Abstract
Oral-facial-digital type 1 syndrome (OFD1 [MIM 311200]) is transmitted as an X-linked dominant condition with lethality in males and is characterized by malformations of the face, oral cavity, and digits, and by a highly variable expressivity even within the same family. Malformation of the brain and polycystic kidneys are commonly associated with this disorder. The locus for OFD1 was mapped by linkage analysis to a 12-Mb interval, flanked by markers DXS85 and DXS7105 in the Xp22 region. To identify the gene responsible for this syndrome, we analyzed several transcripts mapping to the region and found mutations in OFD1 (formerly named “Cxorf5/71-7a”), encoding a protein containing coiled-coil α-helical domains. Seven patients with OFD1, including three with familial and four with sporadic cases, were analyzed. Analysis of the familial cases revealed a missense mutation, a 19-bp deletion, and a single base-pair deletion leading to a frameshift. In the sporadic cases, we found a missense (de novo), a nonsense, a splice, and a frameshift mutation. RNA in situ studies on mouse embryo tissue sections show that Ofd1 is developmentally regulated and is expressed in all tissues affected in OFD1 syndrome. The involvement of OFD1 in oral-facial-digital type I syndrome demonstrates an important role of this gene in human development.
Introduction
Oral-facial-digital syndromes (OFDS) are a heterogeneous group of developmental disorders of which at least nine different forms have been described (Toriello 1993). Oral-facial-digital type 1 (OFD1 [MIM 311200]) syndrome was first reported in 1954 by Papillon-League and Psaume (1954) and was further defined in 1962 by Gorlin and Psaume (1962). This syndrome is transmitted as an X-linked dominant condition with lethality in males (Doege et al. 1964; Wettke-Schafer and Kantner 1983) and is characterized by malformations of the face, oral cavity, and digits. Dysmorphic features affecting the head include facial asymmetry, hypertelorism, micrognathia, broadened nasal ridge, and facial milia. Oral cavity lesions reported for this syndrome are median pseudoclefting of the upper lip (45%), clefts of the palate (>80%) and tongue (30%), and oral frenulae. Thickened alveolar ridges and abnormal dentition, including absent lateral incisors, are additional characteristics of OFD1. The digital abnormalities, which affect the hands (50%–70%) more often than the feet (25%), include syndactyly, brachydactyly, clinodactyly, and, more rarely, pre- or postaxial polydactyly. The CNS may also be involved in as many as 40% of cases. Involvement of the CNS includes mental retardation, hydrocephalus, cerebellar anomalies, porencephaly, and agenesis of the corpus callosum (Towfighi et al. 1985; Connacher et al. 1987). In recent years, with the introduction of high-resolution renal ultrasound scanning, it has become evident that polycystic kidney disease is commonly associated with OFD1 (Connacher et al. 1987; Donnai et al. 1987; Scolari et al. 1997), with reports of patients in whom the renal involvement completely dominates the clinical course of the disease (Coll et al. 1997; Feather et al. 1997a).
These clinical features overlap with those reported in the other nine forms of OFDS (Toriello 1993), although among these, OFD1 can be easily distinguished, because of its X-linked dominant inheritance pattern and because of the polycystic kidney disease, which seems to be specific to type 1. The locus of OFD1 was first mapped by linkage analysis to a 19.8-cM interval, flanked by crossovers with markers DXS996 and DXS7105 in the Xp22 region (Feather et al. 1997b). More recently, the interval was narrowed by Gedeon et al., to a 12-Mb interval of the same region (1999).
Patients, Material, and Methods
Patients
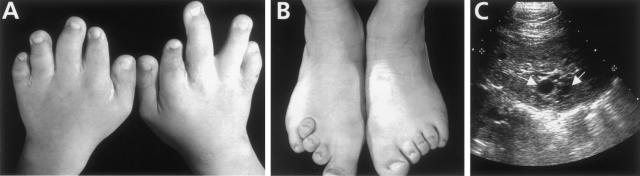
We screened, by direct sequencing, genomic DNA from three familial and four sporadic cases. Although there is overlap with other oral-facial-digital syndromes, the phenotype of OFD1 is highly specific (Toriello 1993), and all patients included in this study were diagnosed with OFD1. In particular, the familial cases were selected on the basis of the X-linked inheritance and/or because of the presence of polycystic kidneys. A summary of the clinical features is given in table 1. An example of the digit anomalies and an ultrasound scan displaying the polycystic lesions reported for this syndrome are provided in figure 1.
Table 1.
Clinical Features of Patients with OFD1 Who Were Included in the Study
|
Anomalies |
||||||
| Family(Casea) | Oral | Facialb | Hand/Foot | Cerebral | Kidney | Miscellaneousb |
| 1 (F) | Hamartomas, dental anomalies, cleft tongue, highly arched palate | Skin milia, dolichocephaly, down-slanting palpebral fissures | … | Suprasellar expansive lesion, mild mental retardation | … | Alopecia, coarse hair |
| 3 (F) | Cleft palate, frenula, lingual hamartomas | Alar hypoplasia | Polydactyly, short second right toe | Cysts of cerebral hemisphere | Polycystic | … |
| 4 (F) | Cleft palate/upper lip | Facial asymmetry | Clinodactyly, syndactyly | Moderate mental retardation, mild cerebral atrophy | Polycystic | Alopecia, dry hair, liver/pancreatic cysts |
| 6 (S) | Frenula, cleft gums, lingual hamartomas | Skin milia | Syndactyly | … | … | … |
| 10 (S) | Cleft lip/palate | NA | Syndactyly | … | … | NA |
| 27 (S) | Cleft palate, dental anomalies, tongue-tied | Skin milia | … | Learning difficulties | Polycystic | … |
| 28 (S) | Cleft palate/upper lip, dental anomalies | Skin milia | Brachydactyly, clinodactyly | Learning difficulties | … | Patchy alopecia |
F = familial; S = sporadic.
NA = not available.
Figure 1.
A and B, Upper- and lower-digit abnormalities in a 12-year-old girl with OFD1. Note brachydactyly and syndactyly. C, Ultrasound scan of kidney of the same individual who is depicted in A and B. She had chronic renal excretory failure and polycystic kidneys: note the cysts (white arrows), up to 1 cm in diameter, located in the parenchyma of the organ.
Mutation Analysis
Primers and conditions used for mutation analysis are available at the TIGEM website. PCRs were performed, by means of the Gentra systems Capture Column Kit, on genomic DNA extracted from peripheral blood leukocytes. Using the ABI Prism Big Dye Terminator Cycle Sequencing Kit (PE Biosystems) and an ABI 377 automated DNA sequencer, we checked PCR products on agarose gel and then sequenced them on both strands. When necessary, PCR amplification products were cloned, by means of the TOPO TA cloning kit supplied by Invitrogen, to separate the mutant and wild-type alleles. Ninety normal X chromosomes from white individuals were analyzed by SSCP for exons 3, 11, and 13, as described (Orita et al. 1989), whereas the same number of chromosomes were analyzed for exon 16 by denaturing high-performance liquid chromatography, using the Wave DNA fragment analysis system (Transgenomic, Inc.), according to the manufacturer’s instructions. Mutation description follows standard nomenclature (Antonarakis 1998). All the patients and controls used in this study were white, and ethical approval was obtained by the universities and research institutes involved in this study.
In Situ Hybridization Studies
RNA in situ studies were performed on mouse tissue sections at E10.5, E12.5, E14.5, and E16.5, by means of radioactive in situ hybridization, according to published protocols (Rugarli et al. 1993). The sense strand of the same fragment did not reveal any detectable signal above the background level (data not shown).
Results
A systematic mutation analysis of genes located in Xp22 in seven female patients with OFD1 was done. Six candidate genes (MSL3L1, MAEG, RAB 9, STK9, M1D1, and pirin) were selected on the basis of their known functions and/or expression patterns and were excluded by mutation analysis (B. Franco, unpublished data; Prakash et al. 1999; Buchner et al. 2000). We then decided to analyze Cxorf5 (alias 71-7a) (GenBank accession numbers Y15164 and Y16355), one of the first transcripts to be assigned to the X chromosome (Kunkel et al. 1983) and included in the OFD1 critical region (de Conciliis et al. 1998). Characterization of the cDNA clones revealed the existence of two alternative transcripts—namely, Cxorf5-1 and Cxorf5-2, which are differentiated by the use of an alternative 3′ splice site. The Cxorf5-1 transcript is predicted to encode a 1011–amino acid protein with a molecular mass of 116 kDA, whereas Cxorf5-2 encodes a predicted protein of 367 amino acids, containing the first 351 residues of the Cxorf5-1 protein and an alternative C-terminal region of 16 amino acids (de Conciliis et al. 1998). Searches of nonredundant DNA and databases with the BLAST algorithm, using both the Cxorf5-1 and the Cxorf5-2, predicted proteins; queries did not reveal significant homology with previously characterized genes and proteins except for the presence of coiled-coil domains (data not shown; de Conciliis et al. 1998). Using genomic sequences available in public databases (GenBank accession number AC003037), we designed primers for PCR amplification of all 23 coding exons, including splice junctions. Cxorf5 was reported to have related loci on both Yq11.22 (Cyorf1) and 5p13 (C5orf1) regions (de Conciliis et al. 1998). Cyorf1 represents a pseudogene (de Conciliis et al. 1998). Concerning C5orf1, Blastn searches of the htgs and gss databases, using the Cxorf5 sequence as query, reveals a genomic sequence corresponding to the C5orf1 locus (AC025449). This sequence is colinear with most of the coding region of the Cxorf5 transcript and does not contain introns (data not shown). On the basis of these results, primers were designed in order to specifically amplify the X chromosome transcript.
Mutation Studies
The patients included in this study were analyzed for the 23 coding exons of the Cxorf5 gene. Mutations were identified in all the patients, and a summary of the results is reported in table 2. Among the familial cases, we identified a missense mutation and two intragenic deletions. In family 1, a 1303 A→C transversion, which changed a polar amino acid (serine) to a basic amino acid (arginine) at codon 434 (S434R), was identified (table 2). DNA from the affected mother and grandmother was analyzed, and the same mutation was recognized, although the father’s DNA displayed a normal pattern (data not shown). In family 3 (Odent et al. 1998), a deletion of a G at nucleotide 312, leading to a frameshift, was identified (table 2). This deletion was confirmed after cloning and analysis of the two distinct alleles. The same mutation was also identified in DNA extracted from an affected female fetus aborted after a 5-wk-old pregnancy from the family described elsewhere (data not shown).
Table 2.
Mutations in Patients with OFD1
| Family(Casea) | Location | Nucleotide Changeb | Effect on Protein |
| 1 (F) | Exon 13 | 1303A→C | S434R |
| 3 (F) | Exon 3 | 312delG | Frameshift |
| 4 (F) | Exon 3 | 294–312delTGGTTTGGCAAAAGAAAAG | Frameshift |
| 6 (S) | Exon 3 | 121C→T | Nonsense mutation |
| 10 (S) | Exon 11 | 1071–1078 del GAAGGATG / 1071–1078 ins TTTTTCCT | KDD 357–359del/FSY 357–359ins |
| 27 (S) | Intron 4 | 312+2del (6) | Abnormal splicing |
| 28 (S) | Exon 16 | 1757delG | Frameshift |
F = familial; S = sporadic.
Mutation description is according to standard nomenclature, with the cDNA sequence of OFD1 used as the reference and with the ATG translation-initiation codon numbered as +1.
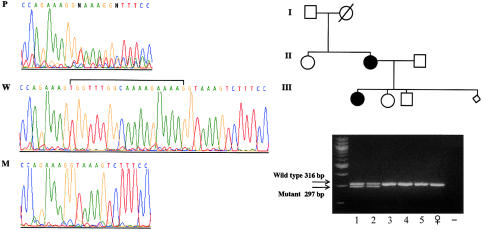
Sequence analysis of family 4 (Scolari et al. 1997) revealed an abnormal pattern, and the study of the two separate alleles identified a deletion of 19 bp in exon 3, leading to a frameshift (fig. 2). This family was studied by SSCP, and although the same abnormal shift was observed in the affected daughter and mother, the father and the unaffected son and daughter displayed a normal pattern. These results were confirmed by sequence analysis and demonstrate that, in this family, the mutation segregates with the disease.
Figure 2.
Analysis of family 4. Left panel, Sequencing chromatograms showing the genomic sequence of the affected individual (P) and of wild-type (W) and mutant (M) alleles. Note the presence of double peaks in the patient’s DNA, downstream of the deletion due to the presence of both normal and out-of-frame alleles. In the wild-type allele, the 19-bp deletion is indicated by a horizontal bracket. Single alleles have been separated by cloning the PCR products. Right panel, Segregation of the 19-nucleotide deletion with the disease phenotype in family 4. After visualization of the PCR products on a 4% agarose gel, two different bands, corresponding to the wild-type (316-bp) and the mutant (297-bp) alleles, appear in the affected individuals (lanes 1 and 2), whereas only the wild-type band is visible in the unaffected individuals (lanes 3–5). In the next-to-last lane, a normal female control is depicted. In the last lane, the PCR-negative control (no DNA) is reported. The lozenge in the genealogical tree indicates an aborted fetus of unknown sex.
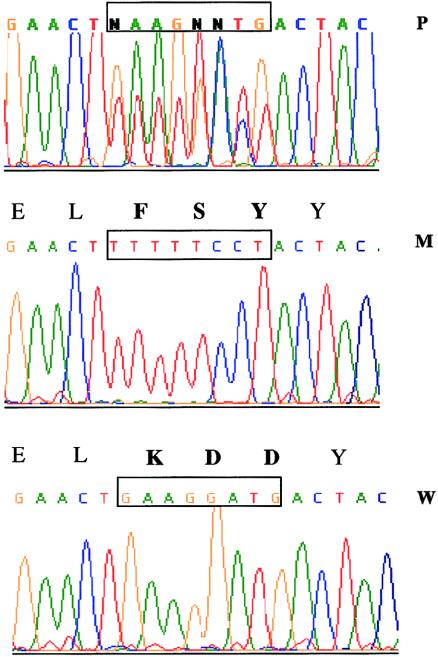
Among the sporadic cases, we identified one missense, one nonsense, one splice, and one frameshift mutation (table 2). In family 6, a 121 C→T transition creates a termination codon TGA (R41X) in exon 3. DNA samples from this patient’s parents were not available for molecular studies. In family 10, a deletion of eight bases, followed by an insertion of eight bases, causes the substitution of three amino acids: basic, acid, and acid (KDD), respectively, with another three amino acids: hydrophobic, polar, and polar (FSY) (fig. 3). We analyzed DNA samples from both parents by direct sequencing, and no abnormalities were found, which demonstrates that this is a de novo mutation. In family 27, a 312+2 deletion of six bases (taaagt) was identified, 312+2del(6), and similar mutations have been shown to disrupt splicing. A deletion of a G at base 1757, resulting in a frameshift, was identified in family 28 (table 2). For both of these latter mutations, maternal DNA was analyzed and revealed a normal pattern; the father’s DNA was not available for the study. Thus, since OFD1 is an X-linked dominant male lethal condition, these can be considered de novo mutations. None of the mutations found in patients with OFD1 were identified in 90 normal X chromosomes, which were analyzed for exons 3, 11, 13, and 16.
Figure 3.
Mutation of OFD1 in family 10. The DNA-sequence fluorogram of the patient’s DNA for exon 11 displays eight double peaks that result from the presence of both the wild-type and the mutant alleles (top). After the PCR product was cloned, the wild-type allele (bottom) and the mutant allele (middle) were analyzed separately. The corresponding amino acid sequence is shown above the nucleotide sequence; the interchange of nine nucleotides results in a change of the three amino acids shown in boldface type.
Expression Studies
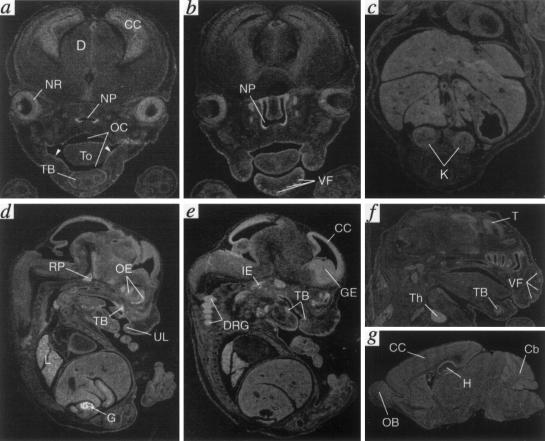
Studies have shown that OFD1 is expressed in all adult tissues examined (pancreas, kidney, skeletal muscle, liver, lung, placenta, brain, and heart) (de Conciliis et al. 1998). To investigate the spatiotemporal expression of this transcript during development, a partial murine cDNA expressed-sequence tag (IMAGE 656917 was identified by BLAST analysis from the Expressed Sequence Tags database, was entirely sequenced, and was used for RNA in situ studies on mouse-embryo tissue sections at different developmental stages. The first site of expression was sharply and exclusively detected at 12.5 days of gestation in the genital ridges.
At later stages (E14.5 and E16.5), Ofd1 expression is not only confined in the developing gonads, but high-to-moderate levels of expression are detected in various craniofacial structures and in the nervous system. In particular, Ofd1 is highly expressed in the epithelium lining the oral and nasal cavities, including the tongue and the gum region, in the developing tooth buds, in the Rathke’s pouch (primordium of the adenohypophysis), and in the vibrissae follicles (fig. 4a–f). The transcript is also highly represented in the following structures: the proliferative and postmitotic layers of the cerebral cortex and ganglionic eminences (anlage of the basal ganglia), the neural retina, the inner ear, and the dorsal root ganglia. Postnatally, the expression in the CNS is diffuse to all the structures but has a higher level in the hippocampus and cerebellum (fig. 4g). Other organs expressing Ofd1 at a lower level are the integumentary system, the lung, the thymus, and the kidney (fig. 4 and data not shown).
Figure 4.
Expression analysis of Ofd1, revealed by in situ hybridization
Discussion
On the basis of the results presented in this paper, we conclude that mutations in Cxorf5 are the cause of OFD1 in the patients we analyzed, and we thus named the transcript OFD1 (Human Gene Nomenclature Committee–approved gene symbol and gene name. Most (6/8) of the mutations identified so far lead to a premature truncation of the protein in its N-terminal region and are therefore predicted to act with a loss of function mechanism. However, due to interference with wild-type proteins in females, we cannot rule out the possibility that truncated OFD1 proteins act with a dominant-negative effect.
OFD1 is the first gene for an X-linked dominant male lethal disorder found to escape X inactivation (de Conciliis et al. 1998). Apparently, in affected females, one normal copy is not sufficient to protect from the disorder. It is conceivable to hypothesize that normal males who carry only one normal copy of OFD1 may display a higher expression of the transcript on the single active X chromosome, but further studies are needed to compare, from a quantitative point of view, the level of expression of this transcript in both sexes. A similar mechanism has been reported for the Clc4 gene, which is X linked and subject to X inactivation in Mus spretus but autosomal in laboratory strains of mice (Rugarli et al. 1995). Quantitative analysis demonstrated that, compared with the single active X-linked locus, each autosomal locus has half the level of Clc4 expression (Adler et al. 1997). An alternative hypothesis is that OFD1 undergoes X inactivation in the tissues affected in OFD1 syndrome at developmental stages when its function is necessary. Therefore, some tissues of affected females at certain stages during development may result in OFD1 functional nullisomy, by inactivation of the normal X. Individual variation in the X-inactivation pattern of this gene may also explain the clinical variability observed in OFD1 syndrome. More detailed X-inactivation studies are needed to test this hypothesis.
Prior study had shown that OFD1 is ubiquitously expressed in adult tissues. Since the clinical features observed in this syndrome are suggestive of a developmental disorder, we decided to study the expression pattern of the Ofd1 gene during development in mouse-embryo tissue sections.
Our studies showed a high-to-moderate level of expression in various craniofacial structures and in the nervous system, and lower levels of expression were also detected in the integumentary system, the lung, the thymus, and the kidney. These sites of expression correlate well with the tissues affected in patients with OFD1.
OFD1 is characterized by craniofacial anomalies, such as facial skin milia, alar hypoplasia (broadened nasal ridge), and facial asymmetry; oral anomalies include hamartomas, clefts of lip and palate, and dental abnormalities. Consistently, expression of Ofd1 is detected in the olfactory and respiratory epithelium of the nasal cavities and nasopharynx, in several ectodermally derived structures of the mouth and palate (upper lips, the surface epithelium of the gum region of the mouth, and tooth primordia), and in the endoderm-derived surface epithelium of the tongue and oropharynx (fig. 4a–f). OFD1 is also characterized by an involvement of the CNS in as many as 40% of the cases with reports of mental retardation, seizures, and other neurological defects (Towfighi et al. 1985; Connacher et al. 1987). In the embryonic nervous system (E14.5 and E16.5), Ofd1 expression is specifically detected in telencephalic primordia of the cerebral cortex and striatum, as well as in cranial and dorsal root ganglia (fig. 4a–f). In postnatal brain (P12.5), Ofd1 transcript is detected in all structures, although with a specifically higher expression in the hippocampal region (fig. 4f). Ofd1 is also expressed at lower levels in the lung, the thymus, the integumentary system (surface ectoderm and vibrissae follicles) and the kidney, which can be related to the alopecia and hair problems and with the kidney abnormalities reported for this syndrome (fig. 4a–f and data not shown).
Since OFD1 shares no sequence homologies—apart from the presence of coiled-coil domains—with other proteins having known functions either in mammals or in lower organisms, it is difficult at this point to predict its function. The presence of several coiled-coil domains suggests that OFD1 acts via a protein-protein interaction mechanism. Identification of OFD1 protein interactors may lead to the discovery of novel genes involved in mammalian development and possibly implicated in other types of OFD syndromes. Furthermore, since OFD1 is often associated with polycystic kidney, it is intriguing to note that PKD1 and PKD2, the genes responsible for autosomal dominant polycystic kidney disease, physically interact through a coiled-coil domain (Qian et al. 1997).
The mouse mutant with X-linked dominant polydactyly (Xpl) maps to the region of the mouse X chromosome, where the murine counterpart of Ofd1 has been localized (Sweet and Lane 1980; de Conciliis et al. 1998). This mutant is characterized by urogenital malformations (hydroureter, hydronephrosis, and cystic or absent kidneys) and polydactyly (Sweet and Lane 1980), and may be considered the possible mouse model of OFD1. We are currently testing whether mutations in Ofd1 may be responsible for the Xpl phenotype. If this is the case, the availability of a mouse model will allow functional studies toward the understanding of the pathogenesis of this syndrome.
In conclusion, the identification of the first gene involved in OFDS will be instrumental in the elucidation of the molecular mechanisms underlying this complex group of developmental disorders and may shed new light on the pathogenesis of polycystic kidney disease.
Acknowledgments
This work is dedicated to the memory of Lisa de Conciliis, who performed the first characterization of the Cxorf5 gene. We acknowledge L. M. Kunkel, for providing us with the 71-7a probes. We thank Dr. Sandro Banfi, for helpful discussion and critical reading of the manuscript, and Claudio Gattuso, for technical assistance. This research was supported by Action Research, the National Kidney Research Fund, the Medical Research Council, the Italian Telethon Foundation, and EC grant QLRT-00791.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Expressed Sequence Tags database, http://www.ncbi.nlm.nih.gov/dbEST/index.html
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for OFD1 locus [accession number AC003037], OFD1 [accession numbers Y15164 and Y16355], and mouse EST [accession number AJ278702])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for OFD1 [MIM 311200]) [PubMed]
- TIGEM, http://www.tigem.it/
References
- Adler DA, Rugarli EI, Lingenfelter PA, Tsuchiya K, Poslinski D, Liggitt HD, Chapman VM, Elliott RW, Ballabio A, Disteche CM (1997) Evidence of evolutionary up-regulation of the single active X chromosome in mammals based on Clc4 expression levels in Mus spretus and Mus musculus. Proc Natl Acad Sci USA 94:9244–9248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE (1998) Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat 11:1–3 [DOI] [PubMed] [Google Scholar]
- Buchner G, Orfanelli U, Quaderi N, Bassi MT, Andolfi G, Ballabio A, Franco B (2000) Identification of a new EGF-repeat-containing gene from human Xp22: a candidate for developmental disorders. Genomics 65:16–23 [DOI] [PubMed] [Google Scholar]
- Coll E, Torra R, Pascual J, Botey A, Ara J, Perez L, Ballesta F, Darnell A (1997) Sporadic orofaciodigital syndrome type I presenting as end-stage renal disease. Nephrol Dial Transplant 12:1040–1042 [DOI] [PubMed] [Google Scholar]
- Connacher AA, Forsyth CC, Stewart WK (1987) Orofaciodigital syndrome type 1 associated with polycystic kidneys and agenesis of the corpus callosum. J Med Genet 24:116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Conciliis L, Marchitiello A, Wapenaar MC, Borsani G, Giglio S, Mariani M, Consalez GG, Zuffardi O, Franco B, Ballabio A, Banfi S (1998) Characterization of Cxorf5 (71-7A), a novel human cDNA mapping to Xp22 and encoding a protein containing coiled-coil α-helical domains. Genomics 51:243–250 [DOI] [PubMed] [Google Scholar]
- Doege T, Thuline H, Priest J, Norby D, Bryant J (1964) Studies of a family with the oral-facial-digital syndrome. N Engl J Med 271:1073–1080 [DOI] [PubMed] [Google Scholar]
- Donnai D, Kerzin-Storrar L, Harris R (1987) Familial orofaciodigital syndrome type I presenting as adult polycystic kidney disease. J Med Genet 24:84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feather SA, Winyard PJ, Dodd S, Woolf AS (1997a) Oral-facial-digital syndrome type 1 is another dominant polycystic kidney disease: clinical, radiological and histopathological features of a new kindred. Nephrol Dial Transplant 12:1354–1361 [DOI] [PubMed] [Google Scholar]
- Feather SA, Woolf AS, Donnai D, Malcolm S, Winter RM (1997b) The oral-facial-digital syndrome type 1 (OFD1), a cause of polycystic kidney disease and associated malformations, maps to Xp22.2-Xp22.3. Hum Mol Genet 6:1163–1167 [DOI] [PubMed] [Google Scholar]
- Gedeon AK, Oley C, Nelson J, Turner G, Mulley JC (1999) Gene localization for oral-facial-digital syndrome type 1 (OFD1:MIM 311200) proximal to DXS85. Am J Med Genet 82:352–354 [PubMed] [Google Scholar]
- Gorlin RJ, Psaume J (1962) Orofaciodigital dysotosis: a new syndrome: a study of 22 cases. J Pediatr 61:520–530 [DOI] [PubMed] [Google Scholar]
- Kunkel LM, Tantravahi U, Kurnit DM, Eisenhard M, Bruns GP, Latt SA (1983) Identification and isolation of transcribed human X chromosome DNA sequences. Nucleic Acids Res 11:7961–7979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odent S, Le Marec B, Toutain A, David A, Vigneron J, Treguier C, Jouan H, Milon J, Fryns JP, Verloes A (1998) Central nervous system malformations and early end-stage renal disease in oro-facio-digital syndrome type 1: a review. Am J Med Genet 75:389–394 [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 86:2766–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillon-Leage M, Psaume J (1954) Une malformation héréditaire de la muquese buccale: brides et freins anomaux. Rev Stomatol 55:209–227 [PubMed] [Google Scholar]
- Prakash SK, Van den Veyver IB, Franco B, Volta M, Ballabio A, Zoghbi HY (1999) Characterization of a novel chromo domain gene in xp22.3 with homology to Drosophila msl-3. Genomics 59:77–84 [DOI] [PubMed] [Google Scholar]
- Qian F, Germino FJ, Cai Y, Zhang X, Somlo S, Germino GG (1997) PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat Genet 16:179–183 [DOI] [PubMed] [Google Scholar]
- Rugarli E, Adler DA, Borsani G, Tsuchiya K, Franco B, Hauge X, Disteche C, Chapman V, Ballabio A. (1995) Different chromosomal localization of the Clcn4 gene in Mus spretus and C57BL/6J mice. Nat Genet 10:466–471 [DOI] [PubMed] [Google Scholar]
- Rugarli EI, Lutz B, Kuratani SC, Wawersik S, Borsani G, Ballabio A, Eichele G (1993) Expression pattern of the Kallmann syndrome gene in the olfactory system suggests a role in neuronal targeting. Nat Genet 4:19–425 [DOI] [PubMed] [Google Scholar]
- Scolari F, Valzorio B, Carli O, Vizzardi V, Costantino E, Grazioli L, Bondioni MP, Savoldi S, Maiorca R (1997) Oral-facial-digital syndrome type I: an unusual cause of hereditary cystic kidney disease. Nephrol Dial Transplant 12:1247–1250 [DOI] [PubMed] [Google Scholar]
- Sweet HO, Lane PW (1980) X-linked polydactyly (Xpl), a new mutation in the mouse. J Hered 71:207–209 [DOI] [PubMed] [Google Scholar]
- Toriello HV (1993) Oral-facial-digital syndromes, 1992. Clin Dysmorphol 2:95–105 [PubMed] [Google Scholar]
- Towfighi J, Berlin CM, Jr., Ladda RL, Frauenhoffer EE, Lehman RA (1985) Neuropathology of oral-facial-digital syndromes. Arch Pathol Lab Med 109:642–646 [PubMed] [Google Scholar]
- Wettke-Schafer R, Kantner G (1983) X-linked dominant inherited diseases with lethality in hemizygous males. Hum Genet 64:1–23 [DOI] [PubMed] [Google Scholar]