatory genes; IS257 is a staphylococcal insertion sequence. Primers for detection of specific PCR products are indicated for Fol33. Vertical bars and boxes downstream of blaI, TIR of transposon Tn552 and a resolution site (res), respectively.
DNA sequencing.
The nucleotide sequences of PCR products and XL-PCR products were determined by using an ABI PRISM BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems) with synthetic oligonucleotide primers on an ABI PRISM 377 automatic sequencer (Applied Biosystems) as recommended by the manufacturer. Before application on the sequencer, the extension products were purified as described by the manufacturer.
Unknown flanking DNA fragments were isolated by a two-step PCR method (A. B. Sørensen, M. Duch, and F. S. Pedersen, Dynalogue customer report 3:2-3, 1999). The unknown DNA flanking regions were amplified with primers coupled to biotin at the 5′ end and degenerate flanking primers (Table 2) in first-step PCR. The PCR products were bound to Dynabead-streptavidin beads and purified with a Dynabeads kilobaseBINDER kit (Dynal, Oslo, Norway). The purified PCR products from the first-step PCR were then used as templates for nested PCR followed by DNA sequencing. Nucleotide sequences were analyzed by using the BLAST website program (National Center for Biotechnology Information, National Institutes of Health), the Sequencher, version 3.0, software package (Gene Codes Corporation, Ann Arbor, Mich.), and the GCG sequence analysis software package, version 8 (Genetics Computer Group, Madison, Wis.).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study have been assigned accession no. AF426833, AF426834, and AF426835 in the GenBank database.
RESULTS
Susceptibility testing.
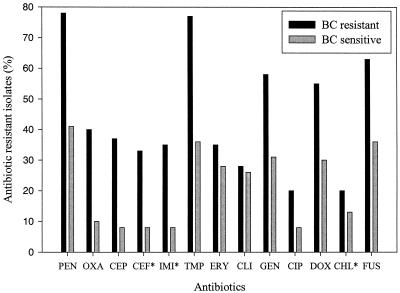
The 238 staphylococcus isolates were screened for QAC (BC) and PEN resistance (Table 1). The strains were categorized as BC resistant or sensitive according to the BC MICs. One hundred eighteen (50%) isolates were considered BC resistant (BC MICs between 3 and 8 μg/ml), and 120 isolates were sensitive to BC (BC MICs ≤ 2 μg/ml). Of the 238 staphylococcus isolates, 143 were found resistant to PEN. One hundred twenty-one CNS isolates from patients with bloodstream infections (n = 60) and from the skin of children under cancer treatment (n = 61) were analyzed for correlation between BC and antibiotic resistance (Fig. 1). This analysis showed that the frequency of antibiotic resistance was higher among the BC-resistant strains. As an example, a large number of BC-resistant strains also showed PEN resistance. On the basis of Fisher's exact test (1), these distributions appear not to be random (P ≤ 0.01 for PEN, oxacillin, cephalothin, cefuroxime, imipenem, TMP, GEN, doxycycline, and fusidic acid). None of the isolates were resistant to vancomycin, and less than 3% of the staphylococcus isolates were resistant to teicoplanin and rifampin (results not shown). The CNS from HIV-positive outpatients and S. aureus from patients with bloodstream infections were excluded from the analyses of the correlation between BC and antibiotic resistance, because the frequencies of antibiotic resistance among these isolates were relatively low (data not shown).
FIG. 1.
Percentages of antibiotic-resistant CNS isolates from patients with bloodstream infections (n = 60) and from the skin of children under cancer treatment (n = 61) among BC-resistant and BC-sensitive isolates. Antibiotics used are PEN, oxacillin (OXA), cephalothin (CEP), cefuroxime (CEF), imipenem (IMI), cotrimoxazole (COT), ERY, clindamycin (CLI), GEN, ciprofloxacin (CIP), doxycycline (DOX), chloramphenicol (CHL), and fusidic acid (FUS). Susceptibility to CEF, IMI, and CHL was tested only among CNS isolates from patients with bloodstream infections (asterisks).
The 118 BC-resistant isolates were screened for susceptibility to a second disinfectant, CHX (Table 3). Twenty-eight of the BC-resistant isolates showed low resistance to CHX (MIC between 1.5 and 3.0 μg/ml), whereas 90 isolates were sensitive (MIC ≤ 1.0 μg/ml). Likewise, of 42 BC-resistant staphylococcus isolates randomly selected among strains for which the BC MIC was ≥4 μg/ml, 11 and 37 isolates were resistant to disinfectant Tego 103G and dye EBR, respectively (Table 3). Systematic cross-resistance between BC and EBR and no systematic cross-resistance between BC and CHX or Tego 103G were found in staphylococci.
TABLE 3.
BC-resistant isolates were screened for susceptibility to disinfectants and dye by a microdilution method
| Bacterium | No. of isolatesa with indicated susceptibilityb to:
|
|||||
|---|---|---|---|---|---|---|
| CHX (118)
|
Tego 103G (42)
|
EBR (42)
|
||||
| R | S | R | S | R | S | |
| S. aureus | 8 | 3 | 3 | 13 | 14 | 2 |
| CNS | 20 | 77 | 8 | 18 | 23 | 3 |
| Total | 28 | 90 | 11 | 31 | 37 | 5 |
Total numbers of BC-resistant strains tested are in parentheses.
R, resistant; S, sensitive.
The MIC tests and disk diffusion test revealed that an isolate from a bloodstream infection, Staphylococcus epidermidis Fol62, was resistant to BC (MIC, 5 μg/ml), EBR (MIC, 160 μg/ml), Tego 103G (MIC, 10 μg/ml), PEN (MIC, 160 μg/ml), AMP (MIC, 160 μg/ml), MET (MIC, 50 μg/ml), TET (MIC, 70 μg/ml), KAN (MIC, 7 μg/ml), ERY (MIC, 80 μg/ml) and TMP (MIC, 8 μg/ml).
DNA hybridizations.
Of the 118 BC-resistant isolates, 78 isolates for which the BC MICs were high (MICs between 4 and 8 μg/ml) were selected for DNA hybridization studies. Plasmid DNA from these isolates was screened for the presence of known gram-positive bacterium qac genes mediating resistance (qacA/B, qacC, qacG, and qacH), β-lactamase genes (blaZ, blaI, and blaR), other antibiotic resistance genes (ermC, cat, dfrA, and tetK), and staphylococcal insertion sequences IS257 and IS256 by Southern blotting and hybridization (Table 4). Sixty-seven of the BC-resistant isolates harbored either qacA/B or qacC on plasmid DNA, and the qacA/B gene was detected by PCR (total DNA was the template) in the remaining 11 isolates. Neither the qacG nor the qacH gene was detected among the isolates. Plasmids from 43 strains also hybridized with β-lactamase probes. In 19 of these strains, qac and blaZ, blaI, and blaR resided on the same plasmid. Only S. epidermidis Fol62 plasmid pMS62 (32 kb) hybridized with other antibiotic resistance gene probes tested (tetK, IS257, ermC, aacA-aphD, and dfrA) in addition to qacA/B, blaZ, blaI, and blaR. tetK and a copy of IS257 were localized adjacent to blaZ. However, no XL-PCR products were obtained when DNA stretches between tetK or qacA/B and antibiotic resistance gene ermC, aacA-aphD, or dfrA were amplified. The determinant for MET resistance (mecA) in S. epidermidis Fol62 was chromosomally encoded.
TABLE 4.
Screening for the presence of disinfectant and various antibiotic resistance genes on plasmid DNA among BC-resistant staphylococci
| Isolate source | Na | Bacterium | No. of isolates with hybridizationb to:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| qacA/B | qacC | blaZc | ermC | tetK | cat | dfrA | IS257 | IS256 | qacA/B + blaZd | |||
| Skine | 25 | CNS | 23 | 1 | 11 | 8 | 4 | 5 | 6 | 13 | 6 | 6 |
| Bloodstream cultures | 26 | S. aureus | 17 | 0 | 14 | 4 | 2 | 5 | 0 | 8 | 2 | 5 |
| 27 | CNS | 25 | 1 | 18 | 12 | 9 | 10 | 9 | 15 | 9 | 8 | |
| Total | 78 | 65 | 2 | 43 | 24 | 15 | 20 | 15 | 36 | 17 | 19 | |
N, number of strains studied.
The qacABC, blaZ, ermC, tetK, cat, and dfrA genes encode QAC, PEN, ERY, TET, chloramphenicol, and TMP resistance, respectively, from plasmids.
Hybridized with blaZ, blaI, and blaR probes.
qacA/B and blaZ on the same plasmid.
Isolates from children treated for cancer.
Genetic organizations of the qac and β-lactamase genes.
From the 19 staphylococcus isolates harboring disinfectant qac and β-lactamase genes on the same plasmids, we selected six isolates for a study of gene organization, isolates Fol24 (pMS24, 30 kb), Fol33 (pMS33, 35 kb), Fol62 (pMS62, 32 kb), Fol89 (pMS89, 40 kb), Fol90 (pMS90, 35 kb), and Fol100 (pMS100, 40 kb) (plasmid designations and approximate sizes are in parentheses). These isolates were all identified as S. epidermidis. Isolates Fol24, Fol33, and Fol62 were from patients having bloodstream infections, and isolates Fol89, Fol90, and Fol100 were from the skin of children treated for cancer. Results obtained by XL-PCR and DNA sequencing indicated different structural arrangements of the β-lactamase genes and the disinfectant resistance qacA/B genes, including variable intergenic distances and different transcriptional orientations of genes located on the same plasmids (Fig. 2).
Staphylococcal plasmids pMS24, pMS33, and pMS89 harbor a complete copy of transposon Tn552, comprising p271, p480, and binL, encoding a potential ATP-binding protein, a transposase, and a resolvase, respectively, in addition to the β-lactamase structural gene (blaZ) and regulatory genes (blaR and blaI). Staphylococcal insertion sequence IS257 was found downstream of the blaZ gene in these plasmids. There were sequences homologous to the right TIR (TIRR) of Tn552 downstream of blaZ, a resL site between blaI and binL, and a sequence homologous to TIRL of Tn552 downstream of p271. Isolates Fol24, Fol33, and Fol89 harbored the qacA/B gene together with their regulatory gene, qacR, downstream of the p271 gene. However, the region between p271 and qacR was different for different isolates. Fol24 and Fol89 had very similar regions and contained the sin gene, encoding a putative recombinase, while the sin gene was absent in Fol33. Plasmid pMS62 harbored the incomplete right half of the transposon Tn552-like bla gene module upstream of the binR and sin genes. Also in pMS62, IS257 was found downstream and adjacent to blaZ. The tetK gene, encoding TET resistance, was located approximately 2.4 kb further downstream.
In Fol90 and Fol100 the orientation of β-lactamase gene clusters was inverted relative to the qacA/B gene cluster, so that qacR was situated downstream of blaZ. In pMS90 the intergenic sequence between blaZ and qacR contained only the inverted repeat of Tn552, while in pMS100 this region contained a sin and an additional noncoding 743 bp of DNA. DNA hybridization and PCR confirmed the presence of staphylococcal insertion sequence IS257 on pMS90 and pMS100.
When the DNA stretches between the β-lactamase genes and disinfectant resistance gene qacA/B were amplified by using primer combinations qac IV and blaI R2 and qac IV and blaZ 2F, both primer sets gave rise to PCR products in strains Fol33 and Fol89 (Fig. 2). DNA sequencing of the PCR products of qac IV and blaZ 2F revealed that the blaZ and the qacR genes were located very close to each other, with the TIR of Tn552 between the genes. These strains apparently harbored the gene clusters in two orientations.
The nucleotide and deduced amino acid sequences derived from complete and incomplete (Tn552-like bla gene module) Tn552 transposons of plasmids pMS24, pMS33, pMS89, and pMS62 were identical to those derived from staphylococcal β-lactamase transposon Tn552 (25). The deduced amino acid sequences derived from qacR of plasmids pMS24, pMS33, pMS62, pMS89, pMS90, and pMS100 and qacA/B of pMS33 were similar to those derived from pST6, pSK156, and pSK23. Similarly, the deduced amino acid sequences derived from the resolvase gene (binR) of pMS62 was 100% identical to that derived from pST6 (AY028779) and 99% similar to that derived from pNVH96 (AJ302698). The recombinase genes (sin) of pMS62 and pMS89 were 100% identical and 99% similar to those of pST6 and pNVH97A (AJ400722), respectively. The sin gene of Fol100 is 100% identical to that of S. epidermidis strain SR1 and 90% similar to the sin genes of Fol62 and Fol89.
DISCUSSION
Little is known about the occurrence and possible genetic linkage of qac and antibiotic resistance in staphylococci. Of the 238 human clinical isolates from Norway investigated in this study, 50% were phenotypically resistant to BC. Plasmid-borne qacA/B and qacC genes were detected in 83 and 3% of the BC-resistant staphylococcus isolates, respectively. The qacA/B genes were also detected in the remaining BC-resistant isolates, indicating that the genes were either chromosomally located or present on large plasmids not obtained by the plasmid isolation procedure. Previous investigators have reported a similar distribution of these three qac resistance genes in clinical S. aureus and CNS (2, 11, 15) although additional staphylococcal disinfectant resistance genes also have been indicated (18). A lower prevalence of QAC resistance (13%) was reported for staphylococci from food and the food-processing industry (7, 8, 9). Here, both qacA/B and qacC as well as qacG and qacH were reported, with qacC as the most prevalent determinant. Overall, it seems likely that the presence, maintenance, and widespread dissemination of multidrug efflux qac genes in staphylococci are a consequence of the selective pressure exerted by the use of antiseptics and disinfectants (11, 18, 20, 26).
Many of the staphylococci included in this study were resistant to different antibiotics, but in general the antibiotic resistance was considered low compared to findings in other studies (10). Interestingly, we observed that staphylococci resistant to BC were generally more often resistant to antibiotics than BC-sensitive isolates (Fig. 1). The results are compatible with selective advantages of isolates carrying both disinfectant and antibiotic resistance genes, and the data indicate that the presence of qac genes in staphylococci results in selection of antibiotic-resistant bacteria (20). Russell also speculated that disinfectant resistance might contribute to antibiotic resistance by coresistance or cross-resistance mechanisms or coselection (26).
This study detected a systematic genetic linkage between resistance to BC and PEN although less than one-half of the plasmid-encoded β-lactamase resistance was linked to disinfectant resistance genes (19 of 43 isolates). Previous investigators have also reported genetic linkage between disinfectant (qac) and antibiotic resistance genes (blaZ, aacA-aphD, dfrA, and ble) on the same staphylococcal plasmids from clinics (3, 14, 21), food environments (27), and an animal clinic (I.-L. Anthonisen et al., unpublished data). To our knowledge, this paper is the first report of closely linked qac, β-lactamase, and tetK genes on a multiresistance plasmid (pMS62). Plasmid pMS62 also harbored ermC, dfrA, and aacA-aphD, encoding resistance to ERY, TMP, and GEN-KAN-tobramycin, respectively. No genetic linkage between qac and these or other antibiotic resistance genes tested was detected. The observed cross-resistance between BC and EBR is compatible with the ability of all known qac resistance determinants to cause the efflux of structurally similar, monovalent cationic agents. A low-level CHX resistance phenotype probably reflects isolates expressing qacA, which also encodes resistance to divalent cations (19), while resistance to the amphoteric disinfectant Tego 103G is probably due to other mechanisms.
The genetic organizations between qac and β-lactamase genes in six plasmids (pMS24, pMS33, pMS62, pMS89, pMS90, and pMS100) were studied (Fig. 2). The plasmids originated from isolates from different sources and distant geographic areas, and their genetic organizations showed similarities with those of other known staphylococcal plasmids, e.g., pSK1, pSK4, pSK23, pSK57, pSK108, pSK156, pNVH97A, pNVH96, pI258, and pI9789 (3, 4, 12, 14, 20). Our results generally confirmed previous findings that β-lactamase-related transposons Tn552 and Tn4002 preferentially insert within TIRL located upstream of the sin gene (4, 21). Insertion of Tn552-like transposons adjacent to qac genes also occurs frequently, as observed in isolates Fol133, Fol89, Fol90, and Fol100 (this study). Overall, this suggests that genetic organizations containing IS257, Tn552, binR, and sin and/or qacR and qacA/B are widely distributed among staphylococci from clinical as well as food environments. Various rearrangements and DNA inversions generating two alternative staphylococcal β-lactamase transposons have been reported (4). We also found the presence of additional organizations of Tn552 and qacA/B within the genomes of strains Fol33 and Fol89 (Fig. 2).
On pMS62, the resolvase- and recombinase-encoding genes (binR and sin, respectively) were present between qac and the incomplete Tn552-like bla gene module as in S. aureus plasmid pS1 (21). The truncated Tn552 observed in pMS62 has probably lost its ability to transpose, as the transposase gene (p480) was not detected.
Staphylococcal insertion sequence IS257 plays a central role in cointegration events (3, 6, 17). Large, conjugative plasmid pSK41 has integrated several small plasmids, where all the cointegrated plasmids are all flanked by copies of IS257 (3). IS257 has been found closely associated with Tn552 (27), qacB (pSK156) (22), qacA (AJ400722), and qacC (11, 12) as well as antibiotic resistance determinants, e.g., dfrA, tetK, and aacA-aphD (3, 6, 12, 13, 14, 27). The presence of IS257 downstream of blaZ on pMS33, pMS24, pMS62, and pMS89, and also detected on pMS90 and pMS100, indicates that IS257 integration events were involved in the formation of the structures of these plasmids. The presence of insertion sequences IS257 and IS256 on some of the plasmids harboring either qacA/B or β-lactamase genes suggests that these plasmids may act as a source for formation of new multiresistance plasmids (5, 13, 17).
Taken together the above observations indicate that a number of different recombination events have occurred to yield the various genetic organizations. To investigate the stability of the plasmids harboring disinfectant and antibiotic resistance genes, we tried to cure the isolates for plasmids. None of the strains were cured for the resistance plasmids. This indicated that plasmids are stably maintained in these strains.
Acknowledgments
We thank Britt Solvår Morken at the Agricultural University of Norway, Terje Steinum, Inger-Lill Anthonisen, Marianne Sunde, and Henning Sørum at the Norwegian School of Veterinary Science, Oslo, Norway, and Per Lea at the Norwegian Food Research Institute for assistance and discussions.
This work was supported by a grant from the Norwegian Research Council (no. 117131/112).
REFERENCES
- 1.Agresti, A. 1992. A survey of exact inference for contingency tables. Stat. Sci. 7:131-153. [Google Scholar]
- 2.Behr, H., M. E. Reverdy, C. Mabilat, J. Freney, and J. Fleurette. 1994. Relationship between the level of minimal inhibitory concentrations of five antiseptics and the presence of qacA gene in Staphylococcus aureus. Pathol. Biol. 42:438-444. [PubMed] [Google Scholar]
- 3.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derbise, A., K. G. Dyke, and N. el Solh. 1995. Rearrangements in the staphylococcal beta-lactamase-encoding plasmid, pIP1066, including a DNA inversion that generates two alternative transposons. Mol. Microbiol. 17:769-779. [DOI] [PubMed] [Google Scholar]
- 5.Dyke, K. G., S. Aubert, and N. el Solh. 1992. Multiple copies of IS256 in staphylococci. Plasmid 28:235-246. [DOI] [PubMed] [Google Scholar]
- 6.Firth, N., and R. A. Skurray. 1998. Mobile elements in the evolution and spread of multiple-drug resistance in staphylococci. Drug Resist. Update 1:49-58. [DOI] [PubMed] [Google Scholar]
- 7.Heir, E., G. Sundheim, and A. Holck. 1998. The Staphylococcus qacH gene product: a new member of the SMR family encoding multidrug resistance. FEMS Microbiol. Lett. 163:49-56. [DOI] [PubMed] [Google Scholar]
- 8.Heir, E., G. Sundheim, and A. L. Holck. 1995. Resistance to quaternary ammonium compounds in Staphylococcus spp. isolated from the food industry and nucleotide sequence of the resistance plasmid pST827. J. Appl. Bacteriol. 79:149-156. [DOI] [PubMed] [Google Scholar]
- 9.Heir, E., G. Sundheim, and A. L. Holck. 1999. The qacG gene on plasmid pST94 confers resistance to quaternary ammonium compounds in staphylococci isolated from the food industry. J. Appl. Microbiol. 86:378-388. [DOI] [PubMed] [Google Scholar]
- 10.Leegaard, T. M., E. Vik, D. A. Caugant, L. O. Frøholm, and E. A. Høiby. 1999. Low occurrence of antibiotic resistance in Escherichia coli and staphylococci isolated from blood cultures in two Norwegian hospitals in 1991-92 and 1995-96. APMIS 107:1060-1068. [PubMed] [Google Scholar]
- 11.Leelaporn, A., I. T. Paulsen, J. M. Tennet, T. G. Littlejohn, and R. A. Skurray. 1994. Multidrugs resistance to antiseptics and disinfectants in coagulase-negative staphylococci. J. Med. Microbiol. 40:214-220. [DOI] [PubMed] [Google Scholar]
- 12.Leelaporn, A., N. Firth, I. T. Paulsen, A. Hettiaratchi, and R. A. Skurray. 1995. Multidrug resistance plasmid pSK108 from coagulase-negative staphylococci; relationships to Staphylococcus aureus qacC plasmids. Plasmid 34:62-67. [DOI] [PubMed] [Google Scholar]
- 13.Leelaporn, A., N. Firth, I. T. Paulsen, and R. A. Skurray. 1996. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J. Bacteriol. 178:6070-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyon, B. R., and R. A. Skurray. 1987. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol. Rev. 51:88-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer, S., M. Boos, A. Beyer, A. C. Fluit, and F. J. Schmitz. 2001. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 47:896-897. [DOI] [PubMed] [Google Scholar]
- 16.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Needham, C., W. C. Noble, and K. G. Dyke. 1995. The staphylococcal insertion sequence IS257 is active. Plasmid 34:198-205. [DOI] [PubMed] [Google Scholar]
- 18.Noguchi, N., M. Hase, M. Kitta, M. Sasatsu, K. Deguchi, and M. Kono. 1999. Antiseptic susceptibility and distribution of antiseptic-resistance genes in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 172:247-253. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen, I. T., M. H. Brown, T. G. Littlejohn, B. A. Mitchell, and R. A. Skurray. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. USA 93:3630-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1998. Characterization of the earliest known Staphylococcus aureus plasmid encoding a multidrug efflux system. J. Bacteriol. 180:3477-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulsen, I. T., M. T. Gillespie, T. G. Littlejohn, O. Hanvivatvong, S. J. Rowland, K. G. Dyke, and R. A. Skurray. 1994. Characterisation of sin, a potential recombinase-encoding gene from Staphylococcus aureus. Gene 141:109-114. [DOI] [PubMed] [Google Scholar]
- 22.Reverdy, M. E., M. Bes, Y. Brun and J. Fleurette. 1993. Evolution of resistance to antibiotics and antiseptics of hospital Staphylococcus aureus strains isolated from 1980 to 1991. Pathol. Biol. 41:897-904. [PubMed] [Google Scholar]
- 23.Rice, L. B., and S. H. Marshall. 1992. Evidence of incorporation of the chromosomal β-lactamase gene of Enterococcus faecalis CH19 into a transposon derived from staphylococci. Antimicrob. Agents Chemother. 36:1843-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowland, S. J., and K. G. Dyke. 1989. Characterization of staphylococcal β-lactamase transposon Tn552. EMBO J. 8:2761-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowland, S. J., and K. G. Dyke. 1990. Tn552, a novel transposable element from Staphylococcus aureus. Mol. Microbiol. 4:961-975. [DOI] [PubMed] [Google Scholar]
- 26.Russell, A. D. 2000. Do biocides select for antibiotic resistance? J. Pharm. Pharmacol. 52:227-233. [DOI] [PubMed] [Google Scholar]
- 27.Sidhu, M. S., E. Heir, H. Sørum, and A. Holck. 2001. Genetic linkage between resistance to quaternary ammonium compounds and β-lactam antibiotics in food-related Staphylococccus spp. Microb. Drug Resist. 7:363-371. [DOI] [PubMed] [Google Scholar]
- 28.Sidhu, M. S., S. Langsrud, and A. Holck. 2001. Disinfectant and antibiotic resistance of lactic acid bacteria isolated from the food industry. Microb. Drug Resist. 7:73-83. [DOI] [PubMed] [Google Scholar]