Abstract
Familial hemophagocytic lymphohistiocytosis (FHL) is an autosomal recessive disease of early childhood characterized by nonmalignant accumulation and multivisceral infiltration of activated T lymphocytes and histiocytes (macrophages). Cytotoxic T and natural killer (NK) cell activity is markedly reduced or absent in these patients, and mutations in a lytic granule constituent, perforin, were recently identified in a number of FHL individuals. Here, we report a comprehensive survey of 34 additional patients with FHL for mutations in the coding region of the perforin gene and the relative frequency of perforin mutations in FHL. Perforin mutations were identified in 7 of the 34 families investigated. Six children were homozygous for the mutations, and one patient was a compound heterozygote. Four novel mutations were detected: one nonsense, two missense, and one deletion of one amino acid. In four families, a previously reported mutation at codon 374, causing a premature stop codon, was identified, and, therefore, this is the most common perforin mutation identified so far in FHL patients. We found perforin mutations in 20% of all FHL patients investigated (7/34), with a somewhat higher prevalence, ∼30% (6/20), in children whose parents originated from Turkey. No other correlation between the type of mutation and the phenotype of the patients was evident from the present study. Our combined results from mutational analysis of 34 families and linkage analysis of a subset of consanguineous families indicate that perforin mutations account for 20%–40% of the FHL cases and the FHL 1 locus on chromosome 9 for ∼10%, whereas the major part of the FHL cases are caused by mutations in not-yet-identified genes.
Introduction
Familial hemophagocytic lymphohistiocytosis (FHL [MIM 267700]) is a rare autosomal recessive disorder of infancy and early childhood, characterized by fever, hepatosplenomegaly, and pancytopenia, and with neurological involvement of varying degrees commonly developing during the course of the disease (Janka 1983; Henter et al. 1998). The histopathological hallmark is an infiltration—in spleen, liver, bone marrow, lymph nodes, and the central nervous system—of lymphocytes and macrophages, the latter being frequently engaged in hemophagocytosis (Janka 1983; Henter et al. 1998). Hypercytokinemia with elevated serum levels of inflammatory cytokines—such as tumor necrosis factor α, interferon γ, and interleukin (IL-6)—are striking features of FHL and indicate an uncontrolled activation of T lymphocytes and macrophages (Henter et al. 1991b; Fujiwara et al. 1993; Imashuku et al. 1996). Without treatment, the disease is inevitably fatal. However, with chemotherapy (Henter et al. 1997) in combination with bone marrow transplantation (BMT), the disease can be cured in a large proportion of cases (Jabado et al. 1997; Durken et al. 1999).
Cytotoxic T and natural killer (NK) cell activity is consistently low or absent in FHL patients (Perez et al. 1984; Arico et al. 1988; Eife et al. 1989; Egeler et al. 1996) and also may be suppressed in first-degree relatives (Sullivan et al. 1998). In patients in remission, this defect in cellular cytotoxicity remains after chemotherapeutic treatment and is restored only after successful BMT. By means of homozygosity mapping, two loci for FHL have been disclosed—on 9q21.3-22 (FHL1) and 10q21-22 (FHL2)—and later studies have provided evidence for additional genetic heterogeneity (Dufourcq-Lagelouse et al. 1999a; Ohadi et al. 1999; Graham et al. 2000). Recently, the chromosome 10q21-22 locus was shown to be the gene encoding perforin (Stepp et al. 1999), an important mediator of cellular cytotoxicity. The perforin gene is a simple gene organized in only three exons, of which only exons 2 and 3 are translated (Lichtenheld and Podack 1989). Mutations in the perforin gene were identified in eight unrelated patients with FHL; cultured lymphocytes from affected patients displayed defective cytotoxic activity, and immunostaining revealed little or no perforin in the granules of the patients studied (Stepp et al. 1999). Perforin is secreted from cytotoxic T lymphocytes and NK cells upon conjugation between effector and target cell, and, in the presence of calcium, it is able to penetrate the membrane of the target cell, where it polymerizes to form a cell death–inducing pore (Lichtenheld and Podack 1989; Tschopp and Nabholz 1990; Lowin et al. 1995). Pore formation leads to destruction of target cells by osmotic lysis and by allowing entrance to granzymes, which trigger apoptosis (Darmon et al. 1995; Darmon and Bleackley 1998). Previous studies in perforin-deficient mice infected with lymphocytic choriomeningitis virus or staphylococcal enterotoxin B have demonstrated an uncontrolled expansion of cytotoxic T cells and attendant cytokine-driven mortality, similar to the phenotype evident in FHL, and a role for perforin in the homeostatic regulation of CD8+ lymphocytes thus was inferred (Kägi et al. 1999; Matloubian et al. 1999). The occurrence of perforin mutations in FHL provides further evidence for the importance of perforin in the regulation of homeostasis within the human immune system.
So far, perforin mutations have been described in eight unrelated FHL patients (Stepp et al. 1999). In the present study, we sought to determine the frequency and type of mutations in the perforin gene by direct sequencing of DNA from a large set of well-defined families affected by FHL.
Patients and Methods
Patients
A total of 34 unrelated families, each with one or several children affected with FHL—originating from Sweden (n=3), Sweden/Portugal (n=1), Norway (n=1), Finland (n=1), Iceland (n=1), Germany (n=4), Romania (n=1), Yugoslavia (n=1), Turkey (n=20), and Egypt (n=1)—were investigated for the presence of mutations in the perforin gene. In families with more than one affected child, only one sibling was investigated. The patients included either had a known family history of FHL (at least two affected children in the family) (n=17) or fulfilled the diagnostic criteria for hemophagocytic lymphohistiocytosis developed by the Histiocyte Society (Henter et al. 1991a) and, in addition, either had undergone BMT (n=9) or had died of their disease prior to BMT (n=8). None of these families or individuals was included in the previous report by Stepp et al. (1999). The study was approved by the ethics committee at the Karolinska Hospital.
Mutation Analysis
Genomic DNA was isolated from peripheral blood leukocytes or cultured fibroblasts by standard procedures. The obtained sequences was compared to the published sequence for perforin (Lichtenheld et al. 1988). Note that, for codon 332 and 426, we found a sequence that differed from this one in all individuals analyzed, although it corresponds to the sequence for perforin reported by Shinkai et al. (1989). Primers for amplification of exon 2 and 3 and cycle sequencing are listed in table 1. For dye-labeled primer cycle sequencing, the forward primers contained the −21M13 complement, and the reverse primers contained the M13 complement. For all primer pairs, the annealing temperature were set to 58°C. Genomic DNA (100–200 ng) was amplified in 5 μl 10 × PCR buffer (Perkin Elmer), 4 μl MgCl2 (25 mM), 4 μl dNTP (1.25 mM), 0.5 μl each of forward and reverse primer (10 μM), and 0.9 U of AmpliTaq Gold (Perkin Elmer). PCR amplification conditions were: 15 min at 96°C, and then 30 s at 96°C, 20 s at annealing temperature, and 30 s at 72°C for 40 cycles and 2 min at 72°C. The PCR products were analyzed on agarose gels and then were cycle sequenced with the ABI PRISM Big Dye Primer Cycle Sequencing Ready Reaction Kit (Applied Biosystems). For the cycle sequencing the following conditions were used: 10 s at 96°C, 5 s at 55°C, and 1 min at 70°C (20 cycles) and then, 10 s at 96°C and 1 min 70°C (20 cycles). All PCRs were performed on an MJ Research PTC-200 DNA Engine. The sequencing reactions were analyzed on an ABI Genetic Analyzer 310 (Applied Biosystems).
Table 1.
Perforin Primer Sequences Used for PCR Amplification and Direct Sequencing
| Region and Primer Sequencea | Annealing Site |
| Exon 2: | |
| F 5′-tgtgccctgataatctgtg-3′ | 3171–3190 |
| F 5′-TCCCAGTGGACACACAAAG-3′ | 3449–3667 |
| R 5′-gcagcctccaagtttga-3′ | 3901–3917 |
| Exon 3: | |
| F 5′-tcctagttctgcccacttac-3′ | 4886–4905 |
| F 5′-tctcttctcgcAGTTTCCAT-3′ | 4982–5001 |
| R 5′-GGGTTCCAGGGTGTAGTCCA-3′ | 5471–5480 |
| F 5′-ACTGCCCTGCGCACCTG-3′ | 5164–5180 |
| F 5′-GCCTCCTTCCACCAAACCTA-3′ | 5323–5340 |
| R 5′-GGTTGTTATTGTCCCACACG-3′ | 5811–5828 |
| F 5′-GTCACCACCCAGGACTGCTG-3′ | 5659–5678 |
| F 5′-ATCCAAGCATGGGGCCTG-3′ | 5719–5736 |
| F 5′-ATGACCTCCTTGGCACCTGT-3′ | 5927–5946 |
| R 5′-ggctcccactgtgaga-3′ | 6169–6184 |
Primers located in introns are lowercase type, whereas exonic primers are capitalized.
Linkage Analysis
In selected consanguineous families, polymorphic microsatellite markers mapped to 9q21.3-22, (D9S922, D9S1867, D9S152, D9S1812, D9S257, D9S1680, and D9S283) and to the perforin gene on chromosome 10q21 (D10S537) were analyzed for homozygosity. PCRs for the markers were carried out as single reactions in 96-well plates, using standard protocols. All the forward primers were fluorescently labeled. PCR reactions for each marker were performed separately, with the products being pooled prior to gel electrophoresis on an ABI 377 (96 wells) (PE Applied Biosystems). Resulting genotype data were analyzed using GENESCAN 2.1 and GENOTYPER 2.0 software from ABI (PE Applied Biosystems).
Results
Strategy Outline for Mutation Analysis
The translated exon 2 and exon 3 of the perforin gene, but not the untranslated exon 1, were amplified and sequenced in a total of 34 families. Primarily, one or both parents were sequenced in each family. If a mutation was detected, this was later confirmed in the affected child. In some families, DNA from the parents was unavailable or insufficient, and, instead, DNA from an affected individual was sequenced. In three families, the amount of DNA was restricted and insufficient, probably because of fragmentation of the DNA, and, therefore, we were able to sequence only ∼95% of the perforin gene in these cases.
Perforin Mutations
Perforin mutations were found in seven families (tables 2 and 3). Among these families, four children originating from Turkey were homozygous for a truncating mutation in exon 3: trp374→stop. This mutation has been described elsewhere in two independent cases (Stepp et al. 1999). One child from Turkey was a compound heterozygote with a maternal truncating mutation, tyr219→stop, in exon 3, and a paternal missense mutation, val50→met, in exon 2. One Swedish patient from a consanguineous family was homozygous for a missense mutation, ile224→asp, in exon 3. One 3-bp deletion in exon 3, resulting in loss of the lys285 residue, was found in one child from Turkey. None of these four latter mutations has been described elsewhere.
Table 2.
Mutations in the Perforin Gene
| MutationType andFamily | Protein-SequenceAlterationa | Nucleotide-SequenceAlteration | Exon |
| Nonsense: | |||
| 1 | Trp374stop | 1122 G→A | 3 |
| 7 | Trp374stop | 1122 G→A | 3 |
| 9b | Tyr219stop | 657 C→A | 3 |
| 11 | Trp374stop | 1122 G→A | 3 |
| 17 | Trp374stop | 1122 G→A | 3 |
| Missense: | |||
| 3 | Ile224asp | 671 T→A | 3 |
| 9b | Val50met | 148 G→A | 2 |
| Deletion: | |||
| 33 | Δlys285c | 853–855 del AAG | 3 |
The Trp374stop mutation was initially reported by Stepp et al. (1999).
Compound heterozygote.
Δ = deletion.
Table 3.
Clinical Findings and Mutation Designation in Families with FHL and Perforin Mutations[Note]
| Family andMutation | Countryof Originof Parents | Consanguinity/Family Historya | Age atDiagnosis(Mo)/Sexb | Fever | Splenomegaly | Cytopeniac | H/Hd | Hemophagocytosise | Treatmentbefore BMT(VP/Cs/CsA/HLH-94)f | BMT Status/Outcome |
| Trp374stop: | ||||||||||
| 1 | Turkey | +/+ | 2/M | + | + | + | + | + | VP/Cs/CsA | No BMT, dead |
| 7 | Turkey | −/+ | 1/F | + | + | + | NDg | + | VP/Cs | No BMT, dead |
| 11 | Turkey | +/+ | 39/M | + | + | + | + | + | HLH-94 | BMT, alive and well |
| 17 | Turkey | +/+ | 10/F | + | + | + | + | + | No treatment | No BMT, dead |
| Ile224asp: | ||||||||||
| 3 | Sweden | +/− | 58/F | + | + | + | + | + | VP/Cs/CsA | BMT, alive, mild retardation |
| Tyr219stop/Val50met: | ||||||||||
| 9 | Turkey | −/− | 4/M | + | + | + | + | + | HLH-94 | No BMT, dead |
| K285: | ||||||||||
| 33 | Turkey | −/− | 3/F | + | + | + | + | + | No treatment | No BMT, dead |
Note.— + = finding is present; − = finding is not present.
Family history = at least two affected children in the family.
M = male; F = female.
Affecting ⩾2 of 3 lineages in peripheral blood (hemoglobin <90 g/L; platelets <100 × 109/L; neutrophils <1.0 × 109/L).
H/H = hypertriglyceridemia and/or hypofibrinogenemia (fasting triglycerides ⩾2.0 mmol/L or ⩾3 SD of the normal value for age, fibrinogen ⩽1.5 g/L or ⩽3 SD).
Hemophagocytosis of bone marrow, spleen, or lymph nodes, with no evidence of malignancy.
VP = etoposide (VP-16); Cs = Corticosteroids; CsA = Cyclosporin A; HLH-94 = the treatment protocol HLH-94 of the Histiocyte Society (Henter et al. 1997).
ND = no data available.
The trp374→stop mutation, which also has been described by Stepp et al. (1999) in two unrelated families of Turkish origin, is the most frequent mutation in the perforin gene reported so far. The location of the mutation is within a cysteine-rich epidermal growth factor (EGF) precursor-type domain, between amino acid residues 356 and 386, that is conserved in human, mouse, and rat (Lowin et al. 1995). This domain exhibits a high degree of homology between perforin and the complement factors (Lichtenheld et al. 1988; Podack 1992).
The ile224→asp mutation found in a consanguineous Swedish family was traced in heterozygous form in three generations of unaffected relatives. This mutation was not identified in either heterozygous or homozygous form in any other sequenced allele, indicating that this is a private mutation. The mutation is located within the transmembrane region, which is important for pore formation (Liu et al. 1995). In addition, this isoleucine residue is conserved in human, mouse, and rat (Lowin et al. 1995). The second missense mutation affects val50→met, which also is conserved in human, mouse, and rat (Lowin et al. 1995).
The homozygous 3-bp in-frame deletion of a lysine residue at codon 285 was found in a Turkish child, who died before treatment was initiated (table 3). Unfortunately, blood from the parents was not available for analysis. Neither the missense mutations nor the in-frame deletion were found in ⩾40 independent internal control chromosomes from individuals not displaying the FHL phenotype, and, thus, they were inferred to be pathogenic.
Polymorphisms in the Perforin Gene
One rare polymorphism was detected at the protein level in the perforin gene (table 4). arg123his was considered to be a polymorphism, since this amino acid change was present in one healthy carrier of FHL, who transmitted a mutation (trp374stop) from the other allele to his child (family 17). Since the carrier did not show any signs of the disease, arg123his does not cause FHL. Moreover, the amino acid at codon 123 is not conserved among human, mouse, and rat. Three additional polymorphic sites were identified that did not have any effect on the coding sequence (table 4).
Table 4.
Polymorphisms in the Perforin Gene
| Codon | SequenceAlteration | Effect on Protein | AlleleFrequency(%) |
| 123 | CGT→CAT | Arginine→histidine | 92.5/7.5 |
| 173 | ACG→ACA | Threonine→threonine | 84/16 |
| 274 | GCC→GCT | Alanine→alanine | 88/12 |
| 300 | CAC→CAT | Histidine→histidine | 40/60 |
Linkage Analysis
To determine the relative frequency, in other genes, of mutations that cause FHL, we used linkage analysis in consanguineous cases of FHL. Of the 27 families in which no perforin mutation was detected, 13 were consanguineous families with material available for linkage analysis. Polymorphic DNA markers were studied within the perforin gene region—as well as within the chromosome 9q21.3-22 region linked to FHL in some families (Ohadi et al. 1999).
Of 12 families who were informative for the marker within the perforin gene on chromosome 10, only two were homozygous (data not shown). These two cases were heterozygous for markers within the 9q21.3-22 region (data not shown) and might carry mutations in the perforin gene that escaped detection by DNA sequencing of exon 2 and 3, and, therefore, ∼15%-20% of the consanguineous families might also be linked to the perforin gene. Taking into account the 20% with detected perforin mutations, no more than 35%–40% of the FHL cases would be caused by mutations in this gene.
Only one out of nine informative cases was homozygous for markers at the chromosome 9 locus (data not shown). This case was heterozygous for the marker within the perforin gene (data not shown). This single case may carry a mutation in the FHL 1 locus on chromosome 9. On the basis of this analysis, >50% of the FHL cases would be caused by mutations neither in the perforin gene or the unidentified gene at the chromosome 9 locus.
Discussion
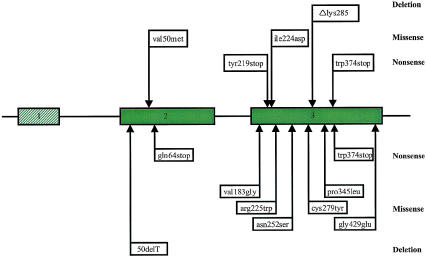
The present work describes the relative frequency and the mutational spectrum in the perforin gene in families afflicted with FHL. We have located homozygous, pathogenic perforin alleles in six families and one compound heterozygote in one family—identifying five different mutations altogether, in a total of seven families, that are predicted to affect the coding sequence and function of the protein (fig. 1) (table 2 and 3). One mutation, trp374→stop, seems to be particularly common. By comparison of the sequences of the four families in our study who originate from Turkey and carry this mutation, we tried to identify single-nucleotide polymorphisms that would indicate a common founder for this mutation, but no such polymorphic sites were found that could elucidate if the mutation is due to a founder effect.
Figure 1.
Schematic representation of the perforin gene, showing the localization of mutations in FHL. The three exons of the perforin gene are represented by boxes, introns by lines. The coding region of the perforin gene comprises exon 2 and 3 and is indicated by coloured boxes. In the upper part of the figure, mutations identified in the present study are indicated. The trp374→stop mutation, initially reported by Stepp et al. (1999), was identified in four families (see table 2). The lower part represents previously described mutations in the perforin gene (Stepp et al. 1999). The mutations have been grouped with regard to whether they are nonsense mutations, missense mutations, or deletions.
The ile224→asp mutation was identified through four generations in a consanguineous Swedish family. The location of the mutation is within a region, with a high degree of conservation among human, mouse, and rat, that encodes one of the transmembrane domains of perforin, and the mutation, therefore, is predicted to give rise to a nonfunctional protein (Liu et al. 1995). Stepp et al. (1999) described a mutation, arg225→trp, in the same region in a compound heterozygote. Cells from this patient showed greatly reduced cytolytic activity, and immunostaining revealed nearly complete absence of perforin (Stepp et al. 1999). The in-frame deletion, Δlys285, was not detected in any of the ⩾40 analyzed control chromosomes from individuals not displaying the FHL phenotype, suggesting that this is not a polymorphism. However, the mutation corresponds to the published mouse and rat sequences. Hence, the possibility can not be excluded entirely that this is a rare polymorphism. In addition, a total of four polymorphisms or sequence variants were identified.
Our mutational analysis detected mutations in the perforin gene in ∼20% of the 34 FHL families. Interestingly, all perforin mutations, except the ile224→asp mutation, were detected in patients with parents who originated from Turkey. Of the subset of families originating from Turkey, perforin mutations were, in fact, detected in ∼30% (6/20). This could be due to the higher prevalence of consanguineous marriages in this group, but it can be speculated that perforin mutations in FHL might be more common in certain geographical areas or ethnic groups.
We found a limited spectrum of missense mutations, compared with those studied elsewhere (Stepp et al. 1999). It can be reasoned that a loss-of-function is supported if it involves an amino acid conserved among human, mouse, and rat. However, not all of the previously described missense mutations involved conserved amino acids, although patients harboring these mutations have been shown to display substantially reduced expression of perforin protein (Stepp et al. 1999). Further assessment of the expression and function of the missense mutations is required to understand how these mutations impair perforin activity.
Heterogeneity has been implied in FHL, and it is reasonable to assume that other genetic defects, in addition to perforin mutations, are involved in this disease (Dufourcq-Lagelouse et al. 1999a; Ohadi et al. 1999; Graham et al. 2000). Our data suggest that perforin mutations might cause 20%–40% of the cases of FHL, if both detected mutations (20%) and subjects with no mutation who were homozygous for the perforin polymorphism (15%–20%) are added. Similarly, the chromosome 9 gene can explain only ∼10% of cases of FHL, whereas the remaining cases, >50%, would be caused by mutations in other, undefined genes.
We have previously demonstrated that immune cells obtained from FHL patients remain susceptible to apoptosis induced in vitro by Fas ligation or the chemotherapeutic drug etoposide (Fadeel et al. 1999). Hence, these stimuli triggered normal activation of intracellular caspases and externalization of the phagocytic ligand, phosphatidylserine, suggesting that the Fas receptor and the intracellular apoptotic signaling machinery remain intact in these patients. However, spontaneous activation of caspase-3-like enzymes was markedly suppressed or absent in a number of patients with FHL prior to the initiation of cytotoxic therapy (Fadeel et al. 1999). On the basis of these observations, we reasoned that FHL might be associated with the lack of an endogenous apoptosis trigger, such as Fas ligand or components of the perforin/granzyme B system, rather than apoptosis resistance per se. Indeed, perforin-gene mutations have now been described in a subset of FHL patients (Stepp et al. 1999; the present study). On the other hand, several lines of evidence, some of which admittedly are circumstantial, suggest that Fas ligand is a less likely candidate gene for FHL. The human Fas ligand has been mapped to chromosome 1q23 (Takahashi et al. 1994) and, therefore, does not correspond to either of the known FHL loci. Moreover, although serum levels of soluble Fas ligand are enhanced in FHL patients, values are normalized upon successful chemotherapeutic treatment (Hasegawa et al. 1998; Fadeel, unpublished observations), thus indicating that these changes are a secondary event, perhaps reflective of excessive immune activation. Finally, using CD4+ and CD8+ cytotoxic T lymphocytes derived from autoimmune lymphoproliferative syndrome patients, Yasukawa et al. (2000) recently have provided evidence that granule exocytosis, and not the Fas/Fas ligand system, is the main pathway of cellular cytotoxicity in humans. At present, we therefore favor the involvement of mutations in genes encoding granzymes A and B—or other, as-yet-unidentified components of lytic granules—in the pathogenesis of FHL in those individuals who do not display perforin-gene mutations.
We were unable to establish a correlation between the different mutations in the perforin gene and the clinical manifestations of FHL. For instance, the age at diagnosis of patients with the trp374→stop mutation varied between one month and three years. This suggests that other factors are also of importance with regard to onset of the disease; such factors may be infections and immunization. Moreover, severe encephalopathy may or may not develop in a patient with a perforin mutation. It can therefore be speculated that initiating infections that affect the CNS may be more devastating to the brain than infections with other organ affection. We were also unable, in this limited patient material, to establish a correlation between the occurrence or absence of mutations in the perforin gene and clinical manifestations. Similarly, in a recent study, neither clinical nor pathological manifestations of FHL differed between patients who displayed linkage to 10q21-22 compared to those who did not (Dufourcq-Lagelouse et al. 1999a). Again, these observations support the notion that other putative genetic defects in non-10q21-22 linked FHL target components of the same (perforin-dependent) signaling pathway. However, subtle differences may exist—for instance, in the pattern of defective cellular cytotoxicity—that would allow for the differentiation of the different subtypes of FHL. Further studies clearly are needed in which a detailed assessment of phenotypic features is performed and is compared to the occurrence and type of perforin mutations. In addition, attention should be focused on the distinction between FHL and other, related disorders of macrophage activation and immune deregulation, such as Chediak-Higashi syndrome, X-linked lymphoproliferative disease, and Griscelli syndrome, for which the causative genetic defects have been characterized (Dufourcq-Lagelouse et al. 1999b).
In conclusion, we have confirmed the occurrence of mutations in the perforin gene in a large subset of FHL patients. We found five different mutations in seven families, four of which were not previously described. However, the detailed mechanisms by which mutations in the perforin gene result in the FHL phenotype still need to be clarified to improve our understanding of the pathogenesis of this fatal disease, and detection of additional pathogenic gene alterations is warranted to further facilitate carrier and prenatal diagnosis.
Acknowledgments
We thank all patients and their families participating in the study for their cooperation and Ulla Grandell and Teresia Pettersson for technical assistance. Presented in part at the Annual Meeting of the Histiocyte Society in Amsterdam, October 2000. This study was supported by the Children's Cancer Foundation of Sweden, the Swedish Medical Research Council, the Swedish Cancer Society, the Märta and Gunnar V. Philipson Foundation, the Ronald McDonald Foundation, and the Histiocytosis Association of America.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genbank, http://www.ncbi.nlm.nih.gov/Genbank (accession number for perforin [NM-005041] and [M28393])
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/genemap98 (for Genemap'98)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for familial hemophagocytic lymphohistiocytosis [MIM 267700], familial hemophagocytic lymphohistiocytosis 1 [MIM 603552], familial hemophagocytic lymphohistiocytosis 2 [MIM 603553], Chediak-Higashi syndrome [MIM 214500], X-linked lymphoproliferative disease [MIM 308240], and Griscelli syndrome [MIM 214450])
References
- Aricò M, Nespoli L, Maccario R, Montagna D, Bonetti F, Caselli D, Burgio GR (1988) Natural cytotoxicity impairment in familial haemophagocytic lymphohistiocytosis. Arch Dis Child 63:292–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon AJ, Bleackley RC (1998) Proteases and cell-mediated cytotoxicity. Crit Rev Immunol 18:255–273 [DOI] [PubMed] [Google Scholar]
- Darmon AJ, Nicholson DW, Bleackley RC (1995) Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature 377:446–448 [DOI] [PubMed] [Google Scholar]
- Dufourcq-Lagelouse R, Jabado N, Le Deist F, Stephan JL, Souillet G, Bruin M, Vilmer E, Schneider M, Janka G, Fischer A, de Saint Basile G (1999a) Linkage of familial hemophagocytic lymphohistiocytosis to 10q21-22 and evidence for heterogeneity. Am J Hum Genet 64:172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourcq-Lagelouse R, Pastural E, Barrat FJ, Feldmann J, Le Deist F, Fischer A, de Saint Basile G (1999b) Genetic basis of hemophagocytic lymphohistiocytosis syndrome. Int J Mol Med 4:127–133 [DOI] [PubMed] [Google Scholar]
- Durken M, Horstmann M, Bieling P, Erttmann R, Kabisch H, Loliger C, Schneider EM, Hellwege HH, Kruger W, Kroger N, Zander AR, Janka GE (1999) Improved outcome in haemophagocytic lymphohistiocytosis after bone marrow transplantation from related and unrelated donors: a single-centre experience of 12 patients. Br J Haematol 106:1052–1058 [DOI] [PubMed] [Google Scholar]
- Egeler RM, Shapiro R, Loechelt B, Filipovich A (1996) Characteristic immune abnormalities in hemophagocytic lymphohistiocytosis. J Pediatr Hematol Oncol 18:340–345 [DOI] [PubMed] [Google Scholar]
- Eife R, Janka GE, Belohradsky BH, Holtmann H (1989) Natural killer cell function and interferon production in familial hemophagocytic lymphohistiocytosis. Pediatr Hematol Oncol 6:265–272 [DOI] [PubMed] [Google Scholar]
- Fadeel B, Orrenius S, Henter J-I (1999) Induction of apoptosis and caspase activation in cells obtained from familial haemophagocytic lymphohistiocytosis patients. Br J Haematol 106:406–415 [DOI] [PubMed] [Google Scholar]
- Fujiwara F, Hibi S, Imashuku S (1993) Hypercytokinemia in hemophagocytic syndrome. Am J Pediatr Hematol Oncol 15:92–98 [DOI] [PubMed] [Google Scholar]
- Graham GE, Graham LM, Bridge PJ, Maclaren LD, Wolff JEA, Coppes MJ, Egeler RM (2000) Further evidence for genetic heterogeneity in familial hemophagocytic lymphohistiocytosis (FHLH). Pediatr Res 48:227–232 [DOI] [PubMed] [Google Scholar]
- Hasegawa D, Kojima S, Tatsumi E, Hayakawa A, Kosaka Y, Nakamura H, Sako M, Osugi Y, Nagata S, Sano K (1998) Elevation of the serum Fas ligand in patients with hemophagocytic syndrome and Diamond-Blackfan anemia. Blood 91:2793–2799 [PubMed] [Google Scholar]
- Henter J-I, Arico M, Egeler RM, Elinder G, Favara BE, Filipovich AH, Gadner H, Imashuku S, Janka-Schaub G, Komp D, Ladisch S, Webb D (1997) HLH-94: a treatment protocol for hemophagocytic lymphohistiocytosis. HLH study Group of the Histiocyte Society. Med Pediatr Oncol 28:342–347 [DOI] [PubMed] [Google Scholar]
- Henter J-I, Arico M, Elinder G, Imashuku S, Janka G (1998) Familial hemophagocytic lymphohistiocytosis: primary hemophagocytic lymphohistiocytosis. Hematol Oncol Clin North Am 12:417–433 [DOI] [PubMed] [Google Scholar]
- Henter J-I, Elinder G, Öst Å, FHL Study Group of the Histiocyte Society (1991a) Diagnostic guidelines for hemophagocytic lymphohistiocytosis. Semin Oncol 18:29–33 [PubMed] [Google Scholar]
- Henter J-I, Elinder G, Söder O, Hansson M, Andersson B, Andersson U (1991b) Hypercytokinemia in familial hemophagocytic lymphohistiocytosis. Blood 78:2918–2922 [PubMed] [Google Scholar]
- Imashuku S, Hibi S, Fujiwara F, Todo S (1996) Hyper-interleukin (IL)-6-naemia in haemophagocytic lymphohistiocytosis. Br J Haematol 93:803–807 [DOI] [PubMed] [Google Scholar]
- Jabado N, de Graeff-Meeder ER, Cavazzana-Calvo M, Haddad E, Le Deist F, Benkerrou M, Dufourcq R, Caillat S, Blanche S, Fischer A (1997) Treatment of familial hemophagocytic lymphohistiocytosis with bone marrow transplantation from HLA genetically nonidentical donors. Blood 90:4743–4748 [PubMed] [Google Scholar]
- Janka GE (1983) Familial hemophagocytic lymphohistiocytosis. Eur J Pediatr 140:221–230 [DOI] [PubMed] [Google Scholar]
- Kägi D, Odermatt B, Mak TW (1999) Homeostatic regulation of CD8+ T cells by perforin. Eur J Immunol 29:3262–3272 [DOI] [PubMed] [Google Scholar]
- Lichtenheld MG, Olsen KJ, Lu P, Lowrey DM, Hameed A, Hengartner H, Podack ER (1988) Structure and function of human perforin. Nature 335:448–451 [DOI] [PubMed] [Google Scholar]
- Lichtenheld MG, Podack ER (1989) Structure of the human perforin gene: a simple gene organization with interesting potential regulatory sequences. J Immunol 143:4267–4274 [PubMed] [Google Scholar]
- Liu CC, Walsh CM, Young JD (1995) Perforin: structure and function. Immunol Today 16:194–201 [DOI] [PubMed] [Google Scholar]
- Lowin B, Peitsch MC, Tschopp J (1995) Perforin and granzymes: crucial effector molecules in cytolytic T lymphocyte and natural killer cell-mediated cytotoxicity. Curr Top Microbiol Immunol 198:1–24 [DOI] [PubMed] [Google Scholar]
- Matloubian M, Suresh M, Glass A, Galvan M, Chow K, Whitmire JK, Walsh CM, Clark WR, Ahmed R (1999) A role for perforin in downregulating T-cell responses during chronic viral infection. J Virol 73:2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohadi M, Lalloz MR, Sham P, Zhao J, Dearlove AM, Shiach C, Kinsey S, Rhodes M, Layton DM (1999) Localization of a gene for familial hemophagocytic lymphohistiocytosis at chromosome 9q21.3-22 by homozygosity mapping. Am J Hum Genet 64:165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez N, Virelizier JL, Arenzana-Seisdedos F, Fischer A, Griscelli C (1984) Impaired natural killer activity in lymphohistiocytosis syndrome. J Pediatr 104:569–573 [DOI] [PubMed] [Google Scholar]
- Podack ER (1992) Perforin: structure, function, and regulation. Curr Top Microbiol Immunol 178:175–184 [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Yoshida MC, Maeda K, Kobata T, Maruyama K, Yodoi J, Yagita H, Okumura K (1989) Molecular cloning and chromosomal assignment of a human perforin (PFP) gene. Immunogenetics 30:452–457 [DOI] [PubMed] [Google Scholar]
- Stepp SE, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, Mathew PA, Henter J-I, Bennett M, Fischer A, de Saint Basile G, Kumar V (1999) Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science 286:1957–1959 [DOI] [PubMed] [Google Scholar]
- Sullivan KE, Delaat CA, Douglas SD, Filipovich AH (1998) Defective natural killer cell function in patients with hemophagocytic lymphohistiocytosis and in first degree relatives. Pediatr Res 44:465–468 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Tanaka M, Inazawa J, Abe T, Suda T, Nagata S (1994) Human Fas ligand: gene structure, chromosomal location and species specificity. Int Immunol 6:1567–1574 [DOI] [PubMed] [Google Scholar]
- Tschopp J, Nabholz M (1990) Perforin-mediated target cell lysis by cytolytic T lymphocytes. Annu Rev Immunol 8:279–302 [DOI] [PubMed] [Google Scholar]
- Yasukawa M, Ohminami H, Arai J, Kasahara Y, Ishida Y, Fujita S (2000) Granule exocytosis, and not the fas/fas ligand system, is the main pathway of cytotoxicity mediated by alloantigen-specific CD4(+) as well as CD8(+) cytotoxic T lymphocytes in humans. Blood 95:2352–2355 [PubMed] [Google Scholar]