Abstract
The activity of a new ketolide, ABT-773, was compared to the activity of the ketolide telithromycin (HMR-3647) against over 600 gram-positive clinical isolates, including 356 Streptococcus pneumoniae, 167 Staphylococcus aureus, and 136 Streptococcus pyogenes isolates. Macrolide-susceptible isolates as well as macrolide-resistant isolates with ribosomal methylase (Erm), macrolide efflux (Mef), and ribosomal mutations were tested using the NCCLS reference broth microdilution method. Both compounds were extremely active against macrolide-susceptible isolates, with the minimum inhibitory concentrations at which 90% of the isolates tested were inhibited (MIC90s) for susceptible streptococci and staphylococci ranging from 0.002 to 0.03 μg/ml for ABT-773 and 0.008 to 0.06 μg/ml for telithromycin. ABT-773 had increased activities against macrolide-resistant S. pneumoniae (Erm MIC90, 0.015 μg/ml; Mef MIC90, 0.12 μg/ml) compared to those of telithromycin (Erm MIC90, 0.12 μg/ml; Mef MIC90, 1 μg/ml). Both compounds were active against strains with rRNA or ribosomal protein mutations (MIC90, 0.12 μg/ml). ABT-773 was also more active against macrolide-resistant S. pyogenes (ABT-773 Erm MIC90, 0.5 μg/ml; ABT-773 Mef MIC90, 0.12 μg/ml; telithromycin Erm MIC90, >8 μg/ml; telithromycin Mef MIC90, 1.0 μg/ml). Both compounds lacked activity against constitutive macrolide-resistant Staphylococcus aureus but had good activities against inducibly resistant Staphylococcus aureus (ABT-773 MIC90, 0.06 μg/ml; telithromycin MIC90, 0.5 μg/ml). ABT-773 has superior activity against macrolide-resistant streptococci compared to that of telithromycin.
Increasing antimicrobial resistance, particularly in Streptococcus pneumoniae, has increased the need for drugs effective against resistant isolates. ABT-773 (A-195773.0) is a novel compound in the ketolide class with significant advantages over the general antimicrobial profile of macrolide antibiotics. Ketolide antibiotics are extremely active against macrolide-resistant streptococci with either low-level (macrolide efflux [Mef]) or high-level (ribosomal methylase [Erm]) resistance to macrolides (1). Ketolides also have excellent in vitro activities against macrolide-resistant organisms with inducible methylase expression, as these compounds appear to be noninducers of methylase (2, 15). ABT-773 has excellent in vitro potency against macrolide-resistant gram-positive pathogens and has entered phase III clinical trials for the treatment of bacterial upper and lower respiratory tract infections (3). In this study, the in vitro activities of ABT-773 against macrolide-resistant and -susceptible respiratory pathogens are compared to those of telithromycin, another ketolide antibiotic, as well as those of a representative of the macrolide class, erythromycin.
(This work was previously presented in part at the 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, Calif., 1999).
MATERIALS AND METHODS
Susceptibility testing.
Testing was performed using the broth microdilution method as described by NCCLS (8). S. pneumoniae and S. pyogenes were tested using Mueller-Hinton broth with 3% lysed horse blood (Becton Dickinson Microbiology Systems, Sparks, Md.). The test medium for Staphylococcus aureus was Mueller-Hinton broth. Quality control was performed according to NCCLS by using S. pneumoniae ATCC 49619 and Staphylococcus aureus ATCC 29213. Incubation was at 35°C for 18 to 24 h in ambient air. Bacterial strains used in this study were clinical isolates collected from 1994 to 2000 in the United States and Europe from several sources, including various surveillance studies and clinical trials.
ABT-773 and telithromycin were synthesized at Abbott Laboratories (Abbott Park, Ill.). Erythromycin, clindamycin, and penicillin were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Resistance determinants.
The resistance mechanisms of macrolide-resistant streptococci and staphylococci were determined by PCR using primers specific for known resistance determinants [mef(A), erm(A), erm(B), erm(C), and msr(A)] as previously described (10, 11). Amplification primers were chosen from sequences deposited in GenBank by using Oligo 5.0 (NBI Software, Plymouth, Minn.) as previously described. The inducible regulation of methylase in Staphylococcus aureus and S. pyogenes was identified by the double disk diffusion test and by susceptibility to clindamycin as previously described (9). All primers in this study were purchased from Sigma-Genosys (The Woodlands, Tex.).
S. pneumoniae isolates which were negative for known macrolide resistance markers were examined for mutations in 23S rRNA domains II and V and ribosomal proteins L4 and L22 to include regions where mutations associated with macrolide resistance have been previously described in streptococci and other species (6, 12-14). Primers for amplification and sequence of the DNA regions that encode the 23S rRNA (rrl) and the ribosomal proteins L4 (rplD) and L22 (rplV) were chosen from publicly available DNA sequences (Streptococcus pneumoniae TIGR4 rDNA, SPSPrrnaD23S6; L4 TIGR4, SP0210; and L22 TIGR4, SPO214) (http://www.tigr.org). Primers used were: 23S rDNA domain V primer set 1 (Escherichia coli numbering) forward primer (bp 1921 to 1938) 5"-GGTCCTAAGGTAGCGAAA-3", reverse primer (bp 2163 to 2179) 5"-GGGTAGTATCCCAACAG-3"; 23S rDNA domain V primer set 2 (E. coli numbering) forward primer (bp 2491 to 2507) 5"-TTTGGCACCTCGATGTC-3", reverse primer (bp 2687 to 2704) 5"-GGCAAGACAACTGGTACA-3"; 23S rDNA domain II (E. coli numbering) forward primer (bp 517 to 534) 5"-CGTGTGCCTACAACAAGT-3", reverse primer (bp 823 to 839) 5"-ACGCTAGCCCTAAAGCT-3"; L4 protein forward primer (bp +37 to +53 [from ATG start]) 5"-GAAGCTGGCCAAGTTGT-3", reverse primer (bp +393 to +413 [from ATG start]) 5"-TCAGCAGTTTTTGGAGCTGTA-3"; and L22 protein forward primer (bp −133 to −117 [from ATG start]) 5"-GACGTAAACACGTACCT-3", reverse primer (bp +457 to +473 [from ATG start]) 5"-CGGATTGCAAGATCTTC-3". PCR was performed for 40 cycles with the following parameters: 94°C for 15 s, 45°C (for rDNA) or 55°C (for rplD and rplV) for 30 s, and 72°C for 30 s followed by 72°C elongation for 5 min. PCR products were purified using a Quantum Prep Spin Column (Bio-Rad, Hercules, Calif.).
Amplification products were sequenced using the Big Dye sequencing kit (Applied Biosystems Inc., Foster City, Calif.). Sequencing reactions were purified with an Auto-Seq G-50 column (Amersham Pharmacia Biotech, Piscataway, N.J.) and run on an ABI 377 automated sequencer. DNA sequences of amplified products from the macrolide-resistant strains were compared to publicly available sequences of target genes and were found to be highly homologous to those of wild-type strains, with the exception of nucleotide substitutions in regions of previously described mutations, which are discussed in Results. The above-mentioned operons (rrl, rplD, and rplV) were also amplified from S. pneumoniae ATCC 49619, a macrolide-susceptible strain, and sequenced for comparison as wild-type controls.
Ribosome binding studies.
Ribosomes were purified from the S. pneumoniae macrolide-susceptible strain ATCC 49619. Mid-log phase (optical density at 600 nm, ∼0.3 to 0.4) cells were collected by centrifugation and washed twice with high salt buffer (10 mM Tris-HCl [pH 7.5], 1 M NH4Cl, 15 mM Mg(AcO)2, 50 mM KCl, 6 mM β-mercaptoethanol) followed by a single wash with low-salt buffer (10 mM Tris-HCl [pH 7.5], 60 mM NH4Cl, 15 mM Mg(AcO)2, 50 mM KCl, 6 mM β-mercaptoethanol). After the final wash, the cells were frozen on EtOH-dry ice and stored at −80°C until further processing. Cells were thawed and broken by two passes of French press at 12,000 lb/in2. The crude lysate was centrifuged at 30,000 × g for 40 min. The pellet was discarded and the clear portion of the supernatant was centrifuged again at 100,000 × g for 2 h. Phenylmethylsulfonyl fluoride (1 mM) (Sigma Chemical Co.) and RNase inhibitor (Prime RNase inhibitor; Brinkmann Instruments, Westbury, N.Y.) were added prior to cell lysis. The pellet from the final centrifugation (ribosome fraction) was resuspended in ribosome binding buffer (10 mM Tris-HCl [pH 7.2], 10 mM NH4Cl, 4 mM MgCl2, 100 mM KCl) and stored at −80°C. Binding affinity was measured by the competition of [14C]ABT-773 with unlabeled erythromycin or telithromycin. [14C]ABT-773 was synthesized by Abbott Laboratories.
RESULTS
ABT-773 was generally one to two twofold dilutions more active than telithromycin against macrolide-susceptible streptococci, with ABT-773 at MIC90s of 0.002 and ≤0.002 μg/ml for S. pneumoniae and S. pyogenes, respectively, versus telithromycin at MIC90s of 0.008 and 0.015 μg/ml, respectively (Table 1; also see Table 3).
TABLE 1.
Activities of ABT-773 and telithromycin against macrolide-susceptible and -resistant S. pneumoniae containing Mef(A) efflux or Erm(B) methylase or having ribosomal mutations
| Organism and drug | Drug concn (μg/ml)
|
|||
|---|---|---|---|---|
| MIC50 | MIC90 | MIC min. | MIC max. | |
| S. pneumoniae susceptible (n = 94) | ||||
| ABT-773 | 0.001 | 0.002 | ≤0.0005 | 0.004 |
| Telithromycin | 0.004 | 0.008 | ≤0.0005 | 0.015 |
| Erythromycin | ≤0.03 | 0.06 | ≤0.03 | 0.5 |
| S. pneumoniae mef(A) (n = 100) | ||||
| ABT-773 | 0.004 | 0.12 | ≤0.002 | 0.5 |
| Telithromycin | 0.06 | 1 | 0.015 | 2 |
| Erythromycin | 2 | 8 | 0.25 | >32 |
| S. pneumoniae erm(B) (n = 104) | ||||
| ABT-773 | 0.004 | 0.015 | ≤0.002 | 1 |
| Telithromycin | 0.015 | 0.12 | ≤0.002 | 0.5 |
| Erythromycin | >32 | >32 | 8 | >32 |
| S. pneumoniae erm(B) + mef(A) (n = 39) | ||||
| ABT-773 | 0.06 | 0.25 | ≤0.002 | 0.5 |
| Telithromycin | 0.12 | 0.25 | ≤0.002 | 0.5 |
| Erythromycin | >128 | >128 | 0.5 | >128 |
| S. pneumoniae ribosomal mutationa (n = 19) | ||||
| ABT-773 | 0.015 | 0.12 | ≤0.008 | 1 |
| Telithromycin | 0.015 | 0.12 | ≤0.008 | 0.5 |
| Erythromycin | 16 | >128 | 0.06 | >128 |
| Clindamycin | 0.5 | 2 | 0.06 | 16 |
14 strains with A2059G; 1 with A2058G; 2 with C2611T; 2 with L4 multiple amino acid substitutions.
TABLE 3.
ABT-773 activities against macrolide-susceptible or -resistant S. pyogenes with resistant strains containing macrolide efflux mef(A) or inducible erm(A) or constitutive erm(B) methylase production
| Organism and drug | Drug concn (μg/ml)
|
|||
|---|---|---|---|---|
| MIC50 | MIC90 | MIC min. | MIC max. | |
| S. pyogenes susceptible (n = 30) | ||||
| ABT-773 | ≤0.002 | ≤0.002 | ≤0.002 | ≤0.002 |
| Telithromycin | 0.008 | 0.015 | 0.004 | 0.015 |
| Erythromycin | 0.06 | 0.06 | ≤0.03 | 0.12 |
| S. pyogenes mef(A) (n = 60) | ||||
| ABT-773 | 0.06 | 0.12 | ≤0.002 | 0.12 |
| Telithromycin | 1 | 1 | 0.015 | 2 |
| Erythromycin | 4 | 8 | 1 | 16 |
| S. pyogenes erm(B) constitutive (n = 30) | ||||
| ABT-773 | 0.12 | 0.5 | 0.004 | 1 |
| Telithromycin | 8 | >8 | 0.12 | >8 |
| Erythromycin | >32 | >32 | >32 | >32 |
| Clindamycin | >32 | >32 | >32 | >32 |
| S. pyogenes erm(A) inducible (n = 16) | ||||
| ABT-773 | 0.002 | 0.002 | 0.002 | 0.004 |
| Telithromycin | 0.015 | 0.015 | 0.008 | 0.03 |
| Erythromycin | 2 | 2 | 1 | 4 |
| Clindamycin | 0.03 | 0.06 | 0.03 | 0.06 |
The improved activity of ABT-773 compared to that of telithromycin was more evident when examining the MICs of the compounds for macrolide-resistant pneumococci, particularly strains with Mef (Table 1). The ABT-773 MIC90 for S. pneumoniae with Mef(A) is 0.12 μg/ml, three twofold dilutions lower than the telithromycin MIC90 of 1.0 μg/ml. The MIC90 of erythromycin for efflux strains is 8.0 μg/ml. Both compounds have excellent activity against S. pneumoniae strains producing Erm(B) methylase, with telithromycin having a fourfold-higher MIC90 than ABT-773 (the MIC90 of ABT-773 is 0.015 μg/ml; the MIC90 of telithromycin is 0.12 μg/ml; the MIC90 of erythromycin is >32 μg/ml). Approximately 5% of macrolide-resistant strains in the United States produce both Mef(A) and Erm(B) (7, 10). The MICs of ABT-773 and telithromycin for 39 clinical isolates of S. pneumoniae with both erm(B) and mef(A) were determined. The MIC90 for both ketolides was 0.25 μg/ml, indicating that against strains with both mechanisms, the activities of ABT-773 and telithromycin are similar to those observed against strains with efflux only.
The susceptible, Mef, and Erm S. pneumoniae strains in Table 1 were also categorized by penicillin susceptibility (Table 2). The activities of both ketolides indicate that they are unaffected by penicillin susceptibility, although the MIC90 values reflect the increases in MIC observed with Mef- and Erm-containing stains in each penicillin susceptibility category.
TABLE 2.
Activities of ABT-773 and telithromycin against S. pneumoniae categorized by penicillin susceptibility
| Organism and drug | Drug concn (μg/ml)
|
|||
|---|---|---|---|---|
| MIC50 | MIC90 | MIC min. | MIC max. | |
| Penicillin-susceptible S. pneumoniae (n = 146) | ||||
| Penicillin | 0.03 | 0.06 | 0.008 | 0.06 |
| ABT-773 | 0.002 | 0.015 | ≤0.0005 | 0.5 |
| Telithromycin | 0.008 | 0.25 | ≤0.0005 | 2 |
| Erythromycin | 0.03 | >32 | 0.03 | >32 |
| Penicillin-intermediate S. pneumoniae (n = 95) | ||||
| Penicillin | 0.5 | 1 | 0.12 | 1 |
| ABT-773 | 0.004 | 0.008 | 0.002 | 1 |
| Telithromycin | 0.03 | 0.06 | 0.002 | 2 |
| Erythromycin | 2 | >32 | 0.03 | >32 |
| Penicillin-resistant S. pneumoniae (n = 58) | ||||
| Penicillin | 2 | 4 | 2 | 8 |
| ABT-773 | 0.004 | 0.03 | 0.002 | 0.12 |
| Telithromycin | 0.03 | 0.25 | 0.002 | 1 |
| Erythromycin | 2 | >32 | 0.03 | >32 |
Nineteen clinical S. pneumoniae isolates in the Abbott culture collection were identified as being resistant to erythromycin and/or clindamycin but lacked Erm methylase or Mef efflux genetic determinants (Table 1). The DNA sequences of rrl, rplD, and rplV encoding 23S rRNA (only domain II and domain V regions were studied) and the ribosomal proteins L4 and L22, respectively, were compared to those of macrolide-susceptible strains. All 19 strains were found to contain mutations either in domain V of 23S rDNA or in L4. These mutations had been previously observed to be associated with macrolide-lincosamide-streptogramin B resistance (6, 12, 13). Ribosomal mutations have been recently described as a rare cause of clinical macrolide resistance in S. pneumoniae (5, 12). No mutations in domain II or L22 were observed. The majority (n = 17) of the isolates in this study had mutations in domain V of the 23S rDNA, A2059G (E. coli numbering system) being the most common (14 of 17). Two isolates having a base change at 2611 (C→T) and one at 2058 (A→G) were also identified. Two isolates had multiple amino acid substitutions in the L4 ribosomal protein, including amino acid substitutions of 69GTG71 to TPS, previously described as being associated with macrolide resistance in S. pneumoniae (12, 13). These two L4 mutants had no changes in their 23S rDNA sequences. The activities of the two ketolides and erythromycin were examined against these strains to determine the effect of ribosome mutation on the activities of the ketolides. Although these isolates had differing susceptibilities to erythromycin and clindamycin, all isolates had ketolide MICs of 1 μg/ml or less (MIC90s of 0.12 μg/ml for ABT-773 and telithromycin). These data suggest that mutations in streptococcal rRNA or ribosomal proteins, most likely selected by macrolide exposure, do not typically result in resistance to ketolides.
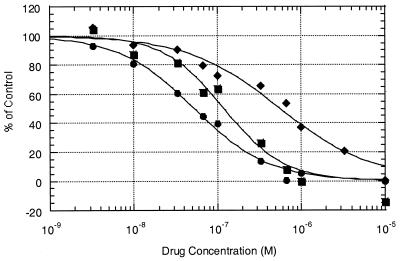
In order to examine the differences between the two ketolides and erythromycin at the ribosomal level, the binding affinities of the compounds to susceptible S. pneumoniae ribosomes were examined. As shown in Fig. 1, the IC50 of erythromycin is 566 nM, the IC50 of telithromycin is 120 nM, and the IC50 of ABT-773 is 52.7 nM. The experiments were repeated at least twice for each competition assay; the results were reproducible. The differences in IC50 between the compounds correlate well with the overall in vitro activities, with ABT-773 having over a twofold increased affinity for ribosomal binding compared to that of telithromycin and a tenfold improvement compared to that erythromycin.
FIG. 1.
Equilibrium competition binding assays with susceptible S. pneumoniae ribosomes. The binding of [14C]ABT-773 to ribosomes in competition with various concentrations (10−5 to 10−9 M) of unlabeled erythromycin (⧫), telithromycin (▪), and ABT-773 (•) is shown. The control is the sample without a competitor drug, which gave an approximately 1,000 dpm count.
ABT-773 is also extremely active against macrolide-resistant S. pyogenes (Table 3). The compound is more potent than telithromycin against strains with Mef or with constitutive Erm(B) methylase (ABT-773 Mef and Erm MIC90s, 0.12 and 0.5 μg/ml, respectively; telithromycin Mef and Erm MIC90s, 1.0 and >8.0 μg/ml, respectively). The ketolides are also very potent against strains with inducibly regulated erm(A), with MIC90s of 0.002 μg/ml and 0.015 μg/ml for ABT-773 and telithromycin, respectively.
The ketolides are active against Staphylococcus aureus strains that are macrolide susceptible, as shown in Table 4, with MIC90s of 0.03 μg/ml and 0.06 μg/ml for ABT-773 and telithromycin, respectively, but lack activity against strains with constitutive production of Erm(A) or Erm(C) methylase. However, due to the apparent inability of ketolides to act as inducers for methylase production, they maintain good in vitro activity against strains with inducible methylase (ABT-773 MIC90, 0.06 μg/ml; telithromycin MIC90, 0.5 μg/ml) (2).
TABLE 4.
ABT-773 activities against macrolide-susceptible and -resistant Staphylococcus aureus
| Organism and drug | Drug concn (μg/ml)
|
|||
|---|---|---|---|---|
| MIC50 | MIC90 | MIC min. | MIC max. | |
| S. aureus susceptible (n = 60) | ||||
| ABT-773 | 0.008 | 0.03 | 0.008 | 0.25 |
| Telithromycin | 0.03 | 0.06 | 0.008 | 0.5 |
| Erythromycin | 0.5 | 1 | 0.12 | 1 |
| S. aureus inducible erm(A) or erm(C)a (n = 47) | ||||
| ABT-773 | 0.03 | 0.06 | 0.004 | 0.12 |
| Telithromycin | 0.12 | 0.5 | 0.03 | 0.5 |
| Erythromycin | >32 | >32 | 0.5 | >32 |
| Clindamycin | 0.12 | 0.25 | 0.06 | 0.5 |
| S. aureus constitutive erm(A) or erm(C) (n = 60) | ||||
| ABT-773 | >32 | >32 | ≤0.008 | >32 |
| Telithromycin | >32 | >32 | 0.06 | >32 |
| Erythromycin | >32 | >32 | 16 | >32 |
| Clindamycin | >32 | >32 | >32 | >32 |
Inducibly resistant strains have inducible expression of either erm(A) or erm(C); constitutively resistant strains have constitutive production of either Erm(A) or Erm(C) methylase.
DISCUSSION
The ketolide class of antibiotics has been shown to be extremely potent against both susceptible and resistant streptococci. The activity of this class is reflected in the excellent in vitro potency of ABT-773 against macrolide-resistant streptococci with either of the two main resistance determinants, Mef [mef(A)] and Erm [erm(B)] (4). Although strains with either mef(A) or erm(B) have slightly higher MICs than susceptible strains, the ABT-773 MIC for macrolide-resistant streptococci rarely exceeds 0.25 μg/ml (3). These lower MICs suggest that ketolides may be useful in the treatment of infections caused by these pathogens. ABT-773 displayed a two- to fourfold greater activity against S. pneumoniae than telithromycin, although the clinical impact of this increased in vitro activity remains to be defined. Recently a new macrolide resistance mechanism has been described in which S. pneumoniae clinical isolates have mutations in the 23S rRNA or ribosomal proteins (5, 12). Although these mutations are extremely rare, they have been found in the United States and Europe. Approximately 1 to 3% of erythromycin-resistant S. pneumoniae isolates were negative for erm(B) and mef(A) amplification in recent multicenter U.S. studies, suggesting that rRNA and/or ribosomal protein mutations may account for ≤3% of macrolide resistance (7, 10). The ABT-773 MICs for 19 clinical isolates with rRNA or protein mutations were ≤1 μg/ml, suggesting ribosomal mutation alone is not likely to result in a substantial decrease in in vitro activity.
The affinity of ABT-773 for S. pneumoniae ribosomes is twofold higher than that of telithromycin and tenfold higher than that of erythromycin, suggesting that increased binding affinity for the ribosome is at least partially responsible for the increased potency. Capobianco et al. previously showed that ABT-773 had a rapid association with the ribosome and a very slow dissociation compared to those of erythromycin (4).
The enhanced activity of ABT-773 against macrolide resistant streptococci is most evident with S. pyogenes. The MIC90 of ABT-773 is three to four twofold dilutions lower than that of telithromycin against strains with efflux or methylase. As the MIC of ABT-773 for these strains does not typically exceed 1.0 μg/ml, this drug may be clinically effective against high-level macrolide-resistant group A streptococci.
Although both drugs have good in vitro activity against susceptible staphylococci, they lack activity against Staphylococcus aureus with high-level constitutive macrolide resistance (erythromycin and clindamycin MICs > 128 μg/ml). However, the apparent inability of ketolides to induce expression of methylase allows excellent in vitro activity (ABT-773 MIC90, 0.06 μg/ml) against strains with inducible methylase production. Clinical efficacy against strains with inducible methylase has yet to be determined.
In summary, the excellent in vitro activity of ABT-773 against macrolide-susceptible and -resistant gram-positive cocci suggests that this compound may be very useful in the treatment of respiratory infections caused by these pathogens, particularly since streptococci for which ABT-773 MICs are >1 μg/ml are extremely rare (<0.1%) (3). Clinical treatment trials are presently under way to determine the safety and efficacy of this compound.
REFERENCES
- 1.Barry, A. L., P. C. Fuchs, and S. D. Brown. 1998. In vitro activities of the ketolide HMR 3647 against recent gram-positive clinical isolates and Haemophilus influenzae. Antimicrob. Agents Chemother. 42:2138-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnefoy, A., A. M. Girard, C. Agouridas, and J. F. Chantot. 1997. Ketolides lack inducibility properties of MLS(B) resistance phenotype. J. Antimicrob. Chemother. 40:85-90. [DOI] [PubMed] [Google Scholar]
- 3.Brueggemann, A. B., G. V. Doern, H. K. Huynh, E. M. Wingert, and P. R. Rhomberg. 2000. In vitro activity of ABT-773, a new ketolide, against recent clinical isolates of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis. Antimicrob. Agents Chemother. 44:447-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capobianco, J. O., Z. Cao, V. D. Shortridge, Z. Ma, R. K. Flamm, and P. Zhong. 2000. Studies of the novel ketolide ABT-773: transport, binding to ribosomes, and inhibition of protein synthesis in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, T. A., L. M. Ednie, D. M. Hoellman, G. A. Pankuch, M. R. Jacobs, and P. C. Appelbaum. 2000. Antipneumococcal activity of ABT-773 compared to those of 10 other agents. Antimicrob. Agents Chemother. 44:1894-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doern, G. V., A. B. Brueggemann, H. Huynh, E. Wingert, and P. Rhomberg. 1999. Antimicrobial resistance with Streptococcus pneumoniae in the United States, 1997-1998. Emerg. Infect. Dis. 5:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.NCCLS. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed., vol. M7-A5. Approved standard. NCCLS, Wayne, Pa.
- 9.Sanchez, M. L., K. K. Flint, and R. N. Jones. 1993. Occurrence of macrolide-lincosamide-streptogramin resistances among staphylococcal clinical isolates at a university medical center. Is false susceptibility to new macrolides and clindamycin a contemporary clinical and in vitro testing problem? Diagn. Microbiol. Infect. Dis. 16:205-213. [DOI] [PubMed] [Google Scholar]
- 10.Shortridge, V. D., G. V. Doern, A. B. Brueggemann, J. M. Beyer, and R. K. Flamm. 1999. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic resistance surveillance study conducted in the United States in 1994-1995. Clin. Infect. Dis. 29:1186-1188. [DOI] [PubMed] [Google Scholar]
- 11.Shortridge, V. D., R. K. Flamm, N. Ramer, J. Beyer, and S. K. Tanaka. 1996. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 26:73-78. [DOI] [PubMed] [Google Scholar]
- 12.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong, L., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633-639. [DOI] [PubMed] [Google Scholar]
- 15.Zhong, P., Z. Cao, R. Hammond, Y. Chen, J. Beyer, V. D. Shortridge, L. Y. Phan, S. Pratt, J. Capobianco, K. A. Reich, R. K. Flamm, Y. S. Or, and L. Katz. 1999. Induction of ribosome methylation in MLS-resistant Streptococcus pneumoniae by macrolides and ketolides. Microb. Drug Resist. 5:183-188. [DOI] [PubMed] [Google Scholar]