Abstract
A total of 1,664 new mtDNA control-region sequences were analyzed in order to estimate Gaelic and Scandinavian matrilineal ancestry in the populations of Iceland, Orkney, the Western Isles, and the Isle of Skye and to investigate other aspects of their genetic history. A relative excess of private lineages in the Icelanders is indicative of isolation, whereas the scarcity of private lineages in Scottish island populations may be explained by recent gene flow and population decline. Differences in the frequencies of lineage clusters are observed between the Scandinavian and the Gaelic source mtDNA pools, and, on a continent-wide basis, such differences between populations seem to be associated with geography. A multidimensional scaling analysis of genetic distances, based on mtDNA lineage-cluster frequencies, groups the North Atlantic islanders with the Gaelic and the Scandinavian populations, whereas populations from the central, southern, and Baltic regions of Europe are arranged in clusters in broad agreement with their geographic locations. This pattern is highly significant, according to a Mantel correlation between genetic and geographic distances (r=.716). Admixture analyses indicate that the ancestral contributions of mtDNA lineages from Scandinavia to the populations of Iceland, Orkney, the Western Isles, and the Isle of Skye are 37.5%, 35.5%, 11.5%, and 12.5%, respectively.
Introduction
The period from the late 8th century through the early 11th century is commonly referred to in European history as the Viking Age. Raiding, trading, and settling, the Vikings expanded east, south, and west from Scandinavia—invading the Baltic region and Russia from Sweden; invading England, France, and as far south as Spain from Denmark; and invading Ireland, north England, Scotland, and the North Atlantic islands from Norway (Jones 1984; Collins 1991). In the westward spread, around 780 a.d., Shetland and Orkney were the first of the North Atlantic islands to be colonized by the emerging Vikings (Davies 1999), but, following the Scottish coastline, the Vikings soon reached the Western Isles. By 800 a.d., the Vikings had reached the Faroe Islands, and, some 60 years later, Iceland was discovered. Whereas Orkney and the Western Isles are known to have had thriving pre-Norse settlements of Picts and Gaels, respectively, Shetland was less densely populated, and the Faroes and Iceland are both held to have been largely uninhabited at the time of the Norse settlements (Berry and Muir 1975; Davies 1999).
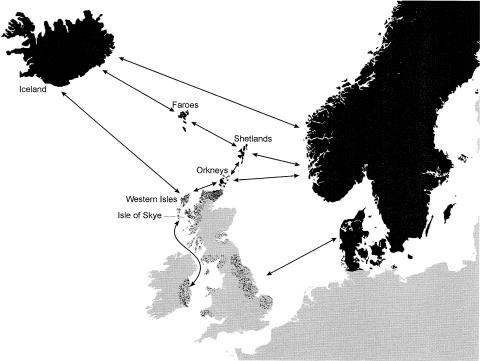
Iceland was the last of the islands to be settled in this rapid expansion of Norse peoples, and historical sources clearly indicate that this episode of colonization in the North Atlantic involved the greatest movement of people (Rafnson 1999). Iceland is still inhabited by Norse-speaking descendants of the first settlers, some of whom are thought to have originated from the British Isles. The legacy of the Viking period is less clear for Orkney, the Western Isles, and the Isle of Skye. Archaeological, linguistic, and historical evidence all indicate that Viking activities had the most far-reaching effect in Orkney, where the indigenous Pictish population may have been entirely replaced by Norse settlers (Graham-Campbell and Batey 1998). The apparent dominance of the Norse language and material culture in Orkney and Shetland contrasts with evidence from the Western Isles, where the coexistence of Vikings and the indigenous Gaelic population led to general bilingualism and a greater level of cultural amalgam (Graham-Campbell and Batey 1998). Scotland regained control over the Western Isles after the battle of Largs in 1263, whereas Orkney and Shetland remained under Norse control until 1469 (Boyce et al. 1973; Berry and Muir 1975; Clegg et al. 1985). In both cases, the return of Scottish rule led to a period of repopulation from the mainland and the dominance of Scottish language and culture (although Norn, the Norse dialect of Orkney and Shetland, survived until the 19th century). Figure 1 shows the geographic location of the North Atlantic islands, the sailing routes of the Vikings, and the areas of Norse influence in the North Atlantic region.
Figure 1.
Map of the North Atlantic region. The blackened area represents regions where Norse cultural and linguistic dominance was complete during the Viking period. The dark speckled areas in the British Isles provide an indication of the core areas of Viking exploits, on the basis of archaeological sites, place names of Norse origin, and raided monasteries and towns (Bjarnason et al. 1973; Graham-Campbell and Batey 1998; Corráin 1999; Keynes 1999). The arrows show some of the main sailing routes of the Vikings in the North Atlantic.
A number of previous studies have attempted to shed light on the genetic affinities of the North Atlantic islanders using classical allozyme genetic markers, but their results have been difficult to interpret. Most studies have focused on the Icelanders, with the aim of calculating the contributions to admixture of the Norse and Gaelic ancestral populations. Estimates have varied considerably—from a 93%–98% Gaelic contribution (Thompson 1973) to an 86% Norse contribution (Wijsman 1984). More recent analyses of mtDNA and Y-chromosome variation in the Icelanders suggests that a majority of the female settlers may have originated from the British Isles, whereas ∼80% of male settlers were Scandinavian (Helgason et al. 2000b, 2000c). Fewer studies have dealt with the other island populations in the North Atlantic. An analysis of classical allozyme markers in a Western Isles population from Lewis found that allele frequencies showed substantial differences from neighboring European populations (Clegg et al. 1985). Natural selection or genetic drift in the Lewis gene pool or gene flow among the other European populations were suggested as possible causes of these differences. In similar studies of Orkney and Shetland, Roberts reported (1985, 1990) that both island populations diverged considerably in allele frequencies from neighboring populations. Although not ruling out selection or drift as potential causes for this divergence, Roberts concluded that the islanders of Orkney and Shetland most likely represented remnants of an aboriginal gene pool that had changed on the British mainland because of later population movements. None of these studies of allozyme variation in the Scottish Islands reported estimates of Norse admixture.
In this study, we examined mtDNA control-region sequences in the North Atlantic island populations of Orkney, the Western Isles, the Isle of Skye, and Iceland, and we compared them to those observed in the rest of the British Isles, Scandinavia, and other regions of Europe. The primary aims were to assess the relative magnitude of diversity and levels of Gaelic and Scandinavian admixture in the mtDNA pools of the North Atlantic island populations and individuals from the northwest coast of Scotland. mtDNA lineages sampled from contemporary populations provide us with direct links to matrilineal ancestors from the Viking age, and thus enables us to examine the extent to which Scandinavian females were involved in Norse settlements on the North Atlantic islands. Many historians believe that the Norse expansion of the Viking Age was primarily a male enterprise (Clover 1988). If this were the case, one would not expect to find close links between mtDNA lineages found in the North Atlantic island populations and Scandinavia. On the basis of available historical, archaeological, and linguistic information (Graham-Campbell and Batey 1998; Corráin 1999; Davies 1999), we would expect that the largest proportion of mtDNA lineages inherited from Norse matrilineal ancestors would be found (in descending order of magnitude) in Iceland, Orkney, the Western Isles, the Isle of Skye, and the coastal region of northwest Scotland. To achieve these aims, we sequenced the mtDNA hypervariable segment 1 (HVS1) from 1,664 individuals from the Scottish islands, the Scottish mainland, England, and Norway. These new data were added to an existing data set of 3,444 Eurasian HVS1 sequences, for a detailed study of mtDNA variation in the North Atlantic region.
Material and Methods
Population Samples
DNA from 891 individuals from all regions of mainland Scotland, 181 from the Western Isles, 49 from the Isle of Skye, and 142 individuals of English matrilineal descent (representing most regions of England) was extracted from blood collected at Blood Transfusion Service donor sessions throughout Scotland. Information about the birth place of the maternal grandmother was sought from each individual. Similarly, DNA from 323 Norwegians was extracted from blood collected at donor sessions at Ullevål hospital in Oslo. Again, the birth place of the maternal grandmother was recorded for each individual, showing the samples to be representative of all geographic areas of Norway. In all cases individuals gave informed consent. Samples from 78 Orkney Islanders were kindly supplied by Dr. J. Bodmer (see Bodmer et al. 1996). The data produced for this study were deposited in GenBank and are available on request from the corresponding author.
The comparative data set from Europe consisted of 3,444 mtDNA HVS1 sequences from the following populations and sources: Iceland (394 from Helgason et al. 2000b, 20 from the Mitochondrial DNA Concordance, 14 from Richards et al. 1996, and 39 from Sajantila et al. 1995), Ireland (23 from the Mitochondrial DNA Concordance and 105 from Richards et al. 2000), Orkney (74 from the Mitochondrial DNA Concordance), the Western Isles (16 from the Mitochondrial DNA Concordance), England and Wales (160 from Richards et al. 1996, 29 from the Mitochondrial DNA Concordance, and 97 from Piercy et al. 1993), Finland (74 from Kittles et al. 1999, 23 from Pult et al. 1994, and 29 from Richards et al. 1996), Estonia (26 from Sajantila et al. 1995), Austria (99 from Parson et al. 1998 and 16 from Handt et al. 1994), France (10 from Mitochondrial DNA Concordance and 50 from Rousselet and Mangin 1998), Italy (49 from Francalacci et al. 1996 and 68 from Stenico et al. 1996), Germany (151 from Richards et al. 1996, 67 from Hofmann et al. 1997, 200 from Lutz et al. 1999, and 109 from Pfeiffer et al. 1999), Denmark (15 from the Mitochondrial DNA Concordance and 31 Richards et al. 1996), Sweden (28 from Kittles et al. 1999 and 32 from Sajantila et al. 1996), Norway (216 from Opdal et al. 1998), Saami (61 from Delghandi et al. 1998 and 115 Sajantila et al. 1995), European Russia (103 from Orekhov et al. 1999 and 112 from Sajantila et al. 1995), Switzerland (76 from Pult et al. 1994), Spain (132 from Corte-Real et al. 1996, 18 from Pinto et al. 1996, 92 from Salas et al. 1998, 11 from Handt et al. 1998, and 45 from Bertranpetit et al. 1995), Portugal (54 from Corte-Real et al. 1996), Bulgaria (30 from Calafell et al. 1996), Turkey (27 from Calafell et al. 1996 and 45 from Comas et al. 1996). Data obtained from the Mitochondrial DNA Concordance Web site originate from Miller’s (1996) analysis of archival blood samples from populations in the North Atlantic region. Of all 253 sequences from Orkney, the Western Isles, Ireland, Denmark, England, France, and Iceland from Miller (1996), 66 had ambiguous sites (probably because of blood degradation) and were excluded from further analysis.
To obtain equivalent sample sizes for comparative analysis, we combined sequences from geographically proximate populations into the following groups: England/Wales, Spain/Portugal, Austria/Switzerland, France/Italy, Finland/Estonia, and Bulgaria/Turkey. In addition to individuals from Iceland, Orkney, the Western Isles, and the Isle of Skye, Norse and Gaelic admixture was estimated in 91 individuals from the coastal region of north and northwest Scotland (mainly Wester Ross, Caithness, and Sutherland). The term Gaelic is used throughout as a convenient label for the combined populations of mainland Scotland and Ireland and should not be interpreted in a strict linguistic sense.
Markers and Protocols
HVS1 was amplified for the 1,664 new samples from the British Isles and Norway, as described elsewhere (Richards et al. 1996), and was sequenced at the University of Florida DNA Sequencing Core Laboratory, by use of ABI Prism Dye Terminator cycle-sequencing protocols developed by Applied Biosystems. The fluorescently labeled extension products were analyzed on an Applied Biosystems Model 373 Stretch DNA sequencer or on a 377 DNA sequencer (Perkin-Elmer). In most cases, sequences were obtained between sites 16010 and 16400. The mtDNA site numbers referred to in this study are those of Anderson et al. (1981). To maximize the number of sequences available for analysis from the North Atlantic region, all analyses were restricted to the 235 nucleotides between positions 16090 and 16324. Recent studies have indicated that European mtDNA pools contain an extensive array of different mtDNA lineages and that large sample sizes are required to capture a representative sample of this variation (Pfeiffer et al. 1999; Helgason et al. 2000b; Richards et al. 2000). The maximization of sample size at the cost of sequence length is further justified by the observation that 82% of the total polymorphism found in sequences available between sites 16010 and 16400 is contained within the 235-bp segment we have analyzed.
Summary Statistics and Sampling Saturation
Gene diversity was estimated as
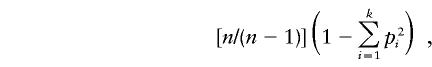 |
where n is the total number of sequences, k the number of distinct lineages, and pi the frequency of the distinct lineages. This parameter represents the probability of two randomly chosen sequences from a sample being nonidentical by state.
Mean pairwise differences between sequences (θπ) were calculated as
 |
where dij is the number of mutational differences between lineages i and j in a sample, k is the number of distinct lineages and pi and pj are the respective frequencies of lineages i and j. θk was estimated using Ewens (1972) sampling formula:
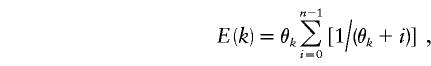 |
where k is the number of distinct lineages observed in a sample size of n. Each of these parameters was calculated using the software package Arlequin 1.1 (Schneider et al. 1997).
The two parameters, θπ and θk, use different aspects of the genetic data to estimate the population mutation parameter 2Nfeμ, where Nfe represents the female effective population size and μ the mutation rate. Helgason et al. (2000b) noted that θk and θπ exhibit quite divergent values for mtDNA control-region sequences in European populations and that θk appears to provide a better reflection of European population sizes during the rapid expansions of the last few centuries. Because our focus is on historical events taking place during the past 1,300 years, we take θk as the more reliable estimator of Nfe, whereas θπ is taken simply to represent the average mutational divergence observed between a population’s mtDNA sequences. Under conditions of neutrality, constant population size, and the infinite-alleles mutation model, and under the assumption that the mutation rate is equal in all populations, differences in θk will reflect differences in Nfe (Ewens 1972). Departures from these assumptions complicate the direct estimation, using Ewens's sampling formula, of Nfe. However, given that such departures are more or less equivalent for all populations, θk can be considered as an effective relative indicator of Nfe.
Recent studies indicate that European populations contain a large number of mtDNA lineages, of which many will remain unsampled, even when sample sizes are >400 (Pfeiffer et al. 1999; Helgason et al. 2000b). Sampling saturation of mtDNA lineages in European populations was assessed using a method based on the above sampling formula by Ewens (1972). Under the assumption of a steady-state distribution of alleles, this formula predicts that the relationship between increasing sample size and the number of new lineages encountered will be one of diminishing returns. We use the observed θk values to estimate the point at which incremental increases of 10 in sample size yields less than one new lineage (see Helgason et al. 2000a, 2000b). The resultant sample sizes represent the points at which the rate of lineage detection is equal for all populations. The ratio of these theoretical sample sizes to the actual sample size for each population can serve as an indicator of relative sampling saturation.
Haplogroup Assignment and Genetic Distances
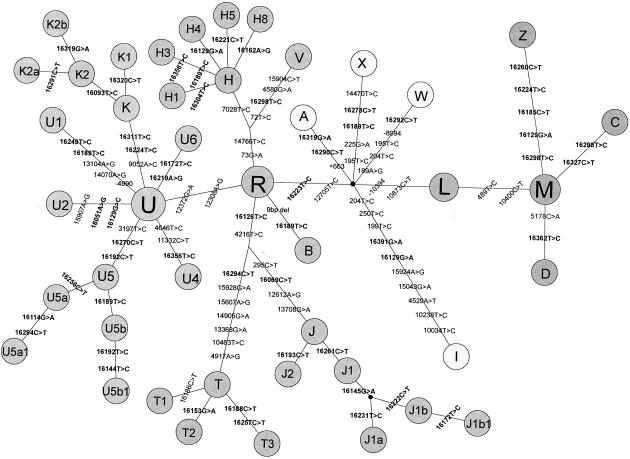
Much of the evolutionary change in the mtDNA pools of European populations during the past 1,300 years and before will have been in the form of lineage redistribution—both within populations, caused by drift, and between populations, caused by migration. Thus, measures of genetic distance based on lineage frequencies may provide important information about the relationships of the populations in the North Atlantic region. In order to reduce statistical noise caused by sampling variance and missing (or unsampled) lineages on the frequency based genetic distance, we collapsed lineages into a smaller number of phylogenetically resolved subclusters. The basic phylogenetic structure of Eurasian mtDNA lineages has been revealed by a number of recent studies (Richards et al. 1998; Macaulay et al. 1999; Helgason et al. 2000b). The skeleton structure is shown in figure 2, along with a number of well-resolved subclusters and known diagnostic substitutions. The maximum amount of character information available for each sequence was used to reduce the full data set of 1,128 lineages to the 42 haplogroups or subclusters identified in figure 2 (excluding the superhaplogroups R, L, and M). Only 35 lineages (46 sequences) could not be assigned to these Eurasian lineage clusters, and almost all of these are rare occurrences of lineages belonging either to African superhaplogroup L or Asian superhaplogroup M. These 35 lineages were assigned to a synthetic subcluster designated “other.” Most of the haplogroups and subclusters shown in figure 2 are now well established in the literature. The following subclusters were identified through the construction of phylogenetic networks using mtDNA HVS1 sequences from populations in the North Atlantic region: H1, H3, H4, H5, H8, K1, K2, K2a, K2b, T2, and T3. Median-joining networks (Bandelt et al. 1999), were generated for the common European haplogroups (H, I, J, K, V, and U5) using the mtDNA HVS1 data set described above. On the basis of these networks, new subclusters were designated when a cluster of lineages emanating from a founder lineage was observed in more than one population. Lineages not assigned to new subclusters retained their basic haplogroup label (see Richards et al. 2000 for a similar approach). Until whole-genome sequencing of mtDNA becomes the norm, allowing the construction of unambiguous phylogenetic trees (see Ingman et al. 2000), any such schemes will necessarily remain arbitrary and the source of debate and confusion (Simoni et al. 2000b; Torroni et al. 2000). In the interim, we must rely on heuristic subclusters, such as those defined in the present study.
Figure 2.
Schematic phylogenetic representation of mtDNA lineage clusters found in European populations, reconstructed on the basis of information obtained from Torroni et al. (1996), Richards et al. (1998), Macaulay et al. (1999), Quintana-Murci et al. (1999), and Helgason et al. (2000b). Lineage clusters are shown as circles, and the connecting lines represent diagnostic substitutions.
Genetic distances between populations based on these subcluster frequencies were calculated using the f distance, which is based on the chord genetic distance introduced by Cavalli-Sforza and Edwards (1967). The resultant matrix of genetic distances between populations was represented in two-dimensional space by means of a multidimensional scaling (MDS) analysis, using the SPSS software package.
ρ Distances
Using the full sequences between sites 16090 and 16324, we estimated the mutational divergence of the North Atlantic mtDNA pools from other European populations using the index ρ. The ρ index is defined as the average number of substitutions between the sequences of one population and the closest founder sequences observed in another population (Forster et al. 1996), and it effectively summarizes the overlap between one mtDNA pool and a potential source mtDNA pool. Unlike an analysis of molecular variance (AMOVA) distance, which summarizes the average mutational distance between all pairs of sequences from two populations (Excoffier et al. 1992), ρ is insensitive to the fact that the divergence between European populations, as measured in mutations at the mtDNA locus, is small relative to the overall mutational time-depth of the European mtDNA phylogeny (see Richards et al. 1998; Simoni et al. 2000a). Thus, an AMOVA analysis shows that <2% of the variance in mutational divergence between all pairs of European mtDNA HVS1 sequences are accounted for by their distribution among different populations (Helgason et al. 2000b).
Admixture Method
To estimate the level of Scandinavian ancestry in the island populations of the North Atlantic and the coastal population of northwest Scotland, we employ a heuristic approach to estimate the admixture proportion that best fits the observed lineage distribution in the admixed and parental populations (see Helgason et al. 2000c). This estimator, designated mρ, is obtained as follows. Given a prior probability of admixture, η, the probability that a randomly chosen lineage observed in the admixed sample is derived from the first source population is given by ηp1/{ηp1+(1-η)p2}, where p1 and p2 are the frequencies of this lineage in the two source populations. For a given value of η, 10,000 random samples are used to obtain the mean and 95% confidence interval for the admixture estimate mρ, conditioned on the prior value of η. The fit to the data is evaluated by Σ(Nmρ-Nη)2/Nη), where N is the size of the admixed sample. The best-fitting model is found by an iterative search over the line 0⩽mρ⩽1 in successively smaller intervals around the best-fitting value for mρ.
To use the information provided by private lineages in the admixed populations, the putative founder lineage(s) for a private lineage is defined as the lineage in the source population(s) that differs by the smallest number of substitutions according to a matrix of mutational distances between lineages. If more than one lineage in the source population(s) meets this criterion as a putative founder lineage (as in the case of a tie), their frequencies are summed to derive p1 and p2, to calculate the conditional probability of origin. We note that the only way to unambiguously determine the genuine founder lineages is by means of the true phylogeny of sequences generated from a sample of all lineages from the populations in question. However, because unambiguous phylogenies cannot be constructed from HVS1 sequence data and because it is likely that many lineages have yet to be sampled from the mtDNA pools of European populations, our method provides an objective heuristic approach to identifying founder lineages that avoids subjective choice of founder lineages “by hand.”
Results
Summary Statistics and Sampling Saturation
Table 1 presents summary statistics for the 5,108 mtDNA HVS1 sequences used in this study, with populations placed in descending order by θk values. As was found elsewhere (Helgason et al. 2000b), θk seems to better reflect current and historical population sizes than does θπ. In most cases, values of θk and θπ differ by more than one order of magnitude and are not significantly correlated. As expected, the North Atlantic island populations exhibit relatively small values of θk, which is indicative of small effective population sizes for females (Nfe). In most respects, the populations from the Western Isles and Orkney exhibit similar levels of genetic diversity to those of the Icelanders. Interestingly, the Irish sample also exhibits very low values of gene diversity and θπ. The Saami and the islanders of Skye have by far the smallest Nfe.
Table 1.
Summary Statistics for HVS1 Sequences from European Populations[Note]
| Population | N | K | S | GD | θk | θπ | PrivateLineages(%) | SamplingRatio |
| France/Italy | 248 | 158 | 97 | .963 | 186.42 | 4.23 | 60.76 | .147 |
| Germany | 527 | 234 | 99 | .97 | 160.68 | 3.70 | 50.00 | .361 |
| Scandinavia | 645 | 243 | 108 | .937 | 141.36 | 3.52 | 44.44 | .504 |
| England/ Wales | 429 | 183 | 91 | .934 | 120.18 | 3.35 | 44.81 | .394 |
| Scotland | 891 | 250 | 102 | .956 | 115.11 | 3.73 | 46.00 | .849 |
| Spain/Portugal | 352 | 154 | 95 | .935 | 103.85 | 3.26 | 45.45 | .371 |
| Bulgaria/Turkey | 102 | 71 | 70 | .977 | 102.25 | 4.34 | 56.34 | .110 |
| Austria/Switzerland | 187 | 93 | 70 | .958 | 72.84 | 3.55 | 37.63 | .279 |
| European Russia | 215 | 90 | 59 | .934 | 57.69 | 3.44 | 32.22 | .406 |
| Western Isles | 197 | 79 | 53 | .968 | 48.43 | 3.75 | 27.85 | .438 |
| Iceland | 467 | 114 | 67 | .966 | 47.76 | 3.96 | 42.98 | 1.061 |
| Orkney Islands | 152 | 67 | 55 | .946 | 45.24 | 3.37 | 27.94 | .362 |
| Ireland | 128 | 61 | 50 | .922 | 45.05 | 2.87 | 29.51 | .305 |
| Finland/Estonia | 202 | 75 | 59 | .949 | 42.74 | 3.49 | 33.33 | .505 |
| Isle of Skye | 49 | 23 | 27 | .935 | 16.30 | 3.70 | 21.74 | .306 |
| Saami | 176 | 30 | 30 | .808 | 10.15 | 3.21 | 46.67 | 1.760 |
Note.— N = sample size; K = no. of lineages; S = no. of variable sites; GD = gene diversity.
The sampling saturation ratio varies considerably among the populations included in this study and indicates that the Saami, Icelanders, and Scots are the most extensively sampled populations for mtDNA variation. In contrast, France/Italy and Bulgaria/Turkey appear to be the least-sampled regions included in this study.
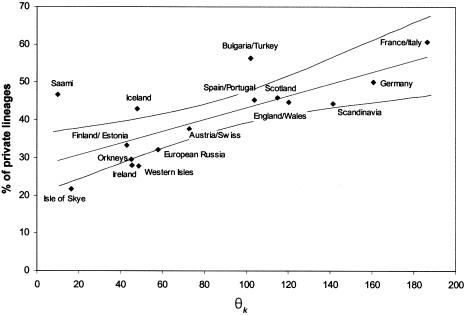
The proportion of private lineages also varies considerably among populations (21.7%–60.7%). In general, the proportion of private lineages sampled from geographically proximate populations should increase as a function of θk, as this parameter reflects the probability of new lineages arising by mutation. Each new lineage will be private to the population in which it appeared until carried through female migration into another population’s mtDNA pool. As is indicated in figure 3, there is a strong correlation between θk and the proportion of private lineages (r=.77; P<.001). A few samples have a relative excess or scarcity of private lineages, of which the Iceland, Saami, Bulgaria/Turkey, Isle of Skye, Western Isles, and Ireland sample are outside the 95% confidence interval for the regression line. An excess of private lineages could have at least three basic causes. The first, where isolation has hindered the migrational flow of new lineages to and from neighboring populations, is perhaps typified by the Icelanders. The second is exemplified by the population sample from Bulgaria/Turkey: considerable gene flow has taken place from regions not included in the comparative database that was used to estimate the proportion of private lineages (in this case, from Asia and Africa). This also is exemplified by the Saami, whose mtDNA pool includes a number of lineages from Asian haplogroups. The third cause would be an excess of private lineages in populations that have been sampled more extensively than others included in the comparison. The relatively thorough sampling of the Icelanders may account partially for their excess of private lineages.
Figure 3.
Scatterplot of θk values and the percentage of private lineages. The least-squares regression line, where r=0.77, is shown. The curved lines show the 95% confidence region around the regression line.
A relative scarcity of private lineages could be indicative of either a very high level of emigration (where few lineages remain private for long, because of rapid outward gene flow to neighboring populations) or immigration (where new lineages arriving into the population would increase θk but not the proportion of private lineages). It is interesting to note that, contrary to the excess of private lineages observed in the Icelanders, the populations of Orkney, the Western Isles, and the Isle of Skye all exhibit a relative scarcity of private lineages. As the Icelanders and Scottish islanders share the same source mtDNA pools, this difference may reflect the well-recorded extensive migration from the Scottish islands to the mainland during the past two centuries, as opposed to the isolation of the Icelanders.
Lineage Sharing and Haplogroup Frequencies
Table 2 shows the pattern of lineage sharing between the North Atlantic island populations and the Scandinavian and Gaelic source mtDNA pools. It is notable that all populations share a higher percentage of their lineages exclusively with the Gaelic source populations. The Icelanders have the highest percentage of lineages that are found in neither source mtDNA pool (again, an indication of greater isolation), and the islanders of Skye have the lowest. As might be expected, the Icelanders have the lowest proportion of lineages shared exclusively with Gaels, and the islanders of Skye have the highest. More surprising is the observation that the islanders of Skye and Orkney share a greater proportion of their lineages with Scandinavians than do the Icelanders. However, if only lineages shared exclusively with either of the two source populations are examined, Iceland (0.38) and Orkney (0.35) are revealed as having the closest relationship to Scandinavia (see table 2).
Table 2.
Pattern of Lineage Sharing between North Atlantic Islands and Source Populations
|
Lineages Shared with(%) |
|||||
| Population | Ka | GaelsOnlyb | ScandinaviansOnlyb | Both | Neither |
| Iceland | 114 | 11.4 (.62) | 7.0 (.38) | 26.3 | 55.3 |
| Orkney | 68 | 16.2 (.65) | 8.8 (.35) | 38.2 | 36.8 |
| Western Isles | 79 | 16.5 (.81) | 3.8 (.19) | 35.4 | 44.3 |
| Isle of Skye | 23 | 30.4 (.78) | 8.7 (.22) | 39.1 | 21.7 |
| NW Scottish coast | 91 | 24.2 (.88) | 3.3 (.12) | 39.6 | 33.0 |
| Scottish Islands | 138 | 18.8 (.74) | 6.5 (.26) | 27.5 | 47.1 |
| North Atlantic islands | 214 | 14.0 (.67) | 7.0 (.33) | 21.0 | 57.9 |
K = number of distinct lineages.
The number in parentheses represents the proportion of lineages shared exclusively with either the Gaels or Scandinavians out of the total number of exclusively shared lineages.
Genetic Distances
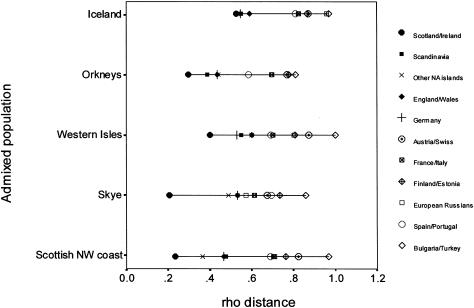
Figure 4 shows ρ distances between island populations of the North Atlantic and other European groups. Note that individuals from the northwest coast of Scotland are treated as a separate population. The ρ distances are largely consistent with expectations based on historical and archaeological records. The Gaels are closest in all cases, followed by a cluster of Scandinavians, other North Atlantic islanders, inhabitants of England and Wales, Germans, and the remaining European populations. The ρ distance to the Gaels is smallest from the Isle of Skye and the northwest coast of Scotland and is greatest for the Icelanders. The least difference between Scandinavians and Gaels is observed for Iceland and then, in descending order, for Orkney, the Western Isles, the northwest coast of Scotland, and the Isle of Skye. Interpreted as a rough indicator of Norse admixture, the relative differences between ρ distances to Gaels and Scandinavians accord with historical evidence of the differential impact of Norse settlement on each of the North Atlantic island populations. However, according to these results, it appears that the Gaelic contribution to the Icelandic mtDNA pool may have been at least as large as that from Scandinavia.
Figure 4.
ρ distances between the mtDNA pools of five North Atlantic populations and those of potential European source populations. ρ distances to the Saami were excluded to maximize clarity in the representation of distances to the other populations. In all cases, the ρ distance to the Saami was ∼1.5.
The small ρ distances to the Germans in all five cases are surprising, as there are no known accounts of recent female gene flow from Germany into the North Atlantic region. This may be accounted for by Germany’s central position in Europe and by the fact that many ancient population movements into the British Isles and Scandinavia originated from or passed through this territory (Collins 1991; Davies 1999). In this case, ancient German ancestral links to both the Norse and Gaelic mtDNA pools would account for the low ρ distances to the admixed North Atlantic island populations. It may also be that ρ values shown in figure 4 are influenced by sample size. ρ distances to putative source populations that have not been adequately sampled will tend to be overestimated (Helgason 2000b; Richards et al. 2000). However, table 1 suggests that, for example, England/Wales, Finland/Estonia, and European Russia have a higher sampling saturation than Germany—and yet the latter two are consistently more distant from the North Atlantic island populations than is Germany. We thus conclude that ρ values are likely to reflect actual relationships between the mtDNA pools of these populations. Moreover, even when an effect caused by the sample size of putative source populations is assumed, this should influence equally ρ distances to the North Atlantic island populations. Thus, the varying configuration of ρ distances would still provide valuable information about the population histories of the North Atlantic island populations.
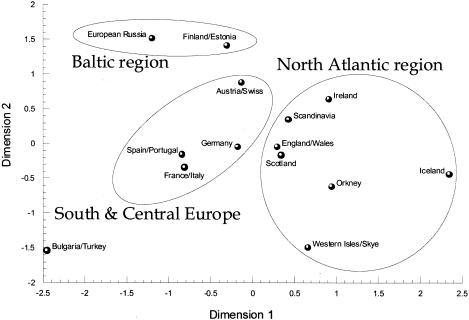
A more complete picture of population relationships in the North Atlantic region and the rest of Europe can be obtained from a multidimensional scaling (MDS) plot of genetic distances based on the frequencies of lineage clusters (fig. 5). Table 3 presents the frequencies of lineage clusters in the populations and groups used in this study (the phylogenetic relationships of these lineage clusters is shown in fig. 2). We have interpreted this plot as showing three nonoverlapping geographic groups of populations: those of the North Atlantic, south and central Europe, and the Baltic region. The North Atlantic island populations clearly have the closest links to the mtDNA pools of the British Isles and Scandinavia. The wide dispersion of the island populations in the top-right area of the diagram agrees with their small effective population size of females and the concomitant effect of genetic drift.
Figure 5.
Multidimensional scaling plot of genetic distances on the basis of haplogroup frequencies. The fourteen dimensions of the genetic distance matrix were reduced to two dimensions, which account for 85% of the genetic variation defined by the original distance matrix.
Table 3.
Haplogroup and Subcluster Frequencies for European Populations
|
Frequency for Population(%) |
|||||||||||||||
| Haplogroup | Austria/Switzerland(N=187) | EuropeanRussia(N=215) | Finland/Estonia(N=202) | France/Italy(N=248) | Germany(N=527) | Iceland(N=467) | Ireland(N=128) | Orkney(N=152) | Scandinavia(N=645) | Scotland(N=891) | Bulgaria/Turkey(N=102) | Spain/Portugal(N=352) | England/Wales(N=429) | WesternIsles/Isleof Skye(N=246) | Saami(N=176) |
| A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .16 | 0 | 0 | .85 | .23 | .41 | 0 |
| B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .11 | 0 | 0 | 0 | 0 | 0 |
| C | 0 | 1.86 | .50 | .40 | .19 | .43 | 0 | 0 | 0 | 0 | 1.96 | 1.14 | 0 | 0 | 0 |
| D | 0 | 1.86 | 0 | .40 | .38 | 0 | 0 | 0 | .16 | 0 | 4.90 | .28 | 0 | 0 | 5.11 |
| H | 47.06 | 33.49 | 36.63 | 45.16 | 38.33 | 28.27 | 41.41 | 40.79 | 39.69 | 38.38 | 31.37 | 50.28 | 40.79 | 27.24 | 1.70 |
| H1 | 3.74 | 5.58 | 2.97 | 2.82 | 4.36 | 8.35 | 2.34 | 6.58 | 3.41 | 3.03 | 1.96 | 2.56 | 4.43 | 1.22 | 1.14 |
| H3 | .53 | 0 | 0 | .40 | 1.14 | .43 | 1.56 | 0 | .62 | .56 | 0 | 0 | .93 | 1.22 | 0 |
| H4 | 1.60 | 0 | 1.49 | 1.61 | 1.52 | 9.64 | 2.34 | .66 | 2.33 | 1.23 | 3.92 | 3.41 | 3.03 | 3.25 | 0 |
| H5 | 1.07 | 0 | 0 | 2.82 | .76 | .43 | 0 | 0 | .62 | .11 | 0 | 1.99 | 1.63 | 0 | 0 |
| H8 | 0 | 3.26 | 2.97 | .81 | 2.85 | .43 | 0 | 2.63 | 1.86 | 2.36 | .98 | .28 | 1.40 | 1.63 | 2.84 |
| I | 2.14 | 1.40 | 2.48 | .81 | 2.28 | 4.71 | 2.34 | 3.29 | 1.86 | 4.38 | 1.96 | .57 | 3.03 | 6.50 | 0 |
| J | 5.35 | 6.51 | 4.95 | 2.42 | 6.83 | 6.85 | 11.72 | 7.89 | 6.82 | 8.64 | 5.88 | 3.69 | 10.72 | 10.57 | 0 |
| J1 | 1.60 | 0 | .50 | 0 | .76 | 0 | 0 | 0 | .47 | .56 | 4.90 | .28 | .47 | 1.22 | 0 |
| J1a | 3.21 | 0 | .50 | .40 | 1.33 | .43 | .78 | 0 | 1.55 | .45 | 0 | .57 | 1.63 | 0 | 0 |
| J1b | 0 | .47 | 0 | .40 | 0 | 0 | 0 | 0 | 0 | .11 | .98 | .57 | 0 | 0 | 0 |
| J1b1 | 0 | 0 | 0 | .40 | .19 | 5.57 | .78 | 1.97 | 1.40 | 3.48 | 0 | 0 | 1.40 | 1.22 | 0 |
| J2 | 0 | .93 | .99 | 2.42 | .19 | 1.28 | .78 | 0 | 0 | 1.12 | 2.94 | .85 | .23 | 1.63 | 0 |
| K | 2.14 | 1.86 | 2.48 | 4.44 | 5.69 | 4.93 | 5.47 | 5.26 | 4.03 | 3.70 | 4.90 | 3.13 | 5.13 | 8.54 | 0 |
| K1 | 0 | 0 | 0 | 0 | .19 | 2.36 | .78 | 1.32 | .47 | .79 | 0 | 0 | .23 | .81 | 0 |
| K2 | 5.88 | .93 | 0 | 1.61 | .76 | .43 | .78 | 0 | .16 | 1.01 | .98 | 1.14 | .47 | .41 | 0 |
| K2a | 0 | 0 | 0 | 0 | 0 | 0 | .78 | 0 | .31 | .56 | 0 | 0 | 0 | 1.22 | 0 |
| K2b | 1.07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .56 | 0 | .28 | .23 | 2.44 | 0 |
| T | 4.81 | 4.65 | 5.45 | 10.89 | 6.26 | 2.14 | 7.03 | 2.63 | 6.51 | 7.63 | 4.90 | 4.55 | 5.36 | 8.94 | 0 |
| T1 | 1.07 | 3.72 | 1.49 | 3.23 | 2.47 | .43 | 2.34 | 3.29 | 1.40 | 2.24 | 4.90 | 1.14 | 2.10 | 3.25 | 0 |
| T2 | .53 | 1.86 | 0 | .40 | .19 | 2.57 | 0 | 0 | .93 | .22 | 0 | .28 | .23 | .41 | 0 |
| T3 | 0 | 0 | 0 | 0 | .19 | 4.93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| U | 0 | 0 | 0 | 0 | .19 | .21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| U1 | .53 | .47 | .50 | 1.21 | .38 | 0 | 0 | 0 | .16 | 0 | 0 | .28 | 0 | 2.03 | 0 |
| U2 | 1.07 | 0 | .99 | .81 | .57 | 0 | .78 | 0 | .16 | .79 | 0 | .28 | .70 | .41 | 0 |
| U3 | .53 | .47 | 0 | .81 | 1.14 | 0 | 0 | 0 | .78 | 1.23 | 5.88 | .57 | .70 | 2.44 | 0 |
| U4 | 3.74 | 8.37 | 3.47 | 2.02 | 2.66 | 2.14 | 2.34 | .66 | 2.17 | 2.47 | 3.92 | 1.99 | 1.63 | .41 | 0 |
| U5 | .53 | 0 | 0 | 1.21 | .19 | .43 | .78 | 2.63 | 1.71 | 1.12 | .98 | .57 | .93 | 2.44 | 0 |
| U5a | 5.88 | 7.91 | 6.44 | 2.82 | 4.93 | 5.57 | 4.69 | 5.92 | 6.82 | 5.05 | .98 | 4.55 | 3.50 | 4.88 | .57 |
| U5a1 | .53 | .93 | 1.49 | 0 | .19 | 0 | 0 | 1.32 | .47 | 0 | 0 | 0 | .70 | 0 | .57 |
| U5b | .53 | 2.33 | 6.44 | 1.21 | 3.80 | 3.43 | .78 | 1.97 | 1.71 | 1.01 | 0 | .85 | 1.40 | .81 | 1.70 |
| U5b1 | 0 | 2.79 | 2.97 | 0 | 0 | 0 | 0 | 0 | 2.33 | 0 | 0 | 0 | 0 | 0 | 42.61 |
| U6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.42 | 0 | 0 | 0 |
| U7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .11 | 0 | 0 | 0 | 0 | 0 |
| V | 2.67 | 4.19 | 6.44 | 2.82 | 5.12 | 1.71 | 7.03 | 1.32 | 5.74 | 4.26 | 0 | 5.97 | 3.73 | 2.03 | 39.77 |
| W | 1.60 | 2.33 | 5.45 | .81 | 2.09 | .21 | 2.34 | 1.97 | 1.55 | .90 | 3.92 | 1.99 | 1.63 | .41 | .57 |
| X | .53 | 0 | 1.49 | 2.02 | .76 | 1.50 | 0 | 7.24 | .62 | 1.68 | 3.92 | 1.70 | .93 | 2.03 | 0 |
| Z | 0 | .47 | 0 | 0 | 0 | .21 | 0 | 0 | .62 | 0 | 0 | 0 | 0 | 0 | 3.41 |
| Other | 0 | 1.40 | .99 | 2.42 | 1.14 | 0 | 0 | .66 | .47 | .11 | 2.94 | 1.99 | .47 | 0 | 0 |
We performed a Mantel test in order to assess the strength and significance of the apparent geographic structure observed in figure 5. Geographic distances were calculated as geodesic distances, using the following coordinates for each population: Austria/Swiss (15°12′E, 48°42′N), European Russia (35°42′E, 57°0′N), Finland/Estonia (25°12′E, 60°6′N), France/Italy (7°24′E, 44°6′N), Germany (10°12′E, 51°0′N), Iceland (18°24′W, 64°42′N), Ireland (7°42′W, 53°23′N), Orkney (2°54′W, 59°17′N), Scandinavia (11°18′E, 59°30′N), Scotland (4°18′W, 56°30′N), Bulgaria/Turkey (29°48′E, 39°17′N), Spain/ Portugal (30°W, 39°42′N), England/Wales (2°6′W, 52°42′N), and the Western Isles/ Isle of Skye (7°6′W, 57°17′N). The product moment correlation between genetic and geographic distances for all the groups in figure 5 was r=0.717, and, of 10,000 random permutations of the distance matrices, none yielded values ⩾0.717. When the North Atlantic island populations are omitted from the matrices used in the Mantel test, we obtain r=0.713, with the same high degree of significance.
Estimates of Admixture
In this section, we apply the heuristic approach to estimation of the relative contributions of the Gaelic and Scandinavian source populations to the mtDNA pools of the island and coastal populations of the North Atlantic. Table 4 shows the estimated ancestral proportions for each of the five admixed populations, along with 95% confidence intervals.
Table 4.
Scandinavian and Gaelic Mitochondrial Ancestry in the North Atlantic Islanders and the Coastal Population of Northwest Scotland
|
mρ (95% CI) for Ancestry |
||
| Population | Scandinavian | Gaelic |
| Iceland | .375 (.215–.570) | .625 (.430–.785) |
| Orkney | .355 (.130–.645) | .645 (.355–.870) |
| Western Isles | .115 (.025–.355) | .885 (.645–.975) |
| Skye | .125 (.025–.405) | .875 (.595–.975) |
| NW Scottish coast | .135 (.035–.355) | .865 (.645–.965) |
The estimate of Scandinavian ancestry ranges from 11.5% in the Western Isles to 37.5% in the Icelanders. For the Scottish island and coastal populations, these findings are consistent with historical, archaeological, and linguistic evidence of Norse settlements in the North Atlantic region. Of the Scottish populations, Orkney evidently has the closest matrilineal links with Scandinavia. The Western Isles, the Isle of Skye, and the coastal population of northwest Scotland all exhibit similarly low levels of Scandinavian mtDNA ancestry. In contrast to expectations based on historical records, the Icelanders have a similar proportion of Scandinavian mtDNA ancestry to that of the Orkney islanders, indicating that the majority of Icelandic matrilines originated from the British Isles.
Discussion
This study of mtDNA variation in the North Atlantic demonstrates the utility of uniparental loci in contributing to the understanding of the origins of human populations not only in broad terms over millennia but also in regional studies well within the historical period. The analysis has revealed important details of the relationships and demographic histories of the North Atlantic island populations. The islanders of Skye are clearly identified as the least diverse of the North Atlantic island populations. The populations of Iceland, Orkney, and the Western Isles exhibit greater levels of genetic diversity, and judging from θk values, had similar female effective population sizes (Nfe). This result appears to conflict with current population sizes on these islands, as the Icelanders presently number just over 270,000, whereas the populations of Orkney and the Western Isles number ∼20,000 and ∼30,000, respectively. However, the Icelandic population has undergone a fivefold expansion in the late 19th and 20th centuries, from a historically stable population size of ∼50,000 (Steffensen 1975). In contrast, the populations of all the Scottish islands have declined over the same period. In 1891, the Icelanders numbered 70,927, whereas Orkney had 30,453 inhabitants and the population of the Western Isles was 44,987. This difference between the populations of Iceland and the Scottish islands was even smaller in earlier centuries. Thus, the reported θk values do seem to reflect recent historical female effective population sizes for all three islands.
On the basis of an expected positive association between θk values and the proportion of private lineages across populations, we observed a relative excess of private lineages in the Icelanders compared with the Scottish island populations. Although this may be explained partially by greater sampling saturation in Iceland, it is also likely to reflect the geographic isolation of the Icelanders, which will have hindered gene flow to and from the island. The relative lack of private lineages in the Scottish islands suggests higher levels of gene flow and accords with the recorded depopulation of the last 200 years, which would have brought many private island lineages to the Scottish mainland.
As is the case with most European populations, there is some overlap between the mtDNA pools of the Scandinavians and Gaels. Of a total of 416 lineages found in these two populations, 73 (17.5%) were shared. Between 35% and 40% of lineages found in the Scottish islands and 26% of those found in Iceland are shared with both the Scandinavian and Gaelic populations. However, there are differences between these mtDNA pools that can be exploited to shed light on the genetic history of the North Atlantic island populations. The inhabitants of the Scottish islands share two to seven times more of their lineages exclusively with Gaels than they do with Scandinavians. This difference is smaller for the Icelanders, but nonetheless indicates a closer link to Gaels.
The pattern of lineage sharing is also informative about interrelationships among the North Atlantic island mtDNA pools. Of the lineages whose distribution is restricted to the North Atlantic islands, only two are not private lineages (one belongs to haplogroup V and has the substitution motif 16124C 16298C 16362C; the other belongs to subcluster T2, with the motif 16093C 16126C 16153A 16294T). In both cases, the lineages are shared between Icelanders and the Western Islanders. Intriguingly, the medieval record of the settlement of Iceland (The Book of Settlements 1972) indicates that the Western Isles were frequently the place of departure for settlement voyages to Iceland and that a number of indigenous Western Islanders accompanied such voyages. In all, the Western Isles are mentioned 22 times in The Book of Settlements, Orkney is mentioned 7 times, the Faroe Islands are mentioned 3 times, and Shetland is mentioned 2 times. Although anecdotal, this apparent link between Iceland and the Western Isles suggests that mtDNA lineages can be used to identify recent migration contributions from closely related populations.
The different estimates of Scandinavian ancestry for mtDNA lineages in Orkney, the Western Isles, the Isle of Skye, and the northwest coastal region of Scotland are consistent with the intensity of Norse activities. Although Norse influence extended to all these places, the Earldom of Orkney (established by the Norwegian king in 900 a.d.) was the political and strategic hub of Norse activities in the North Atlantic region. The sheer number of Norse archaeological sites in Orkney and historical accounts in medieval texts, such as Orkneyinga Saga, testify to the thorough Norse occupation of this island group. Clover (1988) has pointed out that, like many human range expansions, the movement of Norse people during the Viking period was male dominated. Thus, it is recorded that initial Viking activities in the British Isles involved raiding parties of Norse men (Davies 1999; Sawyer 1999). After these early raids and for >50 years before Iceland was discovered and colonized, many of the same Viking men settled and intermarried with existing populations in Shetland, Orkney, the Western Isles, the Isle of Man, and coastal regions of Ireland, Scotland, and northern England (Jones 1984; Davies 1999; Sawyer 1999). In Orkney, it would have been natural for whole families to move from Scandinavia—those of the Earl, his kinsfolk, and their retainers. However, most of the Norse who raided, traded, and settled elsewhere in the British Isles were young and unmarried men, many of whom would not have been closely linked to the political elite in their homeland. For these young men, it was natural to acquire wives from the indigenous populations (Clover 1988). Later on, many of these admixed families took part in the largest movement of people during the Viking era, leaving the coastal settlements in the British Isles for a new life in Iceland.
A recent study has estimated that ∼80% of Icelandic Y chromosomes are of Scandinavian origin (Helgason et al. 2000c). In conjunction with a much lower estimate of 37.5% Scandinavian ancestry of Icelandic mtDNA lineages, this suggests that Iceland was settled by a group consisting of (or descended from) primarily Norse men and Gaelic women. Although this only partially agrees with traditional accounts of Icelandic history (Jóhannesson 1956; Magnússon 1977), it seems plausible. In a detailed study of the medieval Icelandic Book of Settlements, Steffensen (1975) reported that, of the 48 women whose origin is recorded, 16.7% of their genealogical lines are from the British Isles, whereas only 4.7% of the 220 men whose genealogy is recorded are attributed British ancestry. It is known that the settlers mentioned in The Book of Settlements represent only a small part of the total colonizing population, which is thought to have numbered between 8,000 and 20,000 individuals (Steffensen 1975). According to some historians, many of those not recorded in The Book of Settlements originated from the British Isles—for example, slaves and females captured in Viking raids (Steffensen 1975; Clover 1988; Karras 1988; Sigurðsson 1988). That the authors of The Book of Settlements tended to be biased towards including genealogical links to high-ranking Norse ancestors is reflected in the following passage from the medieval text’s introduction: “People often say that writing about the Settlements is irrelevant learning, but we think we can better meet the criticism of foreigners when they accuse us of being descended from slaves or scoundrels, if we know for certain the truth about our ancestry” (The Book of Settlements 1972, p. 6). According to the mitochondrial data, the truth seems to be that a sizeable portion of Icelandic lines of descent are traced back 1,100 years to females whose ancestry was firmly anchored in the British Isles.
In general, our findings indicate a good agreement between mtDNA variation and geography in Europe. In the case of the North Atlantic islands, ρ distances demonstrate that the most closely related mtDNA pools are geographic neighbors from the British Isles and Scandinavia. Genetic distances based on lineage-cluster frequencies demonstrate an even more marked and highly significant geographic structure of European mtDNA variation. These findings contrast with those of Simoni et al. (2000a), who detected only very limited geographic patterns in European mtDNA variation using spatial autocorrelation statistics (but see a different interpretation by Torroni et al. 2000). Although the methods are not strictly comparable, the very different conclusions reached by Simoni et al. (2000a) suggests the need for further examination of this problem. One potentially important difference between the two studies is that the average sample size in Simoni et al.’s (2000a) study was 73 (minimum 15 and maximum 249), whereas, in our analysis of genetic and geographic distances, the average sample size was 352 (minimum 128 and maximum 891). As shown in table 1, sampling saturation varies considerably among populations. Even for populations in which the sample size is >200, there are still likely to be many unsampled lineages, and our knowledge of lineage and lineage cluster frequencies is far from exact (see Pfeiffer et al. 1999; Helgason et al. 2000a, 2000b,). The combined effects of a high mutation rate and large effective population size of females in some European populations suggests much larger sample sizes are required for a comprehensive evaluation of geographic patterns of mtDNA variation.
Acknowledgments
We are indebted to the volunteers who allowed us to use their DNA for this study and to the Scottish Blood Service for their help. We thank Emilce Vega for help with the collection and analysis of samples. This research received support from The Wellcome Trust. A. H. was supported by an Overseas Research Scheme award from the Committee of Vice-Chancellors and Principals of Universities and Colleges in the United Kingdom, 1997–1999.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Web/Genbank/index.html (for study results [accession numbers AY026032–AY026032])
- Mitochondrial DNA Concordance, http://shelob.bioanth.cam.ac.uk/mtDNA (for mtDNA HVS1 sequences)
References
- Anderson S, Bankier AT, Barrell BG, Debruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bodmer JG, DeLuca M, Moses JH, Heyes JM, Marsh SGE (1996) HLA class I and II study in an Orcadian population. Hum Immunol 47:O2658909580 [Google Scholar]
- Boyce AJ, Holdsworth VML, Brothwell DR (1973) Demographic and genetic studies in the Orkney Islands. In: Roberts DF, Sunderland E (eds) Genetic variation in Britain. Taylor and Francis, London [Google Scholar]
- Berry RJ, Muir VML (1975) The natural history of man in Shetland. J Biosoc Sci 7:319–344 [DOI] [PubMed] [Google Scholar]
- Bertranpetit J, Sala J, Calafell F, Underhill PA, Moral P, Comas D (1995) Human mitochondrial-DNA variation and the origin of Basques. Ann Hum Genet 59:63–81 [DOI] [PubMed] [Google Scholar]
- Bjarnason Ó, Bjarnason V, Edwards JH, Friðriksson S, Magnússon M, Mourant AE, Tills D (1973) The blood groups of Icelanders. Ann Hum Genet 36:425–458 [DOI] [PubMed] [Google Scholar]
- Book of Settlements, The: Landnámabók (1972) Pálsson H, Edwards P (trans) University of Manitoba Press, Winnipeg [Google Scholar]
- Calafell F, Underhill P, Tolun A, Angelicheva D, Kalaydjieva L (1996) From Asia to Europe: mitochondrial DNA sequence variability in Bulgarians and Turks. Ann Hum Genet 60:35–49 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Am J Hum Genet 19:233–257 [PMC free article] [PubMed] [Google Scholar]
- Clegg EJ (1985) Blood group variation in the Isle of Lewis. Ann Hum Biol 12:345–361 [DOI] [PubMed] [Google Scholar]
- Clover CJ (1988) The politics of scarcity: notes on the sex ratio in early Scandinavia. Scand Stud 60:147–188 [Google Scholar]
- Collins R (1991) Early medieval Europe 300–1000. Macmillan, Hampshire [Google Scholar]
- Comas D, Calafell F, Mateu E, Perez-Lezaun A, Bertranpetit J (1996) Geographic variation in human mitochondrial DNA control region sequence: the population history of Turkey and its relationship to the European populations. Mol Biol Evol 13:1067–1077 [DOI] [PubMed] [Google Scholar]
- Corráin DÓ (1999) Ireland, Wales, Man and the Hebrides. In: Sawyer P (ed) The Oxford illustrated history of the Vikings. Oxford University Press, Oxford, pp 83–109 [Google Scholar]
- Corte-Real H, Macaulay VA, Richards MB, Hariti G, Issad MS, Cambon-Thomsen A, Papiha S, Bertranpetit J, Sykes BC (1996) Genetic diversity in the Iberian peninsula determined from mitochondrial sequence analysis. Ann Hum Genet 60:331–350 [DOI] [PubMed] [Google Scholar]
- Davies N (1999) The isles: a history. Macmillan, London [Google Scholar]
- Delghandi M, Utsi E, Krauss S (1998) Saami mitochondrial DNA reveals deep maternal lineage clusters. Hum Hered 48:108–114 [DOI] [PubMed] [Google Scholar]
- Ewens WJ (1972) The sampling theory of selectively neutral alleles. Theor Popul Biol 3:87–112 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt HJ (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Francalacci P, Bertranpetit J, Calafell F, Underhill PA (1996) Sequence diversity of the control region of mitochondrial DNA in Tuscany and its implications for the peopling of Europe. Am J Phys Anthropol 100:443–460 [DOI] [PubMed] [Google Scholar]
- Graham-Campbell J, Batey CE (1998) Vikings in Scotland: an archaeological survey. Edinburgh University Press, Edinburgh [Google Scholar]
- Handt O, Meyer S, von Haeseler A (1998) Compilation of human mtDNA control region sequences. Nucleic Acids Res 26:126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handt O, Richards M, Trommsdorff M, Kilger C, Simanainen J, Georgiev O, Bauer K, Stone A, Hedges R, Schaffner W, Utermann G, Sykes B, Paabo S (1994) Molecular genetic analyses of the Tyrolean Ice Man. Science 264:1775–1778 [DOI] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir S, Gulcher J, Stefánsson K, Ward R (2000a) Sampling saturation and the European mtDNA pool: implications for detecting genetic relationships among populations. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population prehistory of Europe. Oxbow Books, Oxford [Google Scholar]
- Helgason A, Sigurðardóttir S, Gulcher JR, Ward R, Stefánsson K (2000b) mtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet 66:999–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir S, Nicholson J, Sykes B, Hill E, Bradley DG, Bosnes V, Gulcher JR, Ward R, Stefánsson K (2000c) Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet 67:697–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Jaksch M, Bezold R, Mertens S, Aholt S, Paprotta A, Gerbitz KD (1997) Population genetics and disease susceptibility: characterization of central European haplogroups by mtDNA gene mutations, correlation with D loop variants and association with disease. Hum Mol Genet 6:1835–1846 [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Jóhannesson J (1956) Íslendinga saga. Almenna Bókafélagið, Reykjavík [Google Scholar]
- Jones G (1984) A history of the Vikings. 2d ed. Oxford University Press, Oxford [Google Scholar]
- Karras RM (1988) Slavery and society in medieval Scandinavia. Yale University Press, New Haven [Google Scholar]
- Keynes S (1999) The Vikings in England, c. 790–1016. In: Sawyer P (ed) The Oxford illustrated history of the Vikings. Oxford University Press, Oxford, pp 48–82 [Google Scholar]
- Kittles RA, Bergen AW, Urbanek M, Virkkunen M, Linnoila M, Goldman D, Long JC (1999) Autosomal, mitochondrial, and Y chromosome DNA variation in Finland: evidence for a male-specific bottleneck. Am J Phys Anthropol 108:381–399 [DOI] [PubMed] [Google Scholar]
- Lutz S, Weisser HJ, Heizmann J, Pollak S (1999) Erratum. Int J Legal Med 112:145–150 [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, BonneTamir B, Sykes B, Torroni A (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnússon SA (1977) Northern Sphinx. McGill University Press, Montreal [Google Scholar]
- Miller KWP (1996) Molecular genetic analysis of human populations in Orkney and the North Atlantic region. Doctoral thesis, Magdalene College, University of Cambridge, Cambridge, United Kingdom [Google Scholar]
- Opdal SH, Rognum TO, Vege A, Stave AK, Dupuy BM, Egeland T (1998) Increased number of substitutions in the D-loop of mitochondrial DNA in the sudden infant death syndrome. Acta Paediatr 87:1039–1044 [DOI] [PubMed] [Google Scholar]
- Orekhov V, Poltoraus A, Zhivotovsky LA, Spitsyn V, Ivanov P, Yankovsky N (1999) Mitochondrial DNA sequence diversity in Russians. Febs Lett 445:197–201 [DOI] [PubMed] [Google Scholar]
- Parson W, Parsons TJ, Scheithauer R, Holland MM (1998) Population data for 101 Austrian Caucasian mitochondrial DNA d-loop sequences: application of mtDNA sequence analysis to a forensic case. Int J Legal Med 111:124–132 [DOI] [PubMed] [Google Scholar]
- Pfeiffer H, Brinkmann B, Huhne J, Rolf B, Morris AA, Steighner R, Holland MM, Forster P (1999) Expanding the forensic German mitochondrial DNA control region database: genetic diversity as a function of sample size and microgeography. Int J Legal Med 112:291–298 [DOI] [PubMed] [Google Scholar]
- Piercy R, Sullivan KM, Benson N, Gill P (1993) The application of mitochondrial-DNA typing to the study of white Caucasian genetic identification. Int J Legal Med 106:85–90 [DOI] [PubMed] [Google Scholar]
- Pinto F, Gonzalez AM, Hernandez M, Larruga JM, Cabrera VM (1996) Genetic relationship between the Canary Islanders and their African and Spanish ancestors inferred from mitochondrial DNA sequences. Ann Hum Genet 60: 321–330 [DOI] [PubMed] [Google Scholar]
- Pult I, Sajantila A, Simanainen J, Georgiev O, Schaffner W, Paabo S (1994) Mitochondrial DNA sequences from Switzerland reveal striking homogeneity of European populations. Biol Chem Hoppe Seyler 375:837–840 [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 [DOI] [PubMed] [Google Scholar]
- Rafnson S (1999) The Atlantic islands. In: Sawyer P (ed) The Oxford illustrated history of the Vikings. Oxford University Press, Oxford [Google Scholar]
- Richards MB, Macaulay VA, Bandelt HJ, Sykes BC (1998) Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet 62: 241–260 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, Villems R, Thomas M, Rychkov S, Rychkov O, Rychkov Y, Golge M, Dimitrov D, Hill E, Bradley D, Romano V, Cali F, Vona G, Demaine A, Papiha S, Triantaphyllidis C, et al (2000) Tracing European founder lineages in the near eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Richards M, Corte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, Hedges R, Bandelt HJ, Sykes B (1996) Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet 59:185–203 [PMC free article] [PubMed] [Google Scholar]
- Roberts DF (1985) Genetic structure in Orkney. Man (New Ser) 20:131–141 [Google Scholar]
- Roberts DF (1990) Genetic affinities of the Shetland Islanders. Ann Hum Biol 17:121–132 [DOI] [PubMed] [Google Scholar]
- Rousselet F, Mangin P (1998) Mitochondrial DNA polymorphisms: a study of 50 French Caucasian individuals and application to forensic casework. Int J Legal Med 111:292–298 [DOI] [PubMed] [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus ML, Aula P, Beckman L, Tranebjaerg L, Geddedahl T, Isseltarver L, DiRienzo A, Paabo S (1995) Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Res 5:42–52 [DOI] [PubMed] [Google Scholar]
- Sajantila A, Salem AH, Savolainen P, Bauer K, Gierig C, Paabo S (1996) Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc Natl Acad Sci USA 93:12035–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo A (1998) mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet 6:365–375 [DOI] [PubMed] [Google Scholar]
- Sawyer P (1999) The age of the Vikings and before. In: Sawyer P (ed) The Oxford illustrated history of the Vikings. Oxford University Press, Oxford [Google Scholar]
- Schneider S, Kueffer JM, Roessli D, Excoffier L (1997) Arlequin 1.1: software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland [Google Scholar]
- Sigurðsson G (1988) Gaelic influence in Iceland. Menningarsjóður, Reykjavík [Google Scholar]
- Simoni L, Calafell F, Pettener D, Bertranpetit J, Barbujani G (2000a) Geographic patterns of mtDNA diversity in Europe. Am J Hum Genet 66:262–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni L, Calafell F, Pettener D, Bertranpetit J, Barbujani G (2000b) Reconstruction of prehistory on the basis of genetic data. Am J Hum Genet 66:1177–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffenson J (1975) Menning og meinsemdir: ritgerðarsafn um mótunarsögu íslenskrar þjóðar og baráttu hennar við hungur og sóttir. Ísafoldarprentsmiðja, Reykjavík [Google Scholar]
- Stenico M, Nigro L, Bertorelle G, Calafell F, Capitanio M, Corrain C, Barbujani G (1996) High mitochondrial sequence diversity in linguistic isolates of the Alps. Am J Hum Genet 59:1363–1375 [PMC free article] [PubMed] [Google Scholar]
- Thompson EA (1973) The Icelandic admixture problem. Ann Hum Genet 37:69–80 [DOI] [PubMed] [Google Scholar]
- Torroni A, Richards M, Macaulay V, Forster P, Villems R, Nørby S, Savontaus ML, Huoponen K, Scozzari R, Bandelt HJ (2000) mtDNA haplogroups and frequency patterns in Europe. Am J Hum Genet 66:1173–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman EM (1984) Techniques for estimating genetic admixture and applications to the problem of the origin of the Icelanders and the Ashkenazi Jews. Hum Genet 67:441–448 [DOI] [PubMed] [Google Scholar]