Abstract
Human tissues acquire somatic mitochondrial DNA (mtDNA) mutations with age. Very high levels of specific mtDNA mutations accumulate within individual cells, causing a defect of mitochondrial oxidative metabolism. This is a fundamental property of nondividing tissues, but it is not known how it comes about. To explore this problem, we developed a model of mtDNA replication within single human cells. Using this model, we show that relaxed replication of mtDNA alone can lead, through random genetic drift, to the clonal expansion of single mutant events during human life. Significant expansions primarily develop from mutations acquired during a critical period in childhood or early adult life.
Somatic mitochondrial DNA (mtDNA) mutations in humans accumulate with age. mtDNA mutations are generally not detected in young, healthy control subjects, but older individuals typically harbor a wide range of different mtDNA deletions in postmitotic tissues, such as skeletal muscle, myocardium, and brain (Cortopassi and Arnheim 1990; Corral-Debrinski et al. 1992a, 1992b). The overall amount of mutant mtDNA is generally very low (<2%), but individual cells may contain high percentage levels of a single mutant species (Brierley et al. 1998). When the proportion of mutant mtDNA exceeds a critical threshold level, this leads to a defect of mitochondrial oxidative phosphorylation (Schon et al. 1997). This may compromise the function of the whole organ, particularly if the affected cells perform an integral role in a complex network, as in the central nervous system. Our model of random mutation and random intracellular drift of mtDNA explains how these clonal expansions can occur.
mtDNA accumulates somatic mutations throughout human life (Wallace 1992). Unlike nuclear DNA, mtDNA is continuously turning over, independent of the cell cycle (relaxed replication [Bogenhagen and Clayton 1977]). It was originally suggested that deleted mtDNA might accumulate within single cells, because the smaller molecules could replicate faster than larger, wild-type molecules (Wallace 1992). This explanation is no longer accepted, because the entire mtDNA molecule is replicated in <90 minutes, which is a much shorter time than that between replications (Shadel and Clayton 1997). Since the time taken to replicate wild-type mtDNA is not rate limiting, the faster replication of deleted molecules should not influence the number of deleted and wild-type molecules over a period of time. Furthermore, this mechanism cannot explain the accumulation of mtDNA point mutations seen in patients with mtDNA disease (e.g., see Weber et al. 1997). An alternative hypothesis is that “sick mitochondria,” containing high levels of mutant mtDNA, proliferate in response to the respiratory chain defect and eventually repopulate the cell (Yoneda et al. 1992). Other potential mechanisms have been put forward (de Gray 1997), but these are not supported by the available data (Brierley et al. 1998; Wallace 1999). All these explanations assume a selective mechanism leading to the preferential replication (replicative advantage) or longevity of mutant mtDNA over wild-type mtDNA, but this is not necessarily the case. It is currently not possible technically to measure the amount of mutant mtDNA within intact living cells at multiple time points. We therefore developed a model of cellular mitochondrial genetics to explore the possibility that random genetic drift alone might lead to the clonal accumulation of mutant mtDNA within single postmitotic cells. The model was based on a solid foundation of experimentally derived biological parameters that reflect current understanding of mtDNA replication (Shadel and Clayton 1997).
We simulated relaxed replication in nondividing cells over the maximum accepted human life span of 120 years. Each cell contained an initial population, N, of freely intermixing identical mtDNA molecules. With each time step, molecules were randomly destroyed with a half-life, T1/2. New molecules were copied randomly from the remaining pool to maintain an approximately constant number of mtDNA molecules within each cell (Chinnery and Samuels 1999). mtDNA deletions are thought to arise through slippage mispairing of the replicating molecule (Schon et al. 1989). We simulated somatic copy-error mutation by randomly assigning a replicating molecule a new unique label with a probability, Pmut. All the descendents of this molecule inherited the same label, allowing the fate of each mutation to be monitored for the rest of the simulation. The label had no effect on the rate of destruction or replication of the simulated molecule.
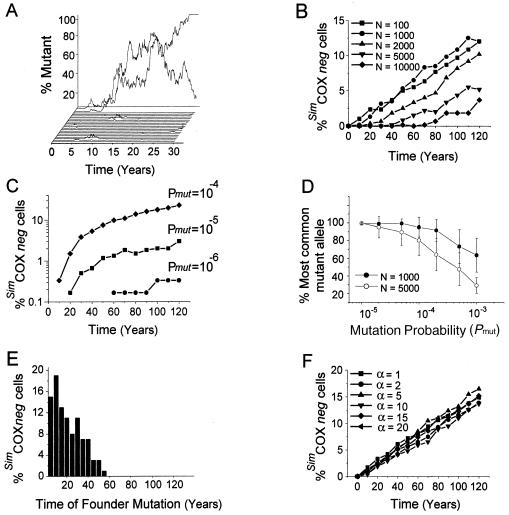
Diploid human cells contain 1,000–100,000 mtDNA molecules that are destroyed with a mean T1/2 of 10 d in postmitotic tissues (Gross et al. 1969). During the 30-year simulation of 600 cells, each containing 1,000 mtDNA molecules being destroyed with a T1/2 of 10 d and having a copy-error probability Pmut of 10−5, we observed rare mutation events (fig. 1A). In some simulated cells, the proportion of mutant mtDNA increased. Most of these expansions were lost over a short period of time, but occasionally the proportion of mutant mtDNA increased further to very high levels within a cell. These high levels were seen to increase and decrease randomly over the course of the simulations. We explored the mechanism further by studying the frequency distribution of the amount of mutant mtDNA in another 600 cells, making observations at 10-year intervals. In the first decade, almost all the simulated cells contained purely wild-type mtDNA or low levels of mutant mtDNA. As time progressed, we observed a gradual increase in the number of cells containing a greater proportion of mutant mtDNA. These findings are exactly what would be predicted for a model of random intracellular drift.
Figure 1.
A, Proportion of mutant mtDNA in single simulated cells. Each simulated cell contained 1,000 mtDNAs and had a half-life, T1/2, of 10 d and a copy-error (mutation) probability of 10−5 (measurements taken monthly for 30 years). For illustration, we have shown the results of 24 simulations from a total of 600. In the 600 cells, one mutant event went to fixation (trace at the back) and three others showed a significant expansion (one shown), but, in most, the mutations were rapidly lost. When these simulations were continued for a full 120 years (not shown on the graph), a mean of only 31.27 mutations occurred per cell, corresponding to the value predicted by the equation Nmut={NmtDNAPmutt ln(2)}/T1/2, where Nmut is the number of mutation events and t is the duration of the simulation in d. Many of these mutations were lost almost instantaneously (41% were lost within one T1/2, and 88% were lost within 10 T1/2), but, in some cells, the proportion of mutant mtDNA increased. B, Proportion of cells containing >60% mutant mtDNA (SimCOXneg cells) accumulates with age. Each line represents 600 simulated cells, followed for 120 years, containing a given number of mtDNA molecules, N, and with a copy-error (mutation) probability 5 x 10−5 and a T1/2 of 10 d (measurements taken every 10 years). The proportion of SimCOXneg cells increases with age. Similar rates of accumulation of SimCOXneg cells were seen for N=100 and N=1,000. Greater numbers of N were associated with a lower number of SimCOXneg cells at each age. C, Effect of Pmut on the proportion of cells containing >60% mutant mtDNA (SimCOXneg cells). Each line represents 600 simulated cells followed for 120 years. For cells containing 1,000 mtDNA molecules and a T1/2 of 10 d, low mutation probabilities (Pmut of 10−7 and 10−6) are associated with a very low or undetectable incidence of SimCOXneg cells throughout life. Higher mutation probabilities lead to a greater number of SimCOXneg cells accumulating throughout a simulated human life. D, Proportion of clonally expanded mutant mtDNA in simulated cells with different copy-error (mutation) probabilities. Each data point represents 1,000–2,000 simulated cells, each containing 1,000 or 5,000 mtDNA molecules (see legend on graph) with a T1/2 of 10 d. The ordinate shows the proportion of mtDNA that was the most populous mutant allele (clonally expanded mtDNA) at 80 years. For moderate mutation probabilities (Pmut of 10−5 to 10−4), the vast majority of mutant mtDNA within a single simulated cell is identical (clonal expansion). For much higher mutation probabilities (Pmut >10−4), a mixture of mutant species is seen within the simulated cells. E, Age when mtDNA copy errors (mutations) occurred, which led to significant clonal expansions by age 80 years. Of 32,000 simulated cells containing 5,000 mtDNA with a copy-error (mutation) probability 10−5 and T1/2 of 10 d, 123 contained >60% mutant mtDNA (SimCOXneg cells) at age 80 years. The percentage distribution of the age at which these mutations occurred is shown in the figure. Seventy-eight percent of the original copy errors occurred in the first three decades of life, and 46% began at age <15 years. F, Effect of mitochondrial proliferation on the rate of accumulation of SimCOXneg cells with age. These simulations were based on the assumption that nondividing cells maintain an optimal number of wild-type mtDNA molecules (Chinnery and Samuels 1999). If the number of wild-type molecules is reduced by mutation, the cell responds by the nonselective proliferation of all mtDNA molecules to redress the balance. The maximum level of mtDNA proliferation, α, times the normal mtDNA number, was based on the maximum degree of mitochondrial proliferation measured in skeletal muscle fibers from patients harboring high levels of mutant mtDNA (see Chinnery and Samuels 1999). We studied the effect of varying α by simulating 1,000 postmitotic cells containing 100 mtDNA molecules with a T1/2 of 10 d over a human life span of 120 years, with a Pmut of 5×10-5. Proliferation had no significant effect on the rate of accumulation of SimCOXneg cells with age.
To determine the biological significance of these simulations, we explored the parameters that governed the behavior of the model using biologically plausible values based on experimental observation. Skeletal muscle fibers develop a biochemical defect of cytochrome c oxidase (COX) when the level of an mtDNA deletion exceeds a critical threshold level of 50%–60% (Hayashi et al. 1991). Thus, by studying the number of cells that contained >60% mutant mtDNA, we were simulating the accumulation of COX-negative cells (SimCOXneg cells). It is important to note that for these simulations, altering the threshold value would not alter the overall behavior of the model, although it would alter the time course.
We then repeated the simulation over a range of different values for N, the number of mtDNA molecules within the cell (fig. 1B). Reducing the value of N to <1,000 had no significant effect on the number of SimCOXneg cells at any time point. Higher values of N reduced the number of SimCOXneg cells. For subjects aged <60 years, there were no cells with 10,000 mtDNAs that were SimCOXneg, but, for those aged >60 years, we saw a slow increase in the number of these cells, reaching 1%–4% between 80 and 120 years (fig. 1B). Altering Pmut had a marked effect on the number of SimCOXneg cells (fig. 1C). A very high Pmut (10−4) led to the rapid accumulation, in the first few decades of life, of SimCOXneg cells. With more conservative values for Pmut (10−6), cells crossed the 60% threshold only after the sixth decade (fig. 1C).
We were interested in determining whether SimCOXneg cells contained multiple mtDNA mutations or a single clonally expanded species for different values of N (fig. 1D). When Pmut was very high (10−3), cells contained a mixed population of mutations. By contrast, when Pmut was <10−4, the majority (>80%) of mutated molecules within the cell were the same, indicating that the cells were repopulated with multiple copies of a single mutated mtDNA molecule originating from one mutation event.
These findings demonstrate that relaxed replication alone can cause marked intracellular drift in postmitotic cells during a human life span: a single mutated mtDNA molecule may expand clonally to reach very high levels in the absence of any replicative advantage. Close scrutiny of individual simulated cells led us to recognize that large clonal expansions took considerable time. We identified 123 cells containing clonal expansions from a population of 32,000 cells, and we measured the time when the original copy-error occurred. After simulation of 80 years of a subject's life, for SimCOXneg cells containing 5,000 mtDNAs, 78% of the original copy error occurred in the first three decades of the subject's life, and 46% began at age <15 years (fig. 1E). This suggests that biologically relevant somatic mtDNA mutations probably occur during a critical period throughout childhood and early adulthood. Mutations acquired in later adult life do not have sufficient time to reach significant levels by random genetic drift alone.
These observations contrast with the “vicious cycle” hypothesis, which predicts that all mtDNA mutations accumulate in aged individuals because an age-related decline in respiratory chain efficiency leads to the increased production of oxygen free radicals, further damaging mtDNA and thereby further compromising respiratory chain function (Wallace 1999). If the accumulation of mutant mtDNA were due to repeated new mutations, then COX-negative cells in older individuals would contain a huge range of different mutations and not the single clonal expansions that have been observed (Brierley et al. 1998). It is important to note, however, that our model and the vicious cycle hypothesis are not mutually exclusive. The clonal expansion of mtDNA defects may lead to a mitochondrial respiratory chain defect, the increased generation of free radicals, and the subsequent generation of new mtDNA mutations. However, on the basis of the model described in this report, it is unlikely that these new mutations would have any biological significance, because the vicious cycle would begin only after a cell has developed a respiratory chain defect due to the clonal expansion of a single mtDNA mutation (fig. 1D).
It is very difficult to measure the rate of mtDNA mutation experimentally. Our simulations show that many new mutant events will be lost almost instantaneously through intracellular drift (fig. 1A). Even with single-cell techniques, only a small proportion of mutant events will reach detectable levels. Limited data suggest that a conservative estimate of the mutation rate for one specific deletion (the 4.7-kb “common” deletion) is 4.60×10-9 to 1.07×10-7 per mtDNA replication in cultured fibroblasts (Shenkar et al. 1996), but this may not reflect the situation in vivo. Even with the sensitive techniques used by Shenkar et al. (1996), relaxed replication would lead to the rapid loss of new mutant events before they can be detected (see legend to fig. 1). The common deletion is one of many potential copy-error mutations that accumulate with age (Melov et al. 1995; Liu et al. 1998), and an estimated mutation rate in postmitotic tissues of 5×10-5 may be highly conservative (e.g., see Denver et al. 2000). The simulations presented in this report indicate that a mutation rate of 5×10-5 is sufficient to cause a low frequency of cells containing >60% mutant mtDNA to develop during human life purely through random intracellular drift. The rate of somatic mtDNA mutation may be increased by genetic factors, such as thymidine phosphorylase deficiency (Nishino et al. 1999); Ant1 gene mutations (Kaukonen et al. 2000); or environmental factors, such as ischemia (Corral-Debrinski et al. 1992b), inflammation (Blume et al. 1997), or radiation (Kubota et al. 1997). The model presented here predicts that any change in the mutation rate will alter the age-related accumulation of cells containing significant clonal expansions, particularly if this happens during childhood or early adult life.
Biochemically deficient skeletal muscle fibers often show evidence of mitochondrial proliferation (ragged-red fibers) with an associated increase in the amount of mtDNA (Kaufmann et al. 1996). We incorporated pathological mtDNA proliferation into our model, as described elsewhere (Chinnery and Samuels 1999). In brief, when the number of wild-type molecules within a cell was reduced by mutation, it responded by nonselectively proliferating its entire mtDNA content in an attempt to redress the balance. These simulations showed that proliferation had no significant effect on the rate of accumulation of SimCOXneg cells (fig. 1F). Within single cells, proliferation increases the amount of both mutant and wild-type mtDNA, maintaining the overall allele proportions in the affected cell but increasing the mutation load over the whole tissue.
Most postmitotic cells are estimated to contain between 5,000 and 10,000 mitochondrial genomes (Lightowlers et al. 1997). In the model we describe in this report, we assumed that there is thorough intermixing of all mtDNA molecules, but in living cells this may not involve the whole cell. It is unlikely that there would be complete mixing of mitochondrial genomes in larger, highly structured cells, such as skeletal muscle and neurons (Bereiter-Hahn and Voth 1994; Davis and Clayton 1996). Under these circumstances, there would be more mixing between adjacent mtDNAs and less between those many millimeters apart. This would reduce the effective mtDNA population size in the model we describe, making rapid drift more likely.
The simulation data presented here demonstrate that relaxed replication alone may lead to the clonal expansion of somatic mtDNA mutations during human life. Using independently derived parameters based on experimental observation, we predict that ⩽4% of postmitotic human cells will be COX negative by the 80th birthday of subjects. This is in striking agreement with histochemical observations of healthy older human muscle and brain (Muller-Hocker 1990; Brierley et al. 1998; Cottrell et al. 2000), in which ⩽5% of postmitotic cells develop COX deficiency by the eighth decade of life.
Acknowledgments
Support for this work was provided by the Wellcome Trust (to P.F.C. and D.M.T.) and The Medical Research Council UK (to J.L.E. and D.M.T.).
References
- Bereiter-Hahn J, Voth M (1994) Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech 27:198–219 [DOI] [PubMed] [Google Scholar]
- Blume G, Pestronk A, Frank B, Johns DR (1997) Polymyositis with cytochrome oxidase negative muscle fibres: early quadriceps weakness and poor response to immunosuppressive therapy. Brain 120:39–45 [DOI] [PubMed] [Google Scholar]
- Bogenhagen D, Clayton DA (1977) Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell 11:719–727 [DOI] [PubMed] [Google Scholar]
- Brierley EJ, Johnson MA, Lightowlers RN, James OFW, Turnbull DM (1998) Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann Neurol 43:217–223 [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Samuels DC (1999) Relaxed replication of mtDNA: a model with implications for the expression of disease. Am J Hum Genet 64:1158–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC (1992a) Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet 2:324–329 [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Shoffner JM, Lott MT, Wallace DC (1992b) Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat Res 275:169–180 [DOI] [PubMed] [Google Scholar]
- Cortopassi GA, Arnheim N (1990) Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res 18:6927–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell DA, Ince PG, Blakely EL, Johnson MA, Chinnery PF, Hanna MG, Turnbull DM (2000) Neuropathological and histochemical changes in a multiple mitochondrial deletion disorder. J Neuropathol Exp Neurol 59:621–627 [DOI] [PubMed] [Google Scholar]
- Davis AF, Clayton DA (1996) In situ localisation of mitochondrial DNA replication in intact mammalian cells. J Cell Biol 135:883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gray AD (1997) A proposed refinement of the mitochondrial free radical theory of aging. Bioessays 19:161–166 [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK (2000) High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science 289:2342–2344 [DOI] [PubMed] [Google Scholar]
- Gross NJ, Getz GS, Rabinowitz M (1969) Apparent turnover of mitochondrial deoxyribonucleic acid and mitochondrial phospholipids in the tissues of the rat. J Biol Chem 244:1552–1562 [PubMed] [Google Scholar]
- Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I (1991) Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc Natl Acad Sci USA 88:10614–10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann P, Koga Y, Shanske S, Hirano M, DiMauro S, King MP, Schon EA (1996) Mitochondrial DNA and RNA processing in MELAS. Ann Neurol 40:172–180 [DOI] [PubMed] [Google Scholar]
- Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zevianni M, Comi GP, Keranen S, Peltonen L, Suomalainen A (2000) Role of Adenine Nucleotide Translocator 1 in mtDNA Maintenance. Science 289:782–785 [DOI] [PubMed] [Google Scholar]
- Kubota N, Hayashi J, Inada T, Iwamura Y (1997) Induction of a particular deletion in mitochondrial DNA by X rays depends on the inherent radiosensitivity of the cells. Radiat Res 148:395–398 [PubMed] [Google Scholar]
- Lightowlers RN, Chinnery PF, Turnbull DM, Howell N (1997) Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet 13:450–455 [DOI] [PubMed] [Google Scholar]
- Liu VW, Zhang C, Nagley P (1998) Mutations in mitochondrial DNA accumulate differentially in three tissues during aging. Nucleic Acids Res 26:1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S, Shoffner JM, Kaufman A, Wallace DC (1995) Marked increase in the number and variety of mitochondrial DNA rearrangements in aging human skeletal muscle. Nucleic Acids Res 23:4122–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Hocker J (1990) Cytochrome c oxidase deficient fibres in the hind limb muscle and diaphragm of a man without muscular disease: an age-related alteration. J Neurol Sci 100:14–21 [DOI] [PubMed] [Google Scholar]
- Nishino I, Spinazzola A, Hirano M (1999) Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science 283:689–692 [DOI] [PubMed] [Google Scholar]
- Schon EA, Bonilla E, DiMauro S (1997) Mitochondrial DNA mutations and pathogenesis. J Bioenerg Biomembr 29:131–149 [DOI] [PubMed] [Google Scholar]
- Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeviani M, DiMauro S (1989) A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science 244:346–349 [DOI] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA (1997) Mitochondrial DNA maintenance in vertebrates. Ann Rev Biochem 66:409–435 [DOI] [PubMed] [Google Scholar]
- Shenkar R, Navidi W, Tavare S, Dang MH, Chomyn A, Attardi G, Cortopassi G, Arnheim N (1996) The mutation rate of the human mtDNA deletion mtDNA4977. Am J Hum Genet 59:772–780 [PMC free article] [PubMed] [Google Scholar]
- Wallace DC (1992) Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science 256:628–632 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Mitochondrial diseases in mouse and man. Science 283:1482–1488 [DOI] [PubMed] [Google Scholar]
- Weber K, Wilson JN, Taylor L, Brierley E, Johnson MA, Turnbull DM, Bindoff LA (1997) A new mtDNA mutation showing accumulation with time and restriction to skeletal muscle. Am J Hum Genet 60:373–380 [PMC free article] [PubMed] [Google Scholar]
- Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G (1992) Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc Natl Acad Sci USA 89:11164–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]