Abstract
Sclerosteosis is an autosomal recessive sclerosing bone dysplasia characterized by progressive skeletal overgrowth. The majority of affected individuals have been reported in the Afrikaner population of South Africa, where a high incidence of the disorder occurs as a result of a founder effect. Homozygosity mapping in Afrikaner families along with analysis of historical recombinants localized sclerosteosis to an interval of ∼2 cM between the loci D17S1787 and D17S930 on chromosome 17q12-q21. Here we report two independent mutations in a novel gene, termed “SOST.” Affected Afrikaners carry a nonsense mutation near the amino terminus of the encoded protein, whereas an unrelated affected person of Senegalese origin carries a splicing mutation within the single intron of the gene. The SOST gene encodes a protein that shares similarity with a class of cystine knot–containing factors including dan, cerberus, gremlin, prdc, and caronte. The specific and progressive effect on bone formation observed in individuals affected with sclerosteosis, along with the data presented in this study, together suggest that the SOST gene encodes an important new regulator of bone homeostasis.
Introduction
The term “sclerosteose” or “sclerosteosis” was first applied by Hausen (1967), who recognized a disorder that was distinct from osteopetrosis. Sclerosteosis (SOST [MIM 269500]) is a severe sclerosing skeletal dysplasia in which massive bone overgrowth throughout life leads to gigantism, distortion of the facies, and entrapment of the seventh and eighth cranial nerves. Widening of the calvarium of the skull causes elevation of intracranial pressure and predisposes to sudden death from impaction of the brain in the foramen magnum. Syndactyly is a variable manifestation but represents an important diagnostic feature in infancy. Histological examination of bone from affected individuals reveals thicker-than-normal trabeculae and cortices, along with increased bone density (Epstein et al. 1979; Stein et al. 1983). Detailed analyses of markers of bone turnover indicate that the disorder affects bone modeling and remodeling, especially in the skull and diaphyseal region of the long bones. Elevated alkaline phosphatase and increased rates of bone formation suggest a defect in osteoblast function. Osteoclast function is generally within the normal range; however, the effect on osteoclast numbers varies. No endocrinological abnormalities have been noted.
Although the condition is rare, >60 affected persons have now been documented. The vast majority are present in the Afrikaner population of South Africa, in which the phenotype was essentially delineated and in which the autosomal recessive disorder was predicted to be homogeneous (Beighton et al. 1976, 1977; Beighton and Hamersma 1979; Beighton 1988). Other isolated reports have also emanated from the United States (Higinbotham and Alexander 1941; Stein et al. 1983), Switzerland (Peitruschka 1958), Japan (Sigiura and Yasuhara 1975), Brazil (Freire de Paes Alves et al. 1982) and Spain (Bueno et al. 1994).
Sclerosteosis is similar in phenotype to van Buchem disease, which has been documented in <30 affected persons worldwide, the majority of whom come from The Netherlands (van Buchem et al. 1976; Dixon et al. 1982; Fryns and Vandenberghe 1988). Van Buchem disease differs from sclerosteosis by virtue of milder manifestations and the absence of syndactyly. At the respective ends of the spectrum of severity, however, these two disorders do share some overlap. The conditions have been classified together as endosteal hyperostoses, and the question of syndromic identity has been debated for many years (Beighton et al. 1984). Since the Afrikaner and Dutch populations share ancestral links, it is possible that these two autosomal recessive disorders have a fundamental genetic relationship. The gene for van Buchem disease was mapped to chromosome 17q12-q21 (van Hul et al. 1998); this was followed by the localization of the sclerosteosis locus to a large overlapping region in two unrelated families from Brazil and the United States, thus adding support to this hypothesis (Balemans et al. 1999).
We ascertained 22 affected families from the Afrikaner population, with common ancestors dating back to the 17th century. Through homozygosity mapping and positional cloning, we determined that the genetic determinant is indeed homogeneous in this population, and we identified the gene that is mutated in South Africans as well as in an unrelated affected person of Senegalese origin. This novel gene encodes a protein exhibiting striking sequence similarity to a class of secreted factors previously shown to act as antagonists of members of the TGFβ superfamily, including bone morphogenetic proteins (BMPs). This sequence similarity suggests a model for how the sclerosteosis protein, herein termed “sclerostin,” may function in bone metabolism, which is consistent with the phenotype of sclerosteosis. In addition, we discuss the implications of this finding in relation to the molecular basis of van Buchem disease.
Material and Methods
Family Material
The majority of the South African families with sclerosteosis have been under the care of H.H. for more than three decades. During this period they have been assessed and documented by P.B. (in the Department of Human Genetics, University of Cape Town), and, since 1993, several new kindreds have been identified. Typical clinical manifestations include distortion of the facies and mandibular protrusion with entrapment of cranial nerves causing deafness and facial palsy (fig. 1A). Skeletal hyperostosis and sclerosis are evident radiographically, especially in the calvarium and base of the skull (fig. 1B). Individuals 1–18 had been previously documented (Beighton et al. 1976; Beighton and Hamersma 1979), whereas individuals 19–29 are from South African families identified more recently (table 1).
Figure 1.
Clinical manifestations of sclerosteosis. A, An adult Afrikaner with the characteristic mandibular overgrowth, facial palsy, and deafness. B, Lateral radiograph of the skull of an affected adult Afrikaner. Gross hyperostosis of the calvarium and base are evident.
Table 1.
Summary of Affected Individuals with Composite of Sclerosteosis Family Pedigrees[Note]
| AffectedIndividual | Kindred | Sex | Sibship | ParentalConsanguinity |
| 1 | A | F | ○○● | + |
| 2 | B | F | ●●○○□ | − |
| 3 | B | F | ||
| 4 | C | M | ○○□▪▪○ | − |
| 5 | D | F | ●▪ | + |
| 6 | D | M | ||
| 7 | E | F | ●□▪ | − |
| 8 | E | M | − | |
| 9 | F | F | □● | ? |
| 10 | G | F | ○○○□● | + |
| 11 | H | M | ▪○●○□ | − |
| 12 | H | F | ||
| 13 | I | F | ●▪ | − |
| 14 | I | M | ||
| 15 | J | F | ○□○□□● | + |
| 16 | K | F | ●● | + |
| 17 | K | F | ||
| 18 | L | M | ▪○ | − |
| 19 | M | M | ▪○□□□□▪ | ? |
| 20 | M | M | ||
| 21 | N | F | □●○ | + |
| 22 | O | F | □○● | − |
| 23 | P | F | □●○ | − |
| 24 | Q | M | ○▪ | − |
| 25 | R | F | ▪□ | − |
| 26 | S | M | ▪ | − |
| 27 | T | F | □● | − |
| 28 | U | M | ▪□ | − |
| 29 | V | M | ▪○ | − |
Note.— 29 affected individuals from 22 kindreds (A-V) are indicated by blackened symbols, along with their respective unaffected sibs in unblackened symbols. Circles represent females, squares represent males. In kindred C, DNA was available for analysis from only one (individual 4) of two affected brothers.
After informed consent was obtained, genomic DNA was isolated from venous blood samples by standard procedures. A genome scan on pools of DNA isolated from venous blood was performed as described elsewhere (Wildenberg et al. 1995; Wang et al. 1997), using M13-tailing of forward primers and Li-Cor DNA 400 automated infrared instruments for microsatellite detection and analysis. We used primers from the high-density Cooperative Human Linkage Center (Weber version 6) human genome screening set (Research Genetics) for the initial scan. The affected Senegalese individual was documented elsewhere (Tacconi et al. 1998). Genomic DNA isolated from venous blood was used for direct sequence analysis. Through M. Hayden, we obtained genomic DNA samples from seven Dutch individuals affected with van Buchem disease.
Linkage Analysis
Linkage analysis was performed using the FASTLINK programs (Cottingham et al. 1993; Schäffer et al. 1994). Two-point linkage analysis between the microsatellite markers and the disorder was performed using the MLINK option of the Linkage Analysis Package (version 5.1) (Lathrop and Lalouel 1984) under the assumption that sclerosteosis is an autosomal recessive disorder with a disease frequency of 1/50,000. Allele frequencies of the markers were set at 1/n, where n was the number of alleles identified in the affected families.
Physical Mapping and Mutation Detection
Bacterial artificial chromosome (BAC) clones were obtained from the California Institute of Technology BACs (CITB) human BAC B and C libraries (Research Genetics). In most cases, we sequenced directly the ends of BAC clone inserts by following the protocols for template preparation provided by The Institute for Genome Research (TIGR) and for Big Dye terminator cycle sequencing as recommended by PE Applied Biosystems. Ordering of smaller contigs relative to each other was greatly aided by mapping of microsatellite markers and BAC end-derived sequence-tagged sites (STSs) on the Stanford TNG mapping panel, essentially following the procedure outlined at the Stanford Human Genome Center (SHGC) Web site. Clones chosen for sequencing across the insert were purified over a CsCl gradient, “shotgun” cloned into M13 or pUC19, and subjected to Big Dye primer sequencing using ABI 377s. We used the SPUTNIK program to detect new microsatellite sequences, and we used BLAST and GENSCAN (Burge and Karlin 1997) to identify all candidate genes in the critical interval. For mutation detection, we used Primer3 to identify PCR primers flanking each known and predicted exon to allow amplification and direct sequencing from genomic DNA. Sequence comparisons were performed using the Staden XGAP analysis package. Control DNAs were derived from 48 unrelated, unaffected South African individuals; the NIGMS Human Variation Collection, panels HD01–HD09; and the NIGMS DNA Polymorphism Discovery Resource, panels 1–3. NIGMS DNAs represent 10 different ethnicities (Coriell Cell Repositories).
In Vitro Splicing Assay
A 4.3-kb EcoRV-XhoI fragment from BAC 202l9 (P50) subcloned into pBSII SK(+) (Stratagene) served as template for site-directed mutagenesis (Muta-Gene Kit, BioRad Corp.) to generate mut1-SOST, mut2-SOST, and SDmut-SOST. The mutagenic primers were 5′-CTCTCCTCCACCCCAAACCTTTGGTCTCAAAGG-3′, 5′-GGACCGGCCCGTCGCTCTCCACCCCAGCC-3′, and 5′-CTCTCCTCCACCCCATACATTTGGTCTCAAAGGG-3′, respectively (where the underlined bases indicate deviation from the nonmutant sequence). In addition, the mutagenic primer 5′-CCAGTGGGAGCTGCATGGGCGGCCGCTACCAGCCAGAGGAGG-3′ was used to introduce a NotI site immediately upstream of the initiating methionine codon of SOST, so that the four gene bodies could be subcloned as NotI-XhoI fragments into pPS1286, a mammalian expression vector containing the Rous Sarcoma virus long terminal repeat (RSV LTR) and the polyadenylation signal from SV40 (P.J.S.). Transient transfections of COS-1 and CHO cells with all four constructs were performed by both electroporation and DEAE-Dextran, with similar results. Two days after transfection, total RNA was isolated from cells with Tri Reagent (Molecular Research Center, Inc.). Northern blot analysis was performed using standard methods and reagents from NorthernMAX-gly kit (Ambion). For RT-PCR analysis, dT-primed first-strand cDNA was synthesized using SuperScriptII (Gibco BRL), and subsequent PCR was performed with primers corresponding to the first and last predicted exons of P50gsc3: 5′-CCGGAGCTGGAGAACAACAAG-3′ and 5′-GCACTGGCCGGAGCACACC-3′. To obtain sequence of the RT-PCR products, they were purified from agarose gels by QiaQuick columns (Qiagen) and sequenced directly with the same primers used for PCR.
Sequencing SOST Gene Region in van Buchem Disease Samples
We performed direct sequencing of ∼40 overlapping amplicons spanning a 22-kb region of genomic DNA from patients with van Buchem disease, including the 5.0-kb SOST gene body (two exons) as well as ∼12 kb of 5′ and ∼5 kb of 3′ flanking sequence.
Complete SOST cDNA
We performed RT-PCR on RNA derived from human whole long bone, peripheral blood lymphocytes (PBLs), and bone marrow, using primers corresponding to the first and last GENSCAN-predicted exons—for example, the primer pair described in the splicing assay above, as well as the following pair, located at the 5′ and 3′ ends of the predicted ORF: 5′-ATGCAGTCCCACTGGCCC-3′ and 5′-CTAGTAGGCGTTCTCCAG-3′. Best results were obtained with the Advantage GC Genomic kit (Clontech) with 1 M GC Melt reagent included and the following conditions: 94°C for 3 min, followed by 30 cycles of 94°C for 35 s, 65°C for 1 min, and 72°C for 1 min. Full-length cDNAs were obtained from the Human Universal cDNA Library (HUCL) (Stratagene) by screening filters with the following 32γ-ATP end-labeled oligonucleotide: 5′-CCGGAGCTGGAGAACAACA-3′.
SOST Gene Orthologs
We obtained total RNA from cow (Bos taurus) and vervet monkey (Cercopithecus aethiops) whole long bone by pulverizing fresh material in liquid nitrogen and extracting RNA with Tri Reagent (Molecular Research Center, Inc.). The complete coding region of the vervet Sost gene was obtained by RT-PCR, using human primers 5′-CCTCCTCTGGCTGGTACC-3′ and 5′-AATCAAACCACGCGCAGAGG-3′; the majority of the bovine coding region was obtained using human primers 5′-GGCCAGGGGTGGCAGGCGTTC-3′ and 5′-CTAGTAGGCGTTCTCCAGC-3′. In both cases, first-strand cDNA was synthesized with SuperScript II (Gibco BRL), and the Advantage GC Genomic PCR Kit (Clontech) with 1 M GC Melt reagent was used for PCR under the following conditions: 94°C for 3 min, followed by 30 cycles of 94°C for 35 s, 60°C for 35 s, and 72°C for 35 s.
Three mouse BACs were isolated from the CITB library (Research Genetics) by screening with human primers 5′-GGTGTGCTCCGGCCAGTGC-3′ and 5′-CTTGAGCTCCGACTGGTTGT-3′, chosen because of their similarity to a rat expressed sequence tag (EST) sequence (GenBank accession number AI113131). BAC clone 15g5 was chosen for random shotgun sequencing; the mouse genomic sequence was then compared to human SOST cDNA to identify mouse Sost exons. To obtain a mouse Sost cDNA encoding the entire protein primers corresponding to the 5′ (5′-ATGCAGCCCTCACTAGCCCCG-3′) and 3′ (5′-CTAGTAGGCGTTCTCCAGCTCCGCCTGGTT-3′) ends of the predicted coding exons were used to obtain RT-PCR products from kidney and heart RNA. The complete rat Sost sequence was obtained by RT-PCR from kidney RNA, using primers 5′-ATGCAGCTCTCACTAG-3′ (found in EST AI113131) and 5′-TAGGCGCGGGCTCGCTCTA-3′ (corresponding to sequence immediately downstream of the mouse Sost termination codon). In both cases, a High GC PCR kit (Clontech) was used according to the manufacturer’s recommendations, with the following PCR conditions: 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 2 min.
Real-Time Quantitative RT-PCR
Random-primed first-strand cDNA (SuperScript Preamplification System, Gibco BRL) was used as template in separate-tube amplification reactions. All samples were run in duplicate in each experiment. For cyclophilin expression, we used the TaqMan Pre-Developed Assay Reagent (PDAR) from ABI Biosystems. For SOST and DAD1 expression, primers were custom designed and obtained from ABI Biosystems. SOST primers were 5′-CGGGCGGAGAACGGA-3′ and 5′-CGGCCCATCGGTCACGTA-3′, and internal TaqMan probe was 6FAM-TCGGACACGTCTTTGGTCTCAAA-TAMRA; DAD1 primers were 5′-AGGAGTAGGAGACTAAAAGAATGTTCACTC-3′ and 5′-GTAAGTGCAAATCTGAAGAAAATCCAT-3′, with TaqMan probe 6FAM-TGTCCAATAAGCTGCCATCTCCAGAACTCTT-TAMRA. The TaqMan Universal Master Mix (ABI Biosystems) was used for all the reaction components except primers, probe, and template. The final primer concentrations were 900 nM, and final probe concentrations were 50 nM. Cycling conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The data were collected and analyzed by the ABI Prism 7700 Sequence Detection System Software, version 1.6.4. The quantity (measured in copies/μl) was determined by the standard curve method, in which a standard curve generated by serial dilution (1×, 1:10, 1:100, 1:1,000, and 1:10,000) of a double-stranded fragment of known concentration was run with the unknown samples. The quantity of each unknown was determined by plotting a standard curve (Threshold Cycle (CT) vs. Starting Quantity) and calculating from the CT of each sample the amount amplified. The normalized value was determined by multiplying the relative quantity of SOST for each sample by the relative quantity of cyclophilin or DAD for that sample.
Results
Sclerosteosis Maps to 17q12-q21 in the Afrikaner Population
A composite of 22 affected Afrikaner families, showing the affected and unaffected members of the sibships, is presented in table 1, with a summary of relevant family data. The 22 families are not known to be directly related; nevertheless, because of the high likelihood that, in this relatively isolated and homogeneous population, the condition is the result of a founder mutation, the technique of homozygosity mapping using pooled DNA samples was chosen as an appropriate strategy for mapping (Lander and Botstein 1987). The availability of a large number of affected families for molecular analysis, several of which were consanguineous, increased the likelihood of a successful outcome.
We used a panel of 391 microsatellite markers to genotype DNA pools derived from affected persons (29 total), obligate heterozygotes, and unrelated unaffected population-matched controls. Initially, locus D17S1299 exhibited an alteration in allele distribution between the “affected” pools of DNA and the parent and control pools. We confirmed this linkage to the long arm of chromosome 17 by typing individual DNAs with D17S1299 as well as other closely linked markers such as D17S951, D17S579, D17S920, D17S791, and D17S806 (table 2). These preliminary mapping results (Beighton et al. 1999; see also Gardner 1999) were consistent with the localization of sclerosteosis to 17q12-q21 in two large families from Brazil and the United States (Balemans et al. 1999).
Table 2.
Two-Point LOD Scores for Sclerosteosis and Chromosome 17q12-q21 Markers
| LOD Score at θ= |
||||||
| Marker | .00 | .05 | .10 | .20 | .30 | .40 |
| D17S1299 | −∞ | 4.42 | 3.75 | 2.30 | 1.08 | .28 |
| D17S951 | 5.57 | 4.68 | 3.83 | 2.31 | 1.11 | .33 |
| D17S579 | 5.73 | 4.92 | 4.07 | 2.45 | 1.14 | .29 |
| D17S920 | 5.34 | 4.57 | 3.80 | 2.34 | 1.12 | .29 |
| D17S791 | 6.19 | 5.30 | 4.38 | 2.67 | 1.29 | .37 |
| D17S806 | 6.21 | 5.16 | 4.16 | 2.44 | 1.13 | .30 |
Additional markers (Dib et al. 1996) were used to study historical recombinants on the disease chromosomes in the 29 affected homozygotes. Three kindreds included the most informative recombinants: kindreds A and H each included affected individuals heterozygous at D17S1787 but not at D17S1793, placing the gene distal or telomeric to D17S1787 (individual H-II.1 in fig. 2), whereas a historical recombination event inherited by sibs B-II.1 and B-II.2 placed the gene proximal or centromeric to marker D17S930. Kindreds H and B are presented in figure 2. All 29 affected individuals shared a common haplotype at the marker loci D17S1793, D17S1789, D17S951, and D17S1860 within the critical region bounded by D17S1787 and D17S930, an interval of ∼2 cM (Dib et al. 1996) on the cytogenetic border between the chromosomal bands 17q12–q21.
Figure 2.
Pedigrees of families H and B, showing haplotypes for selected markers at the sclerosteosis locus on chromosome 17q12-21. Alleles are number coded, and the order of the markers from centromere to telomere is according to the Généthon human linkage microsatellite map (Dib et al. 1996). The blackened bars represent the disease haplotype; unblackened and hatched bars represent nondisease haplotypes and indicate parent of origin in the offspring. In H-II.1, the recombination event, which marks the centromeric boundary of the sclerosteosis locus, is shown at marker locus D17S1787. The recombined ancestral haplotype that B-II.1 and B-II.2 have inherited from their father, B-I.1, at D17S930 marks the telomeric boundary of the locus.
Identification of Candidate Genes across the D17S1787–D17S930 Interval
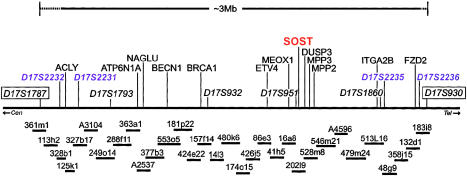
To better characterize the D17S1787–D17S930 interval, we constructed a BAC contig of 3–3.5 Mb, consisting of ∼130 overlapping clones, 32 of which comprised the minimum tiling path (fig. 3). We determined the microsatellite marker order in the 17q12-q21 region to be D17S1787-D17S1793-D17S1802-D17S932-D17S902-D17S965-D17S1789-D17S951-D17S1860-D17S930. In addition, a number of novel microsatellites allowed us to better define the sclerosteosis critical region (fig. 3). Through random shotgun sequencing and more-detailed analysis of BAC clones across the D17S1787–D17S930 interval, we identified ∼70 known genes, along with an estimated 75–100 novel genes, identified either through matching sequences in the Expressed Sequence Tags database or, in some cases, solely on the predictions made by the GENSCAN algorithm (Burge and Karlin 1997).
Figure 3.
Physical map of the sclerosteosis region. Several microsatellite markers from the 17q12-q21 region (e.g., D17S800, D17S1787, D17S1793, D17S855, D17S1789, D17S1860, D17S930, and D17S934) were used simultaneously to screen the human CITB B and C BAC libraries. To resolve discrepancies in marker order observed at the time in publicly available 17q21 maps from the Généthon, CEPH, and GeneBridge Radiation Hybrid mapping efforts, we complemented the construction of BAC contigs with “high-resolution” radiation hybrid analysis on the Stanford TNG mapping panel (Lunetta et al. 1996), using a number of microsatellite markers and novel STSs derived from BAC ends. Genetic analysis of affected families localized the sclerosteosis region to the interval between D17S1787 and D17S930 (boxed). The minimum tiling path across this interval (shown below map) comprised 32 clones; these are labeled according to CITB library ID number, with the exception of clones labeled “Axxxx,” which represent BAC and PAC sequences available in GenBank at the time (full accession numbers are in the format of “AC00xxxx”). A subset of known genes we placed in this interval are indicated on the map; the sclerosteosis candidate gene, SOST, is shown in red and is located within 10 kb of D17S951 on BAC clone 202l9 (lab alias P50). Polymorphic markers are shown in italics; novel microsatellite markers developed as part of this study that further refined the sclerosteosis critical interval are shown in purple. The map is oriented from centromere (cen) to telomere (tel).
Nonsense Mutation in Novel Transcript Detected by Direct Genomic Sequencing
As sequence from the BAC clones became available, we used PCR amplification and direct sequencing of known and predicted exons to search for sclerosteosis-specific polymorphisms. Genomic DNAs from an affected individual, an obligate carrier, and an unrelated unaffected control individual were used as templates. In all, we sequenced ∼1,000 known and novel exons. Potential disease-causing polymorphisms were first tested in all other family members and then, if necessary, in 48 unrelated Afrikaner controls and 90 ethnically diverse individuals from the NIGMS Human Variation Collection, by direct sequence analysis. Only one polymorphism tested in this way did not occur in the unaffected population; this single-nucleotide change introduced a nonsense codon to the coding region of a novel transcript located on BAC 202l9 (lab alias P50).
The novel variant gene (designated “P50gsc3”) predicted by GENSCAN was located between markers D17S951 and D17S1860 (fig. 3 and fig. 4). We found 100% concordance between the polymorphism and the sclerosteosis chromosome when all 29 affected individuals, 33 obligate carriers, and 24 unaffected siblings were analyzed, consistent with P50gsc3 being the determinant gene. We also extended our sequence analysis of unaffected control DNAs to include 270 samples from the NIGMS DNA Polymorphism Discovery Resource. In no instance was the “sclerosteosis variant” observed in any of these control samples. The sclerosteosis-specific C69T transition resulted in a termination codon 23 residues from the amino terminus of the predicted ORF of P50gsc3 (fig. 4).
Figure 4.
Novel predicted P50gsc3 gene contains distinct disease-specific polymorphisms in affected individuals from South Africa and Senegal. The novel gene as predicted by GENSCAN (P50gsc3) includes four coding exons; however, corresponding cDNAs obtained from a number of human tissues differed from the predicted transcript in that they did not include the two internal exons (hatched boxes). Note that the probability scores assigned by GENSCAN to these two internal exons were only .33 and .59, respectively, compared with a score of .99 assigned to both the first and last exons (solid boxes). The single splice donor/splice acceptor pair seen in RT-PCR products did correspond to that of the first and last predicted exons (indicated by ]). A single-base substitution located 69 bp downstream of the predicted translation initiation site (C69T) was found in affected Afrikaners, resulting in termination of translation 23 residues from the amino terminus of P50gsc3. Genomic DNA from the affected Senegalese individual included two substitutions within intronic sequence, located 3 bp (A+3T) and 2,692 bp (A+2692C), respectively, downstream of the first splice-donor site. None of these three polymorphisms was found in ⩾360 unaffected control individuals.
Second Mutation in Sclerosteosis Candidate Gene Affects Splicing
To confirm the identity of the novel mutated gene as the sclerosteosis determinant, we analyzed the sequence of the gene in an unrelated indigenous African person of Senegalese origin, whose condition of sclerosteosis had been reported elsewhere to be identical to that found in affected Afrikaners (Tacconi et al. 1998). Indeed, genotype analysis of this individual revealed a region of homozygosity across the entire D17S1787–D17S930 interval, although with a haplotype distinct from that of affected Afrikaners, consistent with localization of the disorder to this same region. Although no sequence differences existed in the predicted coding region of P50gsc3, we did observe two different polymorphisms in intronic sequences that were homozygous and specific for the Senegalese individual (fig. 4). One of these (an A→T substitution) was located at position +3 of an intron (mut1-SOST), whereas the other (an A→C substitution) was located 67 bp upstream of a splice-acceptor site (mut2-SOST). Neither of these nucleotide differences existed in the sequence of 360 unaffected control DNAs, thus raising the possibility that one or both of them could have a deleterious effect on splicing of the P50gsc3 messenger RNA (Cox et al. 1998; Teraoka et al. 1999).
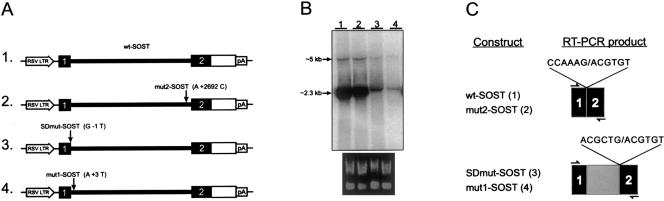
We designed an in vitro splicing assay to determine the effect of the Senegalese-specific polymorphisms on expression of the sclerosteosis candidate gene. A genomic fragment from BAC P50, which contained the coding exons of the gene, served as template for site-directed mutagenesis to introduce either the proximal intronic polymorphism (mut1-SOST) or the distal intronic polymorphism (mut2-SOST). In addition, a sclerosteosis gene in which the first splice-donor site was mutated from AG/GT to AT/GT (SDmut-SOST) served as a positive control. The four different SOST gene bodies (parental wild-type subclone [wt-SOST], mut1-SOST, mut2-SOST, and SDmut-SOST) were then subcloned into a mammalian expression vector in which the ubiquitous RSV LTR controlled expression of the insert (fig. 5A).
Figure 5.
Proximal Senegalese-specific polymorphism affects processing of P50gsc3 transcript in vitro. A, Mammalian expression constructs containing the four forms of the P50gsc3 gene body under the control of the RSV 3’LTR are shown: (1) wt-SOST derived from BAC P50; (2) mut2-SOST, containing the distal A+2692C intronic polymorphism; (3) SDmut-SOST, containing a nucleotide substitution at the donor splice site (G-1T); and (4) mut1-SOST, containing the A+3T intronic polymorphism. Nucleotide positions of the three mutants are numbered relative to the first base of the intron. P50gsc3 coding sequence, including the initiating methionine, is represented by blackened boxes; 3′ UTR (excluding a polyadenylation signal) is represented by unblackened box; heavy horizontal line corresponds to entire 2758 bp intron; the RSV 3′ LTR and polyadenylation signal (pA) elements derived from the expression vector are also indicated. B, After transient transfection of COS-1 cells with the indicated construct, total RNA was recovered and analyzed by northern blot, using a fragment corresponding to the full-length SOST cDNA as a probe. Shown here is one representative blot (the experiment was repeated in triplicate). The quality and concentration of the RNA samples were first ascertained by running 1 μg of each sample on a 1% agarose gel (nondenaturing) and staining with ethidium bromide (below northern blot), then 20 μg of each were used for northern blot analyses. Transcripts deriving from constructs wt-SOST and mut2-SOST were consistently observed at high levels (lanes 1 and 2) and at a size of ∼2.3 kb, indicative of splicing of the intron as shown in 3A. The faint bands at ∼5 kb most likely represent low level of unprocessed transcript. Constructs SDmut-SOST and mut1-SOST consistently resulted in ⩾10-fold lower levels of transcript at a size slightly larger than the processed wt-SOST and mut2-SOST transcripts (lanes 3 and 4). C, When the same RNA samples were subjected to RT-PCR using primers (small arrows) that span the 2758 bp intron, products derived from both the SDmut-SOST and mut1-SOST transfections indicated the use of a cryptic splice-donor site located 214 bases 3′ of that seen in the products from the wt-SOST and mut2-SOST constructs. The sequences at the splice junctions are indicated, the additional 214 bp are shown in gray, and the authentic exonic sequences are solid boxes.
After transient transfection of COS-1 or CHO cells, total RNA was extracted and analyzed both by northern blot and by RT-PCR. Northern blot analyses consistently revealed >10-fold higher levels of transcripts from cells transfected with wt-SOST and mut2-SOST, as compared with cells transfected with mut1-SOST and SDmut-SOST (fig. 5B). The length of these transcripts was ∼2.3 kb, indicative of a splicing event (an unspliced transcript would have an expected length of ∼5 kb). We then subjected RNA from the transfectants to RT-PCR, using primers corresponding to the first and last predicted exons (fig. 5C); sequence analysis of resulting fragments showed that, whereas the wt-SOST and mut2-SOST transcripts used the same splice donor and acceptor as transcripts in unaffected tissue (see below), both the mut1-SOST and the SDmut-SOST transcripts used a cryptic splice donor located 214 bp downstream of the authentic site (fig. 5C). From these experiments we concluded that the proximal intronic polymorphism (mut1-SOST) found in the Senegalese individual resulted in the generation of an improperly spliced message. The relatively low levels of mutant transcripts observed could be the result of nonsense-mediated mRNA decay, as the inclusion of intronic sequence introduced an in-frame nonsense codon (Frischmeyer and Dietz 1999; Hentze and Kulozik 1999).
The identification of two independent mutations in the sclerosteosis candidate gene P50gsc3, the first being a nonsense mutation and the second affecting transcript processing, led us to conclude that sclerosteosis is the result of the loss of function of this novel gene product. In keeping with the interim symbol assigned to the sclerosteosis locus in the OMIM database (MIM 269500), we have designated the gene SOST and will refer to its product as sclerostin.
SOST Gene Not Mutated in van Buchem Disease
We next asked whether sclerosteosis and van Buchem disease have a direct genetic relationship. Analysis of genomic DNA from seven Dutch patients with van Buchem disease across the D17S1787–D17S930 region revealed a common disease haplotype (not shown). We then obtained complete sequence across a 22-kb genomic region, including the 5-kb SOST gene body, from one patient, and we did not detect any mutations in the coding region of SOST. Although five noncoding single-nucleotide polymorphisms (SNPs) in van Buchem disease DNA were detected (relative to the sequence of BAC P50), none fulfilled the criterion for causing the condition, as they all existed within the NIGMS Human Variation Collection control DNAs. One of these was not in Hardy-Weinberg equilibrium; the remaining four potential “van Buchem alleles” were present at frequencies ranging from .12 to .99 and were homozygous in ⩾2 of 94 unaffected individuals. We therefore have no evidence that the two disorders are due to defects in the same gene.
The SOST Gene Is Highly Conserved across Vertebrate Species
We obtained RT-PCR fragments corresponding to the predicted P50gsc3 ORF from osteoblast, thymus, and PBL RNA. In addition to analyzing RT-PCR products, we screened the HUCL library (Stratagene) and isolated two polyadenylated cDNA clones, which included a 639-bp coding sequence and 47 bp of 5′ and 1615 bp of 3′ UTR sequence. A 5′ rapid amplification of cDNA ends (RACE) procedure applied to a human osteoblast cDNA library failed to identify any additional 5′ sequence in the P50gsc3 transcript. The sequence of these cDNAs and RT-PCR products differed from the original P50gsc3 GENSCAN prediction in that they did not include the two internal exons (fig. 4). The single splice donor–splice acceptor pair seen in the cDNAs, however, did correspond to that of the first and last predicted exons.
We next characterized SOST gene orthologs from several other vertebrate species, including rat, mouse, vervet monkey, and cow. BLAST analysis identified two partial rat ESTs (AI113131 and AI556282). PCR primers corresponding to regions of 100% identity between human and rat SOST identified three mouse BACs, and one of these was sequenced to completion. On the basis of the mouse genomic sequence, we then designed primers to obtain RT-PCR products from various mouse tissues (e.g., kidney and heart), and found the same exon structure observed in human cDNAs. The rat coding sequence was subsequently extended by RT-PCR, using mouse primers. Primers from immediately upstream and downstream of the human SOST gene coding sequence (i.e., from the UTRs) amplified vervet cDNA from whole long bone RNA, and a bovine sequence corresponding to the most highly conserved portion of the gene was obtained from kidney RNA. At the amino acid level, the vervet, rat, and mouse sclerostin sequences were 98%, 89%, and 88% identical, respectively, to the human sequence (fig. 6). The partial bovine sclerostin sequence was 94% identical to the human sequence (not shown).
Figure 6.
The SOST gene product is highly conserved across vertebrate species. The vervet (v), rat (r), and mouse (m) sclerostin sequences are 98%, 89%, and 88% identical, respectively, to the human (h) sequence. Conserved residues across all four species are highlighted with a black background; residues shared by three species are highlighted in gray. Putative secretory signal sequence is overlined, and potential sites for asparagine (N)-glycosylation are underlined.
Expression of the SOST Gene
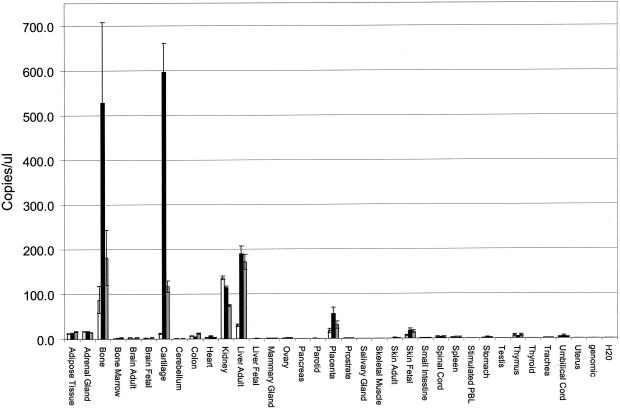
We examined SOST gene expression in a set of 76 human tissues and cell lines by probing the Multiple Tissue Expression Array (Clontech) with a fragment corresponding to the second exon; aorta and fetal kidney exhibited the highest levels of expression (not shown). It is noteworthy, however, that bone-related tissues were not represented on this array. To examine levels of expression more quantitatively in tissues including bone, we used real-time quantitative RT-PCR (ABI 7700 instrument). Overall expression of the SOST gene was relatively low, compared with control genes such as PPI (cyclophilin) and DAD1, but was significant in whole long bone, cartilage, kidney, and liver and was detectable at lower levels in placenta and fetal skin (fig. 7). Semiquantitative RT-PCR analyses of a number of mouse tissues (whole fetus, liver, heart, kidney, brain, thymus, and whole long bone) gave similar results (not shown).
Figure 7.
The SOST gene is expressed in a tissue-specific manner. Levels of SOST gene expression were determined using the standard curve method (separate tube reactions) on an ABI Prism 7700 detection instrument, in which the PPI (cyclophilin) and DAD1 genes served as endogenous references, and the standard curves were generated from a dilution series of a linear plasmid standard of known concentration. Standard curves were derived by plotting the threshold cycle (CT) vs. starting quantity, in this case expressed in copies/μl, and raw values for each of the three genes were then calculated from the CT of each test sample. Mean values from duplicate reactions for SOST, PPI, and DAD1 were used in subsequent calculations. Two independent normalized SOST values were derived from the ratio of raw mean SOST to raw mean PPI value and raw mean SOST to raw mean DAD1 value, for each tissue sample. Expression in the human tissues indicated are represented by the raw mean SOST values (open bars), SOST values normalized to PPI (solid bars), and SOST values normalized to DAD1 (gray bars).
Analysis of Sclerostin Amino Acid Sequence
Amino acid sequence analysis of the SOST gene product revealed the presence of a putative secretion signal and two N-glycosylation sites (fig. 6), suggesting that sclerostin is secreted. BLAST analysis of public sequence and ProDom databases revealed weak but significant similarity to proteins such as gastric mucin, the beta subunit of luteinizing hormone, as well as dan and a protein termed “prdc” (protein related to dan and cerberus). In all instances, the region of similarity corresponded to a cysteine-rich region in sclerostin that covers residues 80–167. Outside this region, the sequence of sclerostin is unique. Analysis of the Pfam hidden Markov model database of structural motifs recognized similarity between the cysteine-rich region of sclerostin and the so-called cystine-knot motif. Although the percent identity at the level of primary amino acid sequence was quite low, the specific pattern of cysteine residues was consistent with sclerostin falling into the class of secreted proteins that contain this structural knot motif, such as the TGFβ superfamily, the Norrie disease protein (NDP), the mucins, and von Willebrand factor (VWF). In particular, sclerostin includes the highly conserved CxGxC and CxC motifs characteristic of growth-factor cystine knots, as well as appropriate spacing of six conserved cysteines thought to configure the knot structure (Meitinger et al. 1993; Isaacs 1995). Other features of the cysteine-rich region of sclerostin indicate that it is most closely related to the subfamily of cystine knot factors that includes dan, cerberus, gremlin, prdc, and caronte (fig. 8).
Figure 8.
Sclerostin shares sequence similarity with the dan family of proteins. The cysteine-rich region of the SOST gene product (residues 80–167) is aligned with the analogous regions of proteins encoded by the human DAN, CER1 (cerberus), DRM (gremlin), mouse Prdc, chicken Car (caronte), human NDP, MUC2, VWF, and TGFβ2 genes. The six canonical cysteine residues forming the core of the knot structure, including the CxGxC motif, are numbered 1–6 (highlighted in black), and cysteines that may be involved in additional stabilizing disulfide bonds are labeled a–d, according to Meitinger et al. (1993) (highlighted in gray). Pairwise alignments of the cystine knot motif of sclerostin to each of these other factors show the following percent identities and number of gaps: DAN (22%, 10); CER1 (24%, 10); DRM (20%, 12); Prdc (23%, 12); Car (22%, 10); NDP (18%, 17); MUC2 (15%, 25); VWF (17%, 22); and TGFβ2 (11%, 24).
Discussion
Because of the relative homogeneity of the Afrikaner population, it had been assumed that individuals affected with sclerosteosis shared a common disease allele. We therefore used the approach of homozygosity mapping (Lander and Botstein 1987) to identify and refine the sclerosteosis locus in affected families. Subsequent physical mapping and mutation analysis resulted in the identification of the determinant gene, which we term “SOST.” In the present study, we describe two mutations in this gene: the first, found in affected Afrikaners, is a nonsense mutation near the amino terminus of the coding region; the second, in an unrelated Senegalese individual, alters processing of the SOST transcript. Both of these mutations are expected to result in a loss of function of the gene product, sclerostin, which is consistent with the recessive nature of the disorder. The failure to find either of these mutations in a large number of control individuals (nonsense mutation absent in >400 controls; splicing mutation absent in >300 controls) confirmed their direct association with the disorder. Discovery of the molecular basis of sclerosteosis may now lead to accurate diagnoses, thus representing an important advance in the genetic management of this severe and often fatal condition.
Results presented here fail to resolve the question of syndromic identity between the similar disorders of sclerosteosis and van Buchem disease. The majority of Dutch patients with van Buchem disease carry a common disease haplotype, suggesting a single founder mutation in this population (van Hul et al. 1998). Because of the ancestry of the Afrikaners of South Africa, it was previously hypothesized that the founder mutation in this population may have derived from a Dutch van Buchem disease carrier (Beighton et al. 1984). We have ruled out this hypothesis by demonstrating that the mutation in affected Afrikaners is not present in Dutch patients with van Buchem disease. In fact, we found no disease-causing alterations within a 22-kb region surrounding the SOST gene. The complete understanding of how the two disorders may be associated must await the determination of the molecular basis of van Buchem disease.
In our analysis of the amino acid sequence of the SOST gene product sclerostin, we noted a cysteine-rich region. Comparative analysis further revealed similarity to the so-called dan family of secreted glycoproteins, which includes dan, cerberus/cer1, gremlin/drm, prdc, caronte, and dante (Hsu et al. 1998; Pearce et al. 1999). These factors have been proposed to include a cystine knot structure, on the basis of their sequence similarity to the knot structures of TGFβ2 (determined by crystallography) and NDP (characterized by molecular modeling) (McDonald and Hendrickson 1993; Meitinger et al. 1993). The predominant feature of the cystine knot motif is a conserved set of six cysteine residues, with the CxGxC and CxC fingerprints defining the subclass of growth factor knots (Isaacs 1995). Growth factors of the TGFβ superfamily function as dimers, mediating a wide variety of biological processes. In addition, it has been demonstrated that the cystine knots of TGFβs are released by cleavage of a proprotein precursor during secretion (Gentry et al. 1988). The presence of cystine knot domains in dan family proteins suggests that this subfamily of proteins also functions as secreted dimers. Indeed, the secretion of dan, cerberus, gremlin, and caronte has been well documented (Belo et al. 1997; Nakamura et al. 1997; Biben et al. 1998; Hsu et al. 1998) and, in the case of Cer1 (mammalian homolog of cerberus), dimerization but not processing of a proprotein has been reported as well (Biben et al. 1998; Pearce et al. 1999).
It is interesting to note that members of the dan family share the ability to antagonize BMP signalling. The BMPs comprise a large family of cytokines, also belonging to the TGFβ superfamily. Although this family of morphogens was originally identified on the basis of bone-inducing activity (Urist 1965), a number of these factors have been shown to be critical for very early stages of embryogenesis and fetal development. Depending on the localization, concentration and timing of expression of a particular BMP, the processes of chemotaxis, mitogenesis, differentiation, and apoptosis have all been ascribed to this class of proteins (Hogan 1996; Sakou 1998). In addition, the requirement for the coordinated expression of negative regulators of BMP activity, such as noggin and chordin, as well as members of the dan family, has been well described (Harland and Gerhart 1997; Sasai and De Robertis 1997; Hsu et al. 1998).
Although the significance of sequence similarity between sclerostin and the dan family of proteins awaits direct biochemical characterization of sclerostin function, the observed similarity does suggest a model for the function of the SOST gene product that is consistent with the phenotype of sclerosteosis—that is, loss of a negative regulator of a TGFβ superfamily member (such as a BMP) important for postnatal formation of bone could result in excess bone deposition throughout life. Interestingly, it has been reported recently that the autosomal dominant disorder Camurati-Engelmann disease results from mutations in the TGFβ1 proprotein that most likely lead to increased and/or inappropriate activity of the mature factor (Janssens et al. 2000; Kinoshita et al. 2000). This disorder shares many radiographic features with sclerosteosis, pointing to a shared or parallel pathway for TGFβ1 and SOST signalling.
We view the sclerosteosis mutation as a genetic model of a potential therapy for conditions characterized by bone loss, notably osteoporosis. Currently there is no effective treatment for the restoration of normal bone mass in individuals with pathologically low skeletal density, and there is a large unmet need for therapy that can increase bone deposition in osteoporosis (Rodan and Martin 2000). The pathogenesis and genetics of sclerosteosis suggest that inhibition of the SOST gene product could lead to increased bone density, thus reversing the effects of this common and debilitating condition.
Note: The SOST gene reported here is identical to the gene termed “BEER,” which is found in U.S. patent application PCT/US99/27990.
Acknowledgments
The authors express their sincere gratitude to the Afrikaner families that participated in this study and to Sister Lecia Bartmann for her kindness and care during her interactions with the affected persons. We acknowledge M. Hayden for providing DNA samples from patients with van Buchem disease; P. Charmley, M. Fajardo, W. Brady, R. Cottingham, J. Howbert, and J. Latham for support throughout this study; the entire Darwin/Chiroscience sequencing group for expert technical support; and D. Winkler and R. Schatzman for critical reading of the manuscript.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- CEPH, http://www.cephb.fr/
- Cooperative Human Linkage Center, http://lpg.nci.nih.gov/CHLC/
- Coriell Cell Repositories, http://locus.umdnj.edu/
- Expressed Sequence Tags database, http://www.ncbi.nlm.nih.gov/dbEST/index.html
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for human SOST cDNA [accession number AF326739], mouse Sost cDNA [accession number AF326740], rat Sost cDNA [accession number AF326741], vervet Sost cDNA [accession number AF326742], bovine Sost cDNA [accession number AF326738], 22 kb human genomic sequence [accession number AF326736], mouse genomic sequence [accession number AF326737], D17S2231 [accession number AF326743], D17S2232 [accession number AF326744], D17S2233 [accession number AF326745], D17S2234 [accession number AF326746], D17S2235 [accession number AF326747], and D17S2236 [accession number AF326748])
- GeneBridge Radiation Hybrid Order Generation, http://www.sanger.ac.uk/Software/RHserver/RHgenebridge.shtml
- Généthon, http://www.genethon.fr/
- NIGMS Human Variation Collection and DNA Polymorphism Discovery Resource, http://locus.umdnj.edu/nigms/
- Online Mendelian Inheritance of Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SOST [MIM 269500])
- Pfam database, http://www.cgr.ki.se/Pfam/
- ProDom database, http://www.dur.ac.uk/~dbl0www/prodom.html
- SPUTNIK, http://abajian.net/sputnik/index.html
- Stanford SHGC RH assay procedure, http://www-shgc.stanford.edu/Mapping/rh/procedure/indexold.html
- TIGR, http://www.tigr.org/
References
- Balemans W, van den Ende J, Freire Paes-Alves A, Dikkers F, Willems P, Vanhoenacker F, de Almeida-Melo N, Alves CF, Stratakis CA, Hill SC, van Hul W (1999) Localization of the gene for sclerosteosis to the van Buchem disease-gene region on chromosome 17q12-q21. Am J Hum Genet 64:1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton P (1988) Sclerosteosis. J Med Genet 25:200–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton P, Barnard A, Hamersma H, van der Wouden A (1984) The syndromic status of sclerosteosis and van Buchem disease. Clin Genet 25:175–181 [DOI] [PubMed] [Google Scholar]
- Beighton P, Davidson J, Durr L, Hamersma H (1977) Sclerosteosis—an autosomal recessive disorder. Clin Genet 11:1–7 [DOI] [PubMed] [Google Scholar]
- Beighton P, Durr L, Hamersma H (1976) The clinical features of sclerosteosis: a review of the manifestations in twenty-five affected individuals. Ann Intern Med 84:393–397 [DOI] [PubMed] [Google Scholar]
- Beighton P, Gardner J, Hamersma H, Fu Y-H, Alisch R, Mulligan J (1999) Syndromic status of sclerosteosis and van Buchem disease. Paper presented at 8th Biennial Southern African Society of Human Genetics Congress, Gordon's Bay, March 9–12, 1999 [Google Scholar]
- Beighton P, Hamersma H (1979) Sclerosteosis in South Africa. S Afr Med J 55:783–788 [PubMed] [Google Scholar]
- Belo J, Bouwmeester T, Leyns L, Kertesz N, Ballo M, Follettie M, De Robertis E (1997) Cerberus-like is a secreted factor with neuralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech Dev 68:45–57 [DOI] [PubMed] [Google Scholar]
- Biben C, Stanley E, Fabri L, Kotecha S, Rhinn M, Drinkwater C, Lah M, Wang C-C, Nash A, Hilton D, Ang S-L, Mohun T, Harvey RP (1998) Murine cerberus homologue mCer-1: a candidate anterior patterning molecule. Dev Biol 194:135–151 [DOI] [PubMed] [Google Scholar]
- Bueno M, Olivan G, Jimenez A, Garragori J, Sarria A, Bueno A, Bueno M Jr, Ramos FJ (1994) Sclerosteosis in a Spanish male: first report in a person of Mediterranean origin. J Med Genet 31:976–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94 [DOI] [PubMed] [Google Scholar]
- Cottingham R, Idury R, Schaffer A (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Cox L, Jett C, Hixson J (1998) Molecular basis of an apolipoprotein[a] null allele: a splice site mutation is associated with deletion of a single exon. J Lipid Res 39:1319–1326 [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dixon J, Cull R, Gamble P (1982) Two cases of van Buchem's disease. J Neurol Neurosurg Psychiatry 45:913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein S, Hamersma H, Beighton P (1979) Endocrine function in sclerosteosis. S Afr Med J 55:1105–1110 [PubMed] [Google Scholar]
- Freire de Paes Alves A, Rubim J, Cardoso L, Rabelo M (1982) Sclerosteosis: a marker of Dutch ancestry? Rev Brasil Genet 5:825–834 [Google Scholar]
- Frischmeyer P, Dietz H (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8:1893–1900 [DOI] [PubMed] [Google Scholar]
- Fryns J, Vandenberghe H (1988) Facial paralysis at the age of 2 months as a first clinical sign of van Buchem disease (endosteal hyperostosis). Eur J Pediatr 147:99–100 [DOI] [PubMed] [Google Scholar]
- Gardner J (1999) Profound childhood deafness in South Africa: a clinical and molecular genetic approach. Ph.D. thesis, University of Cape Town, Cape Town [Google Scholar]
- Gentry L, Lioubin M, Purchio A, Marquardt H (1988) Molecular events in the processing of recombinant type 1 pre-pro-transforming growth factor beta to the mature polypeptide. Mol Cell Biol 8:4162–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R, Gerhart J (1997) Formation and function of Spemann's organizer. Annu Rev Cell Dev Biol 13:611–667 [DOI] [PubMed] [Google Scholar]
- Hausen H (1967) Sklerosteose. In: Opitz H, Schmid F (eds) Handbuch der Kinderheilkunde. Vol. 6. Springer-Verlag, Berlin, pp 351–355 [Google Scholar]
- Hentze M, Kulozik A (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307–310 [DOI] [PubMed] [Google Scholar]
- Higinbotham N, Alexander S (1941) Osteopetrosis, four cases in one family. Am J Surg 53:444–454 [Google Scholar]
- Hogan B (1996) Bone morphogenetic proteins: multi-functional regulators of vertebrate development. Genes Dev 10:1580–1594 [DOI] [PubMed] [Google Scholar]
- Hsu D, Economides A, Wang X, Eimon P, Harland R (1998) The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol Cell 1:673–683 [DOI] [PubMed] [Google Scholar]
- Isaacs N (1995) Cystine knots. Curr Opin Struct Biol 5:391–395 [DOI] [PubMed] [Google Scholar]
- Janssens K, Gershoni-Baruch R, Guanabens N, Migone N, Ralston S, Bonduelle M, Lissens W, Van Maldergem L, Vanhoenacker F, Verbruggen L, Van Hul W (2000) Mutations in the gene encoding the latency-associated peptide of TGFβ1 cause Camurati-Engelmann disease. Nat Genet 26:273–275 [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Saito T, Tomita H, Makita Y, Yoshida K, Ghadami M, Yamada K, Kondo S, Ikegawa S, Nishimura G, Fukushima Y, Nakagomi T, Saito H, Sugimoto T, Kamegaya M, Hisa K, Murray J, Taniguchi N, Niikawa N, Yoshiura K (2000) Domain-specific mutations in TGFβ1 result in Camurati-Engelmann disease. Nat Genet 26:19–20 [DOI] [PubMed] [Google Scholar]
- Lander E, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed] [Google Scholar]
- Lathrop G, Lalouel J (1984) Easy calculations of Lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lunetta K, Boehnke M, Lange K, Cox D (1996) Selected locus and multiple panel models for radiation hybrid mapping. Am J Hum Genet 59:717–725 [PMC free article] [PubMed] [Google Scholar]
- McDonald N, Hendrickson W (1993) A structural superfamily of growth factors containing a cystine knot motif. Cell 73:421–424 [DOI] [PubMed] [Google Scholar]
- Meitinger T, Meindl A, Bork P, Rost B, Sander C, Haasemann M, Murken J (1993) Molecular modelling of the Norrie disease protein predicts a cystine knot growth factor tertiary structure. Nat Genet 5:376–380 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ozaki T, Nakagawara A, Sakiyama S (1997) A product of DAN, a novel candidate tumour suppressor gene, is secreted into culture medium and suppresses DNA synthesis. Eur J Cancer 33:1986–1990 [DOI] [PubMed] [Google Scholar]
- Pearce J, Penny G, Rossant J (1999) A mouse cerberus/Dan-related gene family. Dev Biol 209:98–110 [DOI] [PubMed] [Google Scholar]
- Peitruschka G (1958) Weitere Mitteilungen über die Marmorknochenkrankheit (Albers-Schonbergsche Krankheit) nebst Bemerkungen zur Differentialdiagnose. Klin Monatbl 132:509–525 [PubMed] [Google Scholar]
- Rodan G, Martin T (2000) Therapeutic approaches to bone disease. Science 289:1508–1514 [DOI] [PubMed] [Google Scholar]
- Sakou T (1998) Bone morphogenetic proteins: from basic studies to clinical approaches. Bone 22:591–603 [DOI] [PubMed] [Google Scholar]
- Sasai Y, De Robertis E (1997) Ectodermal patterning in vertebrate embryos. Dev Biol 182:5–20 [DOI] [PubMed] [Google Scholar]
- Schäffer A, Gupta S, Shriram K, Cottingham R (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Sugiura Y, Yasuhara T (1975) Sclerosteosis: a case report. J Bone Surg (Am) 57:273–276 [PubMed] [Google Scholar]
- Stein S, Witkop C, Hill S, Fallon M, Viernstein L, Gucer G, McKeever P, Long D, Altman J, Miller NR, Teitelbaum SL, Schlesinger S (1983) Sclerosteosis: neurogenetic and pathophysiologic analysis of an American kinship. Neurology 33:267–277 [DOI] [PubMed] [Google Scholar]
- Tacconi P, Ferrigno P, Cocco L, Cannas A, Tamburini G, Bergonzi P, Giagheddu M (1998) Sclerosteosis: report of a case in a black African man. Clin Genet 53:497–501 [DOI] [PubMed] [Google Scholar]
- Teraoka S, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, Chessa L, Sanal O, Bernatowska E, Gatti RA, Concannon P (1999) Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet 64:1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist M (1965) Bone: formation by autoinduction. Science 150:893–899 [DOI] [PubMed] [Google Scholar]
- van Buchem F, Prick J, Jaspar H (1976) Hyperostosis corticalis generalisata familiaris (van Buchem disease). Excerpta Medica, Amsterdam [Google Scholar]
- van Hul W, Balemans W, van Hul E, Dikkers F, Obee H, Stokroos R, Hildering P, Vanhoenaker F, van Camp G, Willems PJ (1998) van Buchem disease (hyperostosis corticalis generalisata) maps to chromosome 17q12-q21. Am J Hum Genet 62:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Y, Hawkins-Lee B, Ochoa B, Walker R, She J-X (1997) Homozygosity and linkage disequilibrium mapping of the urofacial (Ochoa) syndrome gene to a 1-cM interval on chromosome 10q23-q24. Am J Hum Genet 60:1461–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildenberg S, Oetting W, Almodovar C, Krumwiede M, White J, King R (1995) A gene causing Hermansky-Pudlak syndrome in a Puerto Rican population maps to chromosome 10q. Am J Hum Genet 57:755–765 [PMC free article] [PubMed] [Google Scholar]