Abstract
Familial dysautonomia (FD; also known as “Riley-Day syndrome”), an Ashkenazi Jewish disorder, is the best known and most frequent of a group of congenital sensory neuropathies and is characterized by widespread sensory and variable autonomic dysfunction. Previously, we had mapped the FD gene, DYS, to a 0.5-cM region on chromosome 9q31 and had shown that the ethnic bias is due to a founder effect, with >99.5% of disease alleles sharing a common ancestral haplotype. To investigate the molecular basis of FD, we sequenced the minimal candidate region and cloned and characterized its five genes. One of these, IKBKAP, harbors two mutations that can cause FD. The major haplotype mutation is located in the donor splice site of intron 20. This mutation can result in skipping of exon 20 in the mRNA of patients with FD, although they continue to express varying levels of wild-type message in a tissue-specific manner. RNA isolated from lymphoblasts of patients is primarily wild-type, whereas only the deleted message is seen in RNA isolated from brain. The mutation associated with the minor haplotype in four patients is a missense (R696P) mutation in exon 19, which is predicted to disrupt a potential phosphorylation site. Our findings indicate that almost all cases of FD are caused by an unusual splice defect that displays tissue-specific expression; and they also provide the basis for rapid carrier screening in the Ashkenazi Jewish population.
Introduction
Familial dysautonomia (FD; also known as “Riley-Day syndrome” or as “hereditary sensory and autonomic neuropathy type III” [MIM 223900]) is an autosomal recessive disorder present in 1/3,600 live births in the Ashkenazi Jewish population. This debilitating disorder is due to the poor development, survival, and progressive degeneration of the sensory and autonomic nervous system (Axelrod et al. 1974). FD was first described in 1949, on the basis of five children who presented with defective lacrimation, excessive sweating, skin blotching, and hypertension (Riley et al. 1949). The following cardinal criteria have evolved for diagnosis of FD: absence of fungiform papillae on the tongue, absence of flare after injection of intradermal histamine, decreased or absent deep-tendon reflexes, absence of overflow emotional tears, and Ashkenazi Jewish descent (Axelrod 1984; Axelrod and Pearson 1984).
The loss of neuronal function in FD has many repercussions, with patients displaying gastrointestinal dysfunction, abnormal respiratory responses to hypoxic and hypercarbic states, scoliosis, gastroesophageal reflux, vomiting crises, lack of overflow tears, inappropriate sweating, and postural hypotension (Riley et al.1949; Axelrod et al.1974; Axelrod 1996). Despite recent advances in the management of FD, the disorder is inevitably fatal, with only 50% of patients reaching age 30 years. The clinical features of FD are due to a genetic defect that causes a striking, progressive depletion of unmyelinated sensory and autonomic neurons (Pearson and Pytel 1978a, 1978b; Pearson et al. 1978; Axelrod 1995). This neuronal deficiency begins during development, since extensive pathology is evident even in the youngest subjects. Fetal development and postnatal maintenance of dorsal-root ganglion (DRG) neurons is abnormal, significantly decreasing their numbers and resulting in DRG of grossly reduced size. Slow progressive degeneration is evidenced by continued neuronal depletion with increasing age. In the autonomic nervous system, superior cervical sympathetic ganglia are also reduced in size, because of a severe decrease in the neuronal population.
Previously, we had mapped the FD gene, DYS, to an 11-cM region of chromosome 9q31 (Blumenfeld et al. 1993), which was then narrowed, by haplotype analysis, to <0.5 cM, or 471 kb (Blumenfeld et al. 1999). There is a single major haplotype that accounts for >99.5% of all FD chromosomes in the Ashkenazi Jewish (AJ) population. The recent identification of several single-nucleotide polymorphisms (SNPs) in the candidate interval has allowed us to further reduce the candidate region, to 177 kb, by revealing a common core haplotype shared by the major and one previously described minor haplotype (Blumenfeld et al. 1999). Two minor haplotypes with apparently independent mutational origins have been found in four AJ patients with FD and in one patient of mixed AJ/non-AJ ancestry, all of whom are heterozygous for the major haplotype. The high frequency of the major mutation, combined with the presence of only two additional haplotypes, makes FD unique among the AJ diseases.
Material and Methods
Samples from Patients with FD
We collected blood samples from two major sources, the Dysautonomia Diagnostic and Treatment Center at New York University Medical Center and the Israeli Center for Familial Dysautonomia at Hadassah University Hospital, with approval from the institutional review boards at these institutions and at Massachusetts General Hospital and Harvard Medical School. Either F.B.A. or C.M. diagnosed all patients, using established criteria. We established Epstein-Barr virus–transformed lymphoblast lines, using standard conditions. Fibroblast cell lines were obtained from the Coriell Cell Repositories. RNA isolated from postmortem brain tissue of patients with FD was obtained from the Dysautonomia Diagnostic and Treatment Center at New York University. We prepared genomic DNA, total RNA, and mRNA, using commercial kits (Invitrogen and Molecular Research Center). Cytoplasmic protein was extracted from lymphoblasts, as described elsewhere (Krappmann et al. 2000).
Identification of IKBKAP, and Mutation Analysis
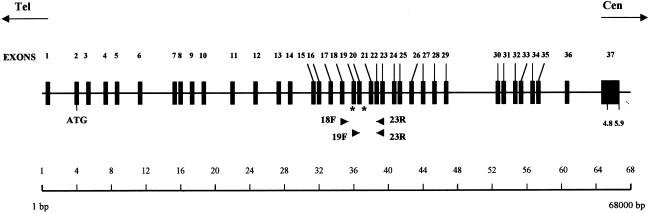
Exon-trapping experiments with cosmids from our physical map of the candidate region yielded five exons that were used to screen a human frontal-cortex cDNA library. Several cDNA clones were isolated and assembled into a novel transcript, encoding a 1,332–amino acid protein, that was later identified as IKBKAP (Cohen et al. 1998). The complete 5.9-kb cDNA sequence of IKBKAP has been submitted to GenBank, under accession number AF153419. For screening of mutations in patients with FD, total lymphoblast RNA was reverse transcribed, and overlapping sections of IKBKAP were amplified by PCR and were sequenced. Evaluation of the splicing defect was performed by use of the following primers: 18F (GCCAGTGTTTTTGCCTGAG), 19F (CGGATTGTCACTGTTGTGC), and 23R (GACTGCTCTCATAGCATCGC) (fig. 1).
Figure 1.
Genomic structure of IKBKAP, showing orientation and placement of the 37 exons within a 68-kb genomic region of chromosome 9q31. The primers used for analysis of the splice defect are denoted as “18F” (exon 18), “19F” (exon 19), and “23R” (exon 23). The asterisks (*) indicate the locations of the two mutations identified; the mutation associated with the major AJ haplotype is located at base pair 6 of intron 20, whereas the mutation association with the minor AJ haplotype is located at base pair 73 of exon 19. The “4.8” and “5.9” designations at exon 37 indicate the lengths (in kb) of the two IKBKAP messages that differ only in the length of their 3′ UTRs.
DNA Sequencing
Sequencing was performed either by use of the AmpliCycle sequencing kit (Applied Biosystems) or on an ABI 377 automated DNA sequencer using the BigDye terminator cycle sequencing kit (Applied Biosystems). The control sequence of the candidate region was obtained by construction of subclone libraries from BACs and sequencing using vector-specific primers. The FD sequence was generated by sequencing of cosmids from a patient homozygous for the major FD haplotype, by use of sequence-specific primers.
Expression Studies
Several human multiple-tissue northern blots (Clontech) were hybridized by use of the following radioactively labeled probes: IKBKAP exon 2, IKBKAP exons 18–20, IKBKAP exon 23, and a 400-bp fragment of the IKBKAP 3′ UTR immediately following the stop codon. Poly (A)+ RNA was isolated from lymphoblast lines form patients and control individuals, was northern blotted, and was hybridized by use of a probe representing the complete coding sequence of IKBKAP. Cytoplasmic protein extracted from lymphoblast cell lines was western blotted and was detected by use of ECL (Amersham) with an antibody raised against a peptide comprising the extreme carboxyl terminus (amino acids 1313–1332) of human IKAP, the protein encoded by IKBKAP (Krappmann et al. 2000).
Results
To identify DYS, we used exon trapping and cDNA selection to clone and characterize all of the genes in the 471-kb candidate region: EPB41L8 (authors' unpublished data) or EHM2 (Shimizu et al. 2000), C9ORF4 (Chadwick et al. 1999a), C9ORF5 (Chadwick et al. 2000), CTNNAL1 (Zhang et al. 1998), a novel gene with homology to the glycine-cleavage system–H proteins (CG-8) (authors' unpublished data), IKBKAP (Cohen et al. 1998), and ACTL7A and ACTL7B (Chadwick et al. 1999b). Since FD is a recessive disorder, the a priori expectation for the mutation was inactivation of one of these genes. Consequently, we screened each of them for mutations, by reverse transcriptase–PCR (RT-PCR) of lymphoblast RNA of patients and by direct sequencing of all coding regions. Although we identified many SNPs, there was no evidence for a homozygous inactivating mutation. We concluded, therefore, that the mutation would be found in noncoding sequence, and we generated the control genomic sequence of the entire 471-kb candidate region, using bacterial artificial chromosomes from our physical map. Direct sequence prediction using GENSCAN and comprehensive searches of the public databases, using BLAST, did not reveal any additional genes in the candidate region, beyond those found by cloning methods. However, SNPs identified during our sequence analysis enabled us to refine our haplotype analysis and to narrow the candidate interval to 177 kb shared by the major haplotype and the previously described minor haplotype 1 (Blumenfeld et al. 1999). This reduced interval contains five genes—CTNNAL1, CG-8, IKBKAP, ACTL7A, and ACTL7B—all previously screened by RT-PCR, without yielding any coding-sequence mutations. We therefore constructed a cosmid library from a patient homozygous for the major haplotype, assembled the minimal-coverage contig for the now-reduced candidate interval, and generated the sequence of the mutant chromosome.
Comparison of the FD and control sequences revealed 152 differences (when simple sequence–repeat markers are excluded), which include 26 variations in the length of dTn tracts, 1 VNTR, and 125 base-pair changes. Each of the 125 base-pair changes was tested in a panel of 50 individuals known, by segregation analysis in families with FD, to carry two non-FD chromosomes. Of the 125 changes tested, only 1 was unique to patients carrying the major FD haplotype. This T→C change is located at base pair 6 of intron 20 in the IKBKAP gene depicted in figure 1 and is shown in figure 2a. IKAP was originally identified as an IκB kinase (IKK) complex–associated protein that can bind both NF-κB–inducing kinase and IKKs, through separate domains, and assemble them into an active kinase complex (Cohen et al. 1998). Recent work, however, has shown that IKAP is not associated with IKKs and that it plays no specific role in cytokine-induced NF-κB signaling (Krappmann et al. 2000); rather, IKAP has been shown to be part of a novel multiprotein complex hypothesized to play a role in general transcriptional regulation.
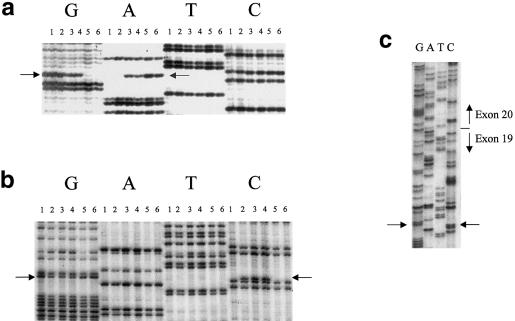
Figure 2.
Demonstration of mutations in IKBKAP. a, Antisense sequence of the T→C mutation (indicated by arrows adjacent to lanes G and A) at base pair 6 of intron 20, which is associated with the major FD haplotype. Lanes 1 and 2, patients with FD who are homozygous for the major haplotype (homozygous GG); lane 3, patient with FD who is heterozygous for the major haplotype and minor haplotype 2 (heterozygous GA); lane 4, patient with FD who is heterozygous for the major haplotype and minor haplotype 3 (heterozygous GA); lanes 5 and 6, control individuals (homozygous AA). b, Heterozygosity for the G→C mutation (indicated by arrows adjacent to lanes G and C) at base pair 73 of exon 19. Lane 1, patient with FD who is homozygous for the major haplotype (homozygous GG); lanes 2–4, patients with FD who are heterozygous for the major haplotype and minor haplotype 2 (heterozygous GC); lane 5, patient with FD who is heterozygous for the major haplotype and minor haplotype 3 (homozygous GG); lane 6, control individual (homozygous GG). c, Sequence of cDNA generated from the RT-PCR of a patient with FD who is heterozygous for the major and minor 2 haplotypes. The arrow points to the heterozygous G→C mutation in exon 19. The boundary of exons 19 and 20 is also indicated, illustrating that this patient expresses wild-type message that includes exon 20, despite the presence of the major mutation on one allele.
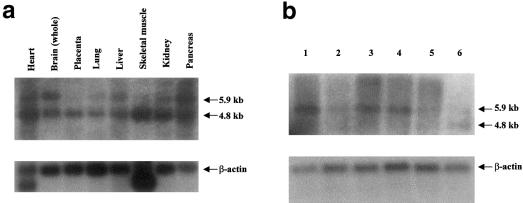
The IKBKAP gene contains 37 exons and encodes a 1,332-amino-acid protein. The full-length 5.9-kb cDNA (GenBank accession number AF153419) covers 68 kb of genomic sequence, with the start methionine encoded in exon 2. IKBKAP previously had been assigned to chromosome 9q34 (GenBank accession number AF044195), but it clearly maps within the FD candidate region of 9q31. Northern blot analysis of IKBKAP revealed two mRNAs—one of 4.8 kb and one of 5.9 kb (fig. 3a and b). The 4.8-kb mRNA has been reported elsewhere (Cohen et al. 1998), whereas the second, 5.9-kb message differs only in the length of the 3′ UTR and is predicted to encode an identical 150-kD protein. As seen in figure 3b, the putative FD mutation does not eliminate expression of the IKBKAP mRNA in lymphoblasts of patient with FD.
Figure 3.
Northern blot analysis of IKBKAP. a, Human multiple-tissue northern blot hybridized with IKBKAP exon 2, showing the presence of two messages, of 4.8 and 5.9 kb (northern blots hybridized with other IKBKAP probes yielded similar patterns). b, Northern blot generated by use of mRNA isolated from lymphoblast cell lines: lanes 1, 2, and 5, patients with FD who are homozygous for the major haplotype; lane 3, individual carrying two definitively non-FD chromosomes; lane 4, patient with FD who is heterozygous for the major haplotype and minor haplotype 2; lane 6, control brain RNA (Clontech). The level of expression of IKBKAP mRNA relative to β-actin mRNA is quite variable in lymphoblasts. No consistent increase or decrease in mRNA levels was observed between patients with FD who are homozygous for the major haplotype, patients with FD who are heterozygous for the major haplotype and minor haplotype 2, and control individuals.
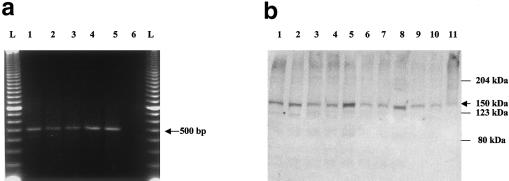
A base-pair change at position 6 of the splice donor site might be expected to result in skipping of exon 20 (74 bp), causing a frameshift and therefore producing a truncated protein; however, initial inspection of our RT-PCR experiments in lymphoblast RNA from patients with FD, using primers located in exons 18 and 23 (fig. 1), showed a normal-length, 500-bp fragment that contained exon 20 (fig. 4a), indicating that lymphoblasts from patients with FD express normal IKBKAP message. The western blot shown in figure 4b demonstrates that full-length IKAP protein is expressed in these lymphoblasts from patients with FD; however, since the antibody used was directed against the carboxyl-terminus of IKAP, it would not be expected to detect any truncated protein, should the latter be present. The presence of apparently normal IKAP in cells of patients with FD is at odds with the expectation of an inactivating mutation in this recessive disease.
Figure 4.
RT-PCR analysis of the exon 20 region of IKBKAP, showing expression of the wild-type message and protein in patients. a, Results of use of primers 18F (exon 18) and 23R (exon 23). Lanes 1 and 2, patients with FD who are homozygous for the major haplotype; lane 3, patient with FD who is heterozygous for the major haplotype and minor haplotype 2; lanes 4 and 5, control individuals; lane 6, water control. b, Results of western blot generated by use of cytoplasmic protein isolated from lymphoblast cell lines of patients with FD and detected with a carboxyl-terminal antibody. Lane 1, patient with FD who is heterozygous for the major and minor haplotype 3; lanes 2, 4, 6, and 8, patients with FD who are homozygous for the major haplotype; lanes 3, 5, 7, and 9, controls individuals; lane 10, patient with FD who is heterozygous for the major and minor haplotype 2; lane 11, Hela cell–line sample.
In the absence of any evidence for a functional consequence of the intron 20 sequence change, the only alteration unique to FD chromosomes, we sought additional genetic evidence to support the view that it represents the FD mutation. The 658 FD chromosomes that carry the major haplotype all show the T→C change. In toto, we have now tested 887 chromosomes that, because of their failure to cause the disorder when present in individuals heterozygous for the major FD haplotype, are definitively non-FD. None of these non-FD chromosomes exhibits the T→C mutation, strongly indicating that none of them is a rare polymorphism. The frequency of the mutation in random AJ chromosomes was 14/1,012 (gene frequency 1/72; carrier frequency 1/36), close to the expected carrier frequency, 1/32 (Maayan et al. 1987).
In view of the strong genetic evidence that this mutation must be pathogenic, we postulated that its effect might be tissue specific. We therefore examined RNA extracted from the brain stem and temporal lobe of a postmortem brain sample from a patient with FD. In contrast to what was seen with FD lymphoblasts, RT-PCR of the FD brain-tissue RNA, using primers in exons 19 and 23 (expected to produce a normal, 393-bp product) revealed a 319-bp mutant product, indicating that there was virtually complete absence of exon 20 from the IKBKAP mRNA (fig. 5, lanes 10 and 11). Since additional FD autopsy material could not be obtained, we performed intensive analyses of additional lymphoblast and fibroblast cell lines, to determine whether exon skipping could be detected. Fibroblast lines from patients who were homozygous for the major FD haplotype yielded variable results. Some primary fibroblast lines displayed approximately equal expression of the mutant and wild-type mRNAs, whereas others displayed primarily wild-type mRNA. In addition, extensive examination of lymphoblast lines from additional patients indicated that the mutant message could sometimes be detected at low levels. An example of both the variability seen in FD fibroblasts and the presence of the mutant message in some FD lymphoblasts is shown in figure 5. In fact, close reexamination of figure 4a shows a trace of the mutant band in two (lanes 1 and 2) of the three FD samples. The absence of exon 20 in the FD brain RNA, as well as the preponderance of wild-type mRNA in fibroblasts and lymphoblasts, indicates that the major FD mutation acts by altering the splicing of IKBKAP, in a tissue-specific manner.
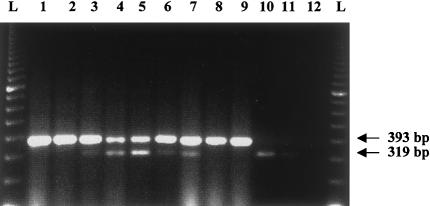
Figure 5.
RT-PCR analysis of the exon 20 region of IKBKAP, showing variable expression of the mutant message in patients with FD patients. The analysis was done by use of primers 19F (exon 19) and 23F (exon 23). Lanes 1 and 2, control fibroblasts; lanes 3–5, fibroblasts of patients with FD who are homozygous for the major mutation; lanes 6 and 7, lymphoblasts of patients with FD who are homozygous for the major mutation; lanes 8 and 9, control lymphoblasts; lane 10, brain stem of patient with FD; lane 11, temporal lobe of patient with FD (showing a faint 319-bp band and no 393-bp band); lane 12, water control. RT-PCR of control brain RNA (Clontech) showed only the 393-bp band (data not shown).
To identify the mutations associated with minor haplotypes 2 and 3 (Blumenfeld et al. 1999), we amplified each IKBKAP exon, including adjacent intron sequence, from genomic DNA. A single G→C change at base pair 2397 (base pair 73 of exon 19), which causes an arginine-to-proline missense mutation (R696P), was identified in all four patients with minor haplotype 2 (fig. 2b). This was subsequently confirmed by RT-PCR in lymphoblast RNA, as shown in figure 2c, for a region that crosses the exon 19/20 border. The PCR product, generated from a patient with FD who is a compound heterozygote with minor haplotype 2 and the major haplotype, clearly shows, on the basis of heterozygosity of the G→C point mutation in exon 19, that RNA is being expressed equally from both alleles. However, the RNA from the major-haplotype allele shows no evidence for skipping of exon 20, which would be expected to produce a mixture of exon 20/21 sequence, beginning at the end of exon 19. This confirms our previous observation that lymphoblasts with the major FD mutation produce a predominance of normal IKBKAP transcript.
The R696P mutation is absent from 500 non-FD chromosomes, and it has been seen in only 1/706 random AJ chromosomes, in an individual who also carries the minor haplotype. This mutation is predicted to disrupt a potential threonine phosphorylation site at residue 699, identified by Netphos 2.0 (Blom et al. 1999), suggesting that it may affect regulation of IKAP. Interestingly, the presence of this minor mutation is associated with a relatively mild disease phenotype, suggesting that a partially functional IKAP protein may be expressed from this allele. No mutation has been identified for minor haplotype 3, which represents the only non-AJ putative FD chromosome.
Discussion
The data presented here establish that mutation in the IKBKAP gene underlies FD. Our findings have direct implications for understanding this devastating disorder, for preventing it, and, potentially, for treating it. The IKAP protein produced from IKBKAP originally had been isolated as part of a large interleukin-1–inducible IKK complex and had been described as a regulator of kinases involved in proinflammatory cytokine signaling (Cohen et al. 1998). However, a recent report questioned this conclusion after demonstrating that cellular IKK complexes do not contain IKAP, by using various protein-protein interaction and functional assays; rather, IKAP appears to be a member of a novel complex containing additional unidentified proteins, of 100, 70, 45, and 39 kD (Krappmann et al. 2000).
IKAP is homologous to the Elp1 protein of Saccharomyces cerevisiae, which is encoded by the IKI3 locus and is required for sensitivity to pGKL killer toxin. The human and yeast proteins exhibit 29% identity and 46% similarity over their entire lengths. Yeast Elp1 protein is part of the RNA polymerase II–associated elongator complex, which also contains Elp2, a WD-40 repeat protein, and Elp3, a histone acetyltransferase (Otero et al. 1999). The human ELP3 gene encodes a 60-kD histone acetyltransferase that shows >75% identity with yeast Elp3 protein, but no 60-kD protein has been found in the human IKAP-containing protein complex. Consequently, it is considered unlikely that IKAP is a member of a functionally conserved mammalian elongator complex (Krappmann et al. 2000). Instead, it has been suggested that the protein may play a role in general gene-activation mechanisms, since overexpression of IKAP interferes with the activity of both NF-κB–dependent and independent reporter genes (Krappmann et al. 2000). Therefore, the FD phenotype may be caused by aberrant expression of genes crucial to the development of the sensory and autonomic nervous systems, secondary to the loss of a functional IKAP protein in specific tissues.
FD is unique among Ashkenazi Jewish disorders, in that one mutation accounts for >99.5% of the disease chromosomes. As in other autosomal recessive diseases with no phenotype in heterozygous carriers, one might have expected to find several different types of mutations producing complete inactivation of the DYS gene in the AJ population. The fact that the major FD mutation does not produce complete inactivation but, rather, allows variable tissue-specific expression of IKAP may explain this lack of mutational diversity. Mutations causing complete inactivation of IKAP in all tissues might cause a more severe or even lethal phenotype. Indeed, CG10535, the apparent Drosophila melanogaster homologue of IKBKAP, maps coincident with a larval recessive lethal locus (l(3)04629), supporting the essential nature of the protein (FlyBase). Thus, the array of mutations that can produce the FD phenotype may be limited if, to permit survival, they must also allow expression of functional or partially functional IKAP in some tissues. With the identification of IKBKAP as DYS, it will now be possible to test this inactivation hypothesis in a mammalian model system.
Despite the overwhelming predominance of a single mutation in patients with FD, the disease phenotype is remarkably variable both within and between families. The nature of the major FD mutation makes it tempting to consider that this phenotypic variability might relate to the frequency of exon 20 skipping in specific tissues and at specific developmental stages, which may be governed by variations in many factors involved in RNA splicing. Even a small amount of normal IKAP protein expressed in critical tissues might permit sufficient neuronal survival to alleviate the most severe phenotypes. This possibility is supported by the relatively mild phenotype associated with the presence of the R696P mutation, which is predicted to permit expression of an altered full-length IKAP protein that may retain some functional capacity. To date, this minor FD mutation has been seen in only four patients heterozygous for the major mutation. Consequently, it is uncertain whether homozygotes for the R696P mutation would display any phenotypic abnormality characteristic of FD. The single patient with minor haplotype 3 and mixed ancestry, whose mutation has yet to be found, is also a compound heterozygote with the major haplotype. The existence of minor haplotype 3 indicates that IKBKAP mutations will be found outside the AJ population, but, as is also the case with the R696P mutation, it is difficult to predict the severity of the phenotype that would result from homozygosity.
The identification of IKBKAP as the DYS gene clearly presents new possibilities for investigating the role that IKAP and associated proteins have in the development and maintenance of the sensory and autonomic nervous systems. What is of more-immediate practical importance, however, is that the discovery of the single-base mutation that characterizes >99.5% of FD chromosomes will permit efficient, inexpensive carrier testing in the AJ population, to guide reproductive choices and to reduce the incidence of FD. The nature of the major mutation also offers some hope for new approaches to the treatment of FD. Despite the presence of this mutation, lymphoblastoid cells from patients with FD are capable of producing full-length wild-type mRNA and normal IKAP protein, whereas, in neuronal tissue, exon 20 is skipped, presumably leading to a truncated product. Investigation of the mechanism that permits lymphoblasts to be relatively insensitive to the potential effect that the mutation has on splicing may suggest strategies to prevent skipping of exon 20 in other cell types. An effective treatment to prevent the progressive neuronal loss of FD may be one that is aimed at facilitating the production of wild-type mRNA from the mutant gene, rather than one aimed at exogenous administration of the missing IKAP protein via gene therapy.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- FlyBase, http://flybase.bio.indiana.edu/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank (for published IKBKAP sequence [accession number AF044195] and 5.9-kb IKBKAP cDNA sequence [accession number AF153419])
- GENSCAN Web Server, http://genes.mit.edu/GENSCAN.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FD (MIM 223900])
References
- Axelrod FB (1984) Familial dysautonomia and other congenital and sensory autonomic neuropathies. In: Blake IB (ed) Cell and molecular biology of neuronal development. Plenum Press, New York, pp 331–340 [Google Scholar]
- ——— (1995) Familial dysautonomia. In: Robertson D, Biaggioni I (eds) Disorders of the autonomic nervous system. Harwood Academic, Luxembourg, pp 217–231 [Google Scholar]
- ——— (1996) Familial dysautonomia. In: Robertson D, Low PA, Polinsky RJ (eds) Primer on the autonomic nervous system. Academic Press, San Diego, pp 242–249 [Google Scholar]
- Axelrod FB, Nachtigal R, Dancis J (1974) Familial dysautonomia: diagnosis, pathogenesis and management. Adv Pediatr 21:75–96 [PubMed] [Google Scholar]
- Axelrod FB, Pearson J (1984) Congenital sensory neuropathies. Diagnostic distinction from familial dysautonomia. Am J Dis Child 138:947–954 [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S (1999) Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362 [DOI] [PubMed] [Google Scholar]
- Blumenfeld A, Slaugenhaupt SA, Axelrod FB, Lucente DE, Maayan C, Liebert CB, Ozelius LJ, Trofatter JA, Haines JL, Breakefield XO, Gusella JF (1993) Localization of the gene for familial dysautonomia on chromosome 9 and definition of DNA markers for genetic diagnosis. Nat Genet 4:160–164 [DOI] [PubMed] [Google Scholar]
- Blumenfeld A, Slaugenhaupt SA, Liebert CB, Temper V, Maayan C, Gill S, Lucente DE, Idelson M, MacCormack K, Monahan MA, Mull J, Leyne M, Mendillo M, Schiripo T, Mishori E, Breakefield X, Axelrod FB, Gusella JF (1999) Precise genetic mapping and haplotype analysis of the familial dysautonomia gene on human chromosome 9q31. Am J Hum Genet 64:1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Gill SP, Leyne M, Mull J, Liebert CB, Robbins CM, Pinkett HW, Makalowska I, Maayan C, Blumenfeld A, Axelrod FB, Brownstein M, Slaugenhaupt SA (1999a) Cloning, genomic organization, and expression of a putative human transmembrane protein related to the Caenorhabditis elegans M01F1.4 gene. Gene 240:67–73 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Leyne M, Gill S, Liebert CB, Mull J, Mezey E, Makalowska I, Robbins C, Frischauf AM, Maayan C, Blumenfeld A, Axelrod FB, Brownstein M, Slaugenhaupt SA (2000) Cloning, mapping, and expression of a novel brain specific transcript in the familial dysautonomia candidate region on 9q31. Mamm Genome 11:81–83 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Mull J, Helbling LA, Gill S, Leyne M, Robbins CM, Pinkett HW, Makalowska I, Maayan C, Blumenfeld A, Axelrod FB, Brownstein M, Gusella JF, Slaugenhaupt SA (1999b) Cloning, mapping and expression of two novel actin genes, actin-like-7A (ACTL7A)and actin-like-7B (ACTL7B) from the familial dysautonomia candidate region on 9q31. Genomics 58:302–309 [DOI] [PubMed] [Google Scholar]
- Cohen L, Henzel WJ, Baeuerle PA (1998) IKAP is a scaffold protein of the IκB kinase complex. Nature 395: 292–297 [DOI] [PubMed] [Google Scholar]
- Krappmann D, Hatada EN, Tegethoff S, Li J, Klippel A, Giese K, Baeuerle PA, Scheidereit C (2000) The IκB kinase (IKK) complex tripartite and contains IKKκ but not IKAP as a regular component. J Biol Chem 275:29779–29787 [DOI] [PubMed] [Google Scholar]
- Maayan C, Kaplan E, Shachar S, Peleg O, Godfrey S (1987) Incidence of familial dysautonomia in Israel 1977–1981. Clin Genet 32:106–108 [DOI] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell 3:109–118 [DOI] [PubMed] [Google Scholar]
- Pearson J, Pytel B (1978a) Quantitative studies of ciliary and sphenopalatine ganglia in familial dysautonomia. J Neurol Sci 39:123–130 [DOI] [PubMed] [Google Scholar]
- ——— (1978b) Quantitative studies of sympathetic ganglia and spinal cord intermedio-lateral gray columns in familial dysautonomia. J Neurol Sci 39:47–59 [DOI] [PubMed] [Google Scholar]
- Pearson J, Pytel BA, Grover-Johnson N, Axelrod F, Dancis J (1978) Quantitative studies of dorsal root ganglia and neuropathologic observations on spinal cords in familial dysautonomia. J Neurol Sci 35:77–92 [DOI] [PubMed] [Google Scholar]
- Riley CM, Day RL, Greely D, Langford WS (1949) Central autonomic dysfunction with defective lacrimation. Pediatrics 3:468–477 [PubMed] [Google Scholar]
- Shimizu K, Nagamachi Y, Tani M, Kimura K, Shiroishi T, Wakana S, Yokota J (2000) Molecular cloning of a novel NF2/ERM/4.1 superfamily gene, Ehm2, that is expressed in high-metastatic K1735 murine melanoma cells. Genomics 65:113–120 [DOI] [PubMed] [Google Scholar]
- Zhang JS, Nelson M, Wang L, Liu W, Qian CP, Shridhar V, Urrutia R, Smith DI (1998) Identification and chromosomal localization of CTNNAL1, a novel protein homologous to alpha-catenin. Genomics 54:149–154 [DOI] [PubMed] [Google Scholar]