Abstract
As seen by the disk diffusion method, the clinical strain of Pseudomonas aeruginosa Pa695, resistant to all extended-spectrum cephalosporins and aminoglycosides, exhibited an unusual synergistic effect between ceftazidime and imipenem. This isolate produced an extended-spectrum β-lactamase (ESBL) with a pI of 5.8 that appeared to be chromosomally encoded. Cloning experiments revealed that this ESBL was encoded by blaGES-1, previously described in an integron from Klebsiella pneumoniae. In P. aeruginosa Pa695, a higher level of resistance to ceftazidime than to ticarcillin was observed, and no synergy between the β-lactamase inhibitors and extended-spectrum cephalosporins was detected, in contrast to the resistance pattern observed in K. pneumoniae. Further sequence analysis demonstrated that the blaGES-1 gene cassette was located in a class 1 integron, which contained another sequence corresponding to the fused aac(3)-Ib and aac(6")-Ib" gene cassettes. The fusion product was functional, as was the product of each gene cloned separately: AAC(3)-I, despite the deletion of the four last amino acids, and AAC(6"), which carried three amino acid changes compared with the most homologous sequence. The AAC(3)-I protein conferred an expected gentamicin and fortimicin resistance, and the AAC(6"), despite the Leu-119→Ser substitution, yielded resistance to kanamycin, tobramycin, and dibekacin, but slightly affected netilmicin and amikacin, and had no apparent effect on gentamicin. The fusion product conveyed a large profile of resistance, combining the AAC(6") activity with a higher level of gentamicin resistance without accompanying fortimicin resistance.
Pseudomonas aeruginosa is intrinsically susceptible to a limited number of antimicrobial agents, mainly including β-lactams (especially ceftazidime and imipenem) and aminoglycosides (particularly tobramycin and amikacin). In addition, strains with an acquired resistance to these antibiotics are widespread. The major enzymatic mechanism of resistance to broad-spectrum cephalosporins in P. aeruginosa is the overproduction of the chromosomally encoded AmpC cephalosporinase (6). Alternatively, this resistance may result from the production of extended-spectrum β-lactamases (ESBLs), mainly belonging to Ambler's class D (4) and rarely to class A, like some TEM and SHV derivative β-lactamases (27). Moreover, uncommon types of class A enzymes have also been reported in this species, notably PER-1 (28) and VEB-1 (25), and a few class B carbapenem-hydrolyzing enzymes, including IMP-1 (18), VIM-1 (19), and VIM-2 (33). The two most-common mechanisms of aminoglycoside resistance in P. aeruginosa are impermeability and production of antibiotic-modifying enzymes, mostly 6"-N-aminoglycoside acetyltransferase of type II [AAC(6")-II] (gentamicin, tobramycin, and netilmicin phenotype) and 2"-O-nucleotidyltransferase of type I [ANT(2")-I] (gentamicin and tobramycin); AAC(3)-I (gentamicin) and AAC(6")-I (tobramycin, netilmicin, and amikacin) are much less frequent (23).
Some genes encoding ESBLs and aminoglycoside-modifying enzymes are located in gene cassettes present in the variable region of integrons. These elements are characterized by the ability to integrate gene cassettes, usually antibiotic resistance genes, by site-specific recombination (7, 14, 34). Among the three major classes of integrons previously described, class 1 is the most frequently encountered. Class 1 integrons consist of a 5"-conserved segment (5"-CS) that contains an intI1 gene coding for an integrase, a recombination site attI1, and generally, a 3"-CS carrying the qacEΔ1 gene, the sul1 gene, and an open reading frame (ORF) of unknown function (ORF 5) (21). Gene cassettes are composed of one coding sequence, and at its 3" end, a so-called 59-base element (59-be), which varies considerably in length, and is bounded by a core site (GTTRRRY) at the recombinant crossover point and an inverse core site (RYYYAAC) at the 3" end of the inserted gene (8, 38). The usual location of integrons on mobile genetic elements such as plasmids and transposons and their ability to integrate gene cassettes explain why they play a major role in the spread of antibiotic resistance (14).
In this work, we report the analysis of a P. aeruginosa strain exhibiting an extended-spectrum β-lactam resistance pattern. Cloning experiments revealed the presence of the blaGES-1 gene cassette within a novel class 1 integron. Further molecular characterization of this integron identified an aac(3)-Ib and aac(6")-Ib" fused gene cassette. The aminoglycoside-resistance pattern of the gene fusion product was analyzed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this work are listed in Table 1. The strain of P. aeruginosa Pa695 was isolated in 1999 from a respiratory sample of a woman hospitalized in an adult intensive care unit (ICU) at the Pellegrin Hospital in Bordeaux, France. This isolate was identified and serotyped by conventional methods (16). P. aeruginosa ATCC 27853 and Escherichia coli JM109 served as controls for MIC determination. A spontaneous rifampin- and nalidixic acid-resistant (Rifr Nalr) mutant of E. coli K-12 and a Rifr mutant of P. aeruginosa ATCC 27853 were used as recipient strains in conjugation assays, and E. coli HB101 in transformation experiments. E. coli DH10B and E. coli JM109 were the host strains for cloning experiments. All bacterial strains were routinely cultured at 37°C on Mueller-Hinton (MH) agar medium (Sanofi-Diagnostics Pasteur, Marnes la Coquette, France), or grown in Luria broth (GibcoBRL, Cergy Pontoise, France) or Trypticase soy broth (Diagnostics Pasteur, Marnes la Coquette, France).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10B | araD139 Δ(ara,leu)7697 deoR endA1 galK1 galU nupG recA1 rpsL F" mcrA Δ(mrr-hsdRMS-mcrBC) φ80 lacZΔM15ΔlacX74 Smr | Gibco BRL |
| E. coli JM109 | endA1 gyrA96 hsdR17 Δ(lac proA) relA recA1 supE44 thi F" (lacIqlacZΔM15 proAB+traD36) | Promega |
| E. coli HB101 | F-mcrB mrr hsdS20(rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 (Smr) supE44 λ− | Gibco BRL |
| In vitro-obtained rifampin-and nalidixic acid-resistant E. coli K-12 | Rifr Nalr | This study |
| In vitro-obtained rifampin-resistant P. aeruginosa ATCC 27853 | Rifr | This study |
| P. aeruginosa Pa695 | Extended-spectrum cephalosporin-and aminoglycoside-resistant clinical isolate | This study |
| Plasmids | ||
| pBK-CMV phagemid | Kanr Neor | Stratagene, Inc. |
| pGEM-T | Ampr | Promega |
| pTK1 | Natural plasmid from K. pneumoniae ORI-1 containing blaGES-1 and aac(6")-Ib" genes | 32 |
| pC18 | Recombinant pBK-CMV plasmid with a 2.86-kb genomic Sau3AI fragment containing blaGES-1 | This study |
| pC23 | Recombinant pBK-CMV plasmid with a 18.5-kb genomic Sau3AI fragment containing blaGES-1 | This study |
| pI18 | 2,390-bp PCR fragment containing the pC18 insert in pGEM-T | This study |
| pA3A6 | 1,104-bp PCR fragment containing the aac(3)-Ib/aac(6")-Ib" gene from pC18 in pGEM-T | This study |
| pA3 | 541-bp PCR fragment containing the aac(3)-Ib gene from pC18 in pGEM-T | This study |
| pA6 | 588-bp PCR fragment containing the aac(6")-Ib" gene from pC18 in pGEM-T | This study |
| pA6In52 | 588-bp PCR fragment containing the aac(6")-Ib" gene from pTK1 in pGEM-T | This study |
Antibiotic susceptibility testing.
Antibiotic susceptibility of P. aeruginosa Pa695 and the E. coli recombinant strains was determined at first by the disk diffusion method on MH agar medium, according to French guidelines (http://www.sfm.asso.fr); disks of 2"- and 6"-N-ethylnetilmicin (100 μg), fortimicin (100 μg), 5-episisomicin (10 μg), and apramycin (100 μg) were kindly provided by Schering-Plough (Herouville Saint Clair, France). MICs of 12 β-lactams and six aminoglycosides were determined by a standard agar dilution method on MH agar plates, using an inoculum of 104 CFU per spot (http://www.sfm.asso.fr). β-Lactams were tested alone or in combination with a fixed concentration of clavulanic acid (2 mg/liter), sulbactam (4 mg/liter), tazobactam (2 mg/liter), or imipenem (0.01 mg/liter).
β-Lactamase extraction and IEF.
β-Lactamases produced by P. aeruginosa Pa695 and E. coli DH10B harboring the recombinant plasmid pC18 were released by ultrasonic treatment, and their pIs were determined by isoelectric focusing (IEF) on an ampholin polyacrylamide gel (pH 3.5 to 10), as described by Matthew et al. (22). Enzyme activities were detected by the iodine procedure in gel, using benzylpenicillin (75 mg/liter) as the substrate.
Plasmid content analysis, conjugation, and transformation experiments.
Transfer of resistance genes to Rifr Nalr E. coli K-12 and Rifr P. aeruginosa ATCC 27853 was attempted by a filter mating technique. Plasmid DNA extraction was carried out for the clinical strain of P. aeruginosa using the three following methods: an alkaline-lysis method (1), a technique using alkaline sodium dodecyl sulfate at elevated temperatures (15), and the Qiagen (Courtaboeuf, France) plasmid DNA midi kit. The putative plasmid DNA extract from P. aeruginosa Pa695 was electroporated into E. coli HB101 with selection on ampicillin (100 mg/liter)-containing MH plates. Plasmid extraction for E. coli recombinant clones was performed with a simple boiling-lysis procedure (36).
PCR experiments.
The detection of β-lactamase genes was performed under standard PCR conditions (36), using published or laboratory designed sets of primers (Table 2). The aminoglycoside resistance genes aac(3)-Ib from pC18 and aac(6")-Ib" from pC18 or pTK1 were separately amplified with the primer sets 5"AAC3-3"AAC3 and 5"AAC6"-3"AAC6", respectively (Table 2). The fused form of the aminoglycoside resistance genes aac(3)-Ib/aac(6")-Ib" from pC18 was amplified with the primer set 5"AAC3-3"AAC6". A ribosome binding site (AGGAGGT) was included in the forward primer to allow gene expression during the cloning experiments, and the reverse primer contained a stop codon. The amplicons were revealed by electrophoresis on a 1.5% agarose gel and a subsequent exposure to UV light in the presence of ethidium bromide.
TABLE 2.
Oligonucleotides used as primers for PCR amplification of β-lactam and aminoglycoside resistance genes
| Gene(s) | Primer | Sequence (5" to 3")a | Reference |
|---|---|---|---|
| blaTEM-1 | TEM-A2 | GTATCCGCTCATGAGACAATA | 39 |
| TEM-ext | TCTAAAGTATATATGAGTAAAC | ||
| blaSHV-1 | OS0 | CTCGCCTTTATCGGCCCTCAC | 2 |
| OS5 | CGGCCACGCGGGTTAGCG | ||
| blaOXA-10, -11, -14, -16, -17 | OPR1 | GTCTTTCGAGTACGGCATTA | 44 |
| OPR2 | ATTTTCTTAGCGGCAACTTAC | ||
| blaPER-1 | PER-A | ATGAATGTCATTATAAAAGC | 28 |
| PER-1B | AATTTGGGCTTAGGGCAGAA | ||
| blaVEB-1 | VEB-F | CGACTTCCATTTCCCGATGC | 25 |
| VEB-B | GGACTCTGCAACAAATACGC | ||
| blaGES-1 | GES-1A | ATGCGCTTCATTCACGCAC | 32 |
| GES-1B | CTATTTGTCCGTGCTCAGG | ||
| blaOXA-13 | OXA-13.1 | GCCGCATATGTAATTACTGC | 24 |
| OXA-13.2 | ATTTTCTTAGCGGCAACTTAT | ||
| blaOXA-18 | OXA-18F | ATTTCAACGGTTTGCCTTACG | 30 |
| OXA-18B | TTGGCATCGGAAAGCGAACC | ||
| aac(3)-Ib | 5" AAC3 | CATCATAGGAGGTTGTTTGATGTTATGGAGCAG | This study |
| 3" AAC3 | TTATTATGGATCAATGTCGAAGTGCA | This study | |
| aac(6")-Ib" | 5" AAC6" | CATCATAGGAGGTGATCCAATGACCAACAGCAACGATTCCG | This study |
| 3" AAC6" | CCTCGATGGAAGGGTTAGGC | This study |
The RBS and start and stop codons are underlined in the primers where they were included.
Cloning experiments and recombinant plasmid analysis.
Total DNA of P. aeruginosa Pa695 was extracted as previously described (33), partially restricted by Sau3AI, and ligated into the BamHI-restricted pBK-CMV phagemid (Stratagene, La Jolla, Calif.). E. coli DH10B strains harboring the recombinant plasmids were selected on MH agar plates containing amoxicillin (30 mg/liter) and kanamycin (30 mg/liter). A double-restriction digestion analysis with HindIII and PstI enzymes allowed precise mapping of recombinant plasmids by electrophoresis on a 0.8% agarose gel. The PCR products of the aminoglycoside resistance genes, either fused or separated, were ligated into the pGEM-T vector (Promega, Charbonnières, France). In order to analyze the expression of the fused gene within the integron environment, a PCR product corresponding to the insert in recombinant plasmid pC18 was also ligated into vector pGEM-T (pI18). E. coli JM109 strains carrying the recombinant plasmids were selected on MH agar plates containing tobramycin (4 mg/liter) for aac(6")-Ib" and aac(3)-Ib/aac(6")-Ib" genes. For the aac(3)-Ib gene, since the conferred gentamicin resistance was at a low level and the pGEM-T vector contained a blaTEM gene, selection was done on ampicillin (100 mg/liter), and then plasmid analysis after PstI and HindIII restriction allowed identification of the clone harboring the insert in the right sense.
DNA sequencing.
The 2.86-kb cloned DNA fragment from recombinant plasmid pC18 was sequenced on both strands using the dideoxy-chain termination method with the D Rhodamine dye terminator kit (Perkin-Elmer, Courtaboeuf, France). Further sequence analysis was performed on PCR products using laboratory-designed sequencing primers and parts of a larger recombinant plasmid, pC23, containing an 18.5-kb genomic Sau3AI fragment. Sequences were analyzed with an automatic sequencer ABI 377 (Perkin Elmer), using the Sequencing Analysis software and compared to each other and to homologous sequences using the Sequence Navigator software. The nucleotide and the deduced protein sequences were analyzed using the software available over the Internet at the National Center of Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper are available in the GenBank nucleotide database under the accession number AF355189.
RESULTS AND DISCUSSION
Clinical case.
P. aeruginosa Pa695, of serotype P11, was isolated from the sputum of a 46-year-old woman hospitalized in 1999 at the Pellegrin Hospital in Bordeaux, France. This patient had been admitted in the adult ICU for vascular brain damage 1 month previously. Two days after her hospitalization she had a pulmonary infection due to Streptococcus pneumoniae and Haemophilus influenzae, but she recovered under antimicrobial therapy with intravenous amoxicillin (6 g)-clavulanic acid, lasting 17 days, and with pristinamycin (6 g), lasting 15 days. Four weeks later, a control sputum sample revealed the presence of P. aeruginosa Pa695 (107 CFU/ml) and a TEM-24 (as verified by sequencing)-producing strain of Enterobacter aerogenes (2 × 107 CFU/ml). In the absence of fever, the patient did not receive any antibiotic. She had not recently traveled to French Guiana or Greece, where GES-1 β-lactamase was previously found (11, 32). No patient coming from these countries was concomitantly hospitalized in the same ICU during the same period of time.
By the disk diffusion method, P. aeruginosa Pa695 was seen to be resistant to all potentially active β-lactam agents except for imipenem and aztreonam. Surprisingly, a synergistic effect was seen between imipenem and ceftazidime or cefsulodin as observed with OXA-13 in P. aeruginosa Pae391 (24), but not between clavulanic acid and ceftazidime. These results suggested the presence of an ESBL of an unusual type. No other P. aeruginosa or enterobacterial isolate with a similar ESBL resistance profile was isolated among patients of the same unit and hospital. Pa695 was additionally resistant to all aminoglycosides and all fluoroquinolones.
Characterization of the β-lactam resistance pattern, the bla gene, and its genetic environment.
Preliminary PCR amplification experiments of known ESBLs in P. aeruginosa (TEM, SHV, OXA derivatives, PER-1, and VEB-1) failed to give positive results. Conjugation experiments between Pa695 and E. coli K-12 Rifr Nalr or P. aeruginosa ATCC 27853 Rifr, did not yield any transconjugant. Despite repeated attempts, plasmid DNA analysis of P. aeruginosa Pa695 did not show any plasmid, and transformation by electroporation of plasmid DNA extract into E. coli HB101 was unsuccessful. Thus, the bla gene seemed to be chromosomally located in this strain. After cloning experiments two recombinant plasmids, pC18 (containing a 2.86 kb insert) and pC23 (containing an 18.5-kb insert), were selected for subsequent analysis and sequencing.
β-Lactam MICs for P. aeruginosa Pa695 (Table 3) showed an intermediate susceptibility to ticarcillin (32 mg/liter), cefepime (16 mg/liter), and ceftazidime (32 mg/liter) and a low-level resistance to cefotaxime (64 mg/liter). In contrast, it was susceptible to aztreonam and imipenem. The addition of β-lactamase inhibitors (clavulanic acid, sulbactam, or tazobactam at 2, 8, or 4 mg/liter, respectively) or imipenem at 0.01 mg/liter did not modify or only slightly decreased the MICs of the tested penicillins and cephalosporins, except for piperacillin plus tazobactam (eightfold decrease in MIC). IEF analysis revealed that P. aeruginosa Pa695 and E. coli DH10B(pC18) produced a β-lactamase with a pI of 5.8.
TABLE 3.
MICs of β-lactams for the clinical strain of P. aeruginosa Pa695 and the reference strain of P. aeruginosa ATCC 27853
| β-Lactam(s)a | MIC (mg/liter) for:
|
|
|---|---|---|
| P. aeruginosa Pa695 | P. aeruginosa ATCC 27853 | |
| Amoxicillin | >512 | 512 |
| Amoxicillin + CLA | >512 | 512 |
| Ticarcillin | 32 | 8 |
| Ticarcillin + CLA | 16 | 8 |
| Ticarcillin + SUL | 16 | 8 |
| Ticarcillin + TZB | 16 | 8 |
| Ticarcillin + IPM | 16 | 8 |
| Piperacillin | 64 | 2 |
| Piperacillin + TZB | 8 | 2 |
| Cephalothin | >512 | >512 |
| Cefuroxime | 512 | 256 |
| Cefoxitin | 512 | >512 |
| Cefotaxime | 64 | 16 |
| Ceftazidime | 32 | 2 |
| Ceftazidime + CLA | 32 | 2 |
| Ceftazidime + SUL | 32 | 2 |
| Ceftazidime + TZB | 32 | 2 |
| Ceftazidime + IPM | 16 | 2 |
| Cefsulodin | 64 | 2 |
| Cefsulodin + IPM | 32 | 2 |
| Cefepime | 16 | 1 |
| Cefepime + CLA | 8 | 1 |
| Cefepime + TZB | 8 | 1 |
| Aztreonam | 4 | 4 |
| IPM | 1 | 2 |
Abbreviations: CLA, clavulanic acid (2 mg/liter); SUL, sulbactam (4 mg/liter); TZB, tazobactam (2 mg/liter); IPM, imipenem (0.01 mg/liter).
Sequence analysis of a 4,808-bp DNA fragment was performed at first with the recombinant plasmid pC18 and then with PCR-amplified fragments from the longer insert of the recombinant plasmid pC23, using laboratory-designed primers. The nucleotide sequence of the ESBL-encoding gene differed by a single silent mutation at position 591 from blaGES-1, recently described in Klebsiella pneumoniae (32), and its amino acid sequence differed by two substitutions from IBC-1, an ESBL reported in Enterobacter cloacae (11). GES-1 is known to inactivate most β-lactams except for aztreonam and imipenem and to be inhibited by clavulanic acid, sulbactam, and tazobactam and strongly inhibited by imipenem (32). These features were recognized in E. coli DH10B(pC18) (data not shown). By MIC determination, GES-1 in Pa695 seemed to affect ticarcillin less than ceftazidime, and the β-lactamase inhibitors such as imipenem had a very slight inhibitory effect (Table 3). Indeed, the ESBL inhibition might be masked by the expression of the chromosomally encoded cephalosporinase of Pa695, either partially derepressed or induced by clavulanate and imipenem. By the disk diffusion method the synergistic effect between cefsulodin and imipenem could lead to a confusion with the OXA-13 enzyme. However, the main difference is the ceftazidime resistance conveyed by GES-1 β-lactamase.
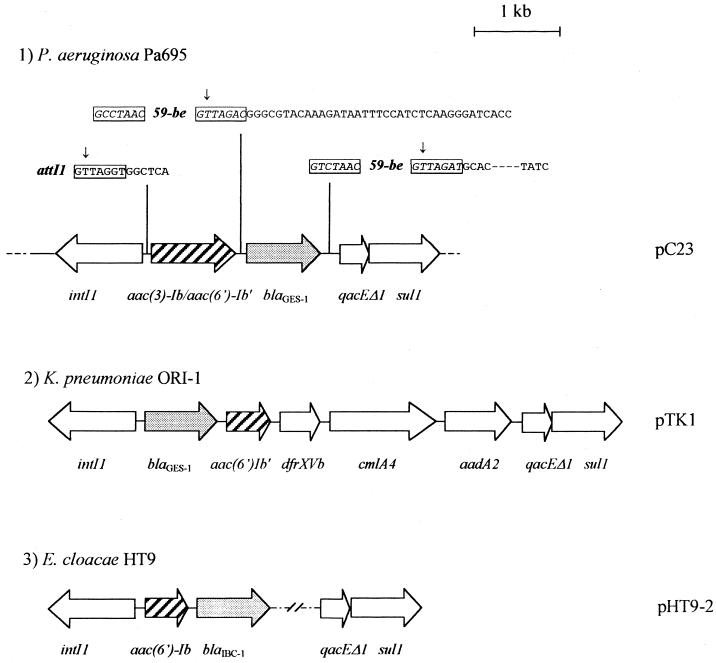
The blaGES-1 gene was found in a cassette located in a class 1 integron (Fig. 1). Indeed, the 5"-CS end contained the intI1 integrase gene, the attI1 recombination site, and the promoter region including the P1 promoter (also called Pc) (regions −35[TGGACA] and −10[TAAACT]) under a hybrid 1 form with a weak activity (20) and different from that present in In52, the blaGES-1 integron in K. pneumoniae (32). At the 3"-CS end, the quaternary ammonium compound-resistance gene qacEΔ1, and the sulfonamide resistance gene sul1 were identified, as reported in most class 1 integrons (13, 34). The blaGES-1 gene cassette contained a core site (GTTAGAC), an inverse core site (GTCTAAC), and a 59-be site of 110 bp different from that of the truncated blaGES-1 gene cassette previously reported in In52 but identical to the 59-be in the blaIBC-1 gene cassette (Fig. 1).
FIG. 1.
Schematic representations of the different blaGES-1- and blaIBC-1-containing integrons. (1) Part of the structure of the recombinant plasmid pC23 encoding blaGES-1 in P. aeruginosa Pa695. The horizontal arrows indicate the translation orientation. The solid lines represent the sequenced fragment from P. aeruginosa Pa695 with the different genes boxed, and the dotted lines indicate the unanalyzed sequence. The conserved core and inverse core sites are boxed, and the cassette boundaries are represented by vertical arrows. Dashes are used to indicate where the reported sequence was identical to already-published sequences. (2) Structure of the blaGES-1 gene cassette-containing integron from K. pneumoniae ORI-1 (33). (3) Structure of the blaIBC-1 gene cassette-containing integron from E. cloacae HT9 (12).
The finding in P. aeruginosa of the blaGES-1 gene, previously described in Enterobacteriaceae, underlines the interspecies spread of this integron-located ESBL gene cassette. GES-1 is another Ambler class A β-lactamase, besides PER-1, VEB-1, TEM-4, TEM-24, TEM-42, and SHV-2a, which have been previously described in this species. Finally, the blaGES-1 gene in Pa695 was the first description of a nonimported case in France, suggesting the worldwide spread of this enzyme.
Aminoglycoside resistance conferred by the integron.
Sequencing of pC18 and pC23 revealed that the integron contained another cassette made up of a 1,005-bp coding region which consisted of the aac(3)-Ib gene fused with the sequence of an aac(6")-Ib" gene. The last 15 nucleotides of the aac(3)-Ib sequence were missing, and the sequence continued with a leucine codon instead of a valine at the beginning of the aac(6")-Ib" sequence (Fig. 2). Cassette fusion may occur by deletion events with end points of the genes in two adjacent gene cassettes leading to the presence of one or two truncated genes or by partial or total loss of the 59-be of the first cassette (34). In the present study the fused cassette contained the truncated aac(3)-Ib gene and the complete aac(6")-Ib" gene. The core site of the fused cassette was identical to that identified for the aac(3)-Ib cassette (37) and the 59-be located at the 3" end of this ORF was identical to that found for some aac(6")-Ib" cassettes (31, 32). Thus, the fused cassette had a core site (GTTAGGT) and an inverse core site (GCCTAAC) presenting a 1-bp mismatch (Fig. 1).
FIG. 2.
Sequence comparison of the fusion point of the aac(3)-Ib/aac(6")-Ib" fused cassette in Pa695 and the ends of the original aac(3)-Ib and aac(6")-Ib" gene cassettes. The data were compiled from the sequences obtained from plasmids pSCH6006 (37), pTK1 (32), pLMM562 (5), pMT222 (41), and the one analyzed here. The possible start codons are underlined, and the stop codons are indicated by asterisks. The deleted region of the aac(3)-Ib gene is shown in boldface type. The conserved core sites are boxed, and the vertical arrow indicates the site of fusion between the aac(3)-Ib and aac(6")-Ib" genes. The deduced amino acid sequence is designated in single-letter code below the nucleotide sequence.
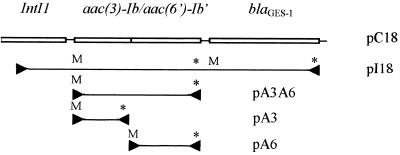
By the disk diffusion method, E. coli DH10B(pC18) exhibited an unusual high-level resistance to tobramycin, compared with a relative susceptibility to gentamicin, netilmicin, and amikacin. Aminoglycoside MICs (Table 4) were consistent with this observation. Since E. coli DH10B is streptomycin resistant due to a chromosomal mutation, and pBK-CMV harbors a kanamycin-neomycin resistance gene, the pC18 insert was cloned in the pGEM-T vector (pI18) (Fig. 3) and expressed in E. coli JM109. E. coli JM109(pI18) exhibited a similar resistance pattern compared with E. coli DH10B(pC18) except for streptomycin and neomycin susceptibility and a lower level of kanamycin resistance. To assess the functionality of the separate aac(3)-Ib and aac(6")-Ib" genes, and of the fused aac(3)-Ib/aac(6")-Ib" gene, the corresponding PCR products were cloned in the pGEM-T vector (Fig. 3) used to transform E. coli JM109. The three types of clones were obtained by the same method, and in all cases the pGEM-T promoter allowed the expression of the cloned gene.
TABLE 4.
Aminoglycoside resistance patterns of various strains
| Strain | MIC (mg/liter) ofa:
|
Additional aminoglycoside resistance marker(s) | |||||
|---|---|---|---|---|---|---|---|
| STR | KAN | GEN | TOB | AMK | NET | ||
| Pa695 | 32 | 512 | 512 | 256 | 32 | >512 | 2"NET, EPI, DIB |
| E. coli DH10B(pC18) | >512 | >512 | 2 | 16 | 8 | 16 | 2"NET, EPI, NEO, DIB |
| E. coli JM109(pI18) | 2 | 64 | 2 | 16 | 4 | 8 | 2"NET, EPI, DIB |
| E. coli JM109(pA3A6) | 2 | 128 | 2 | 32 | 4 | 8 | 2"NET, EPI, DIB |
| E. coli JM109(pA3) | 2 | 2 | 2 | 0.5 | 1 | 0.5 | FOR |
| E. coli JM109(pA6) | 2 | 64 | 0.5 | 32 | 4 | 4 | 2"NET, EPI, DIB |
| E. coli JM109(pA6In52) | 2 | 128 | 2 | 8 | 1 | 4 | 2"NET, EPI, DIB |
| E. coli JM109 | 2 | 2 | 0.2 | 0.5 | 1 | 0.5 | |
Abbreviations: STR, streptomycin; KAN, kanamycin; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; NET, netilmicin; 2"NET, 2"-N-ethylnetilmicin; EPI, 5-episisomicin; NEO, neomycin; DIB, dibekacin; FOR, fortimicin.
FIG. 3.
Schematic representation of cloning experiments of the integron and its aac(3)-Ib, aac(6")-Ib" and aac(3)-Ib/aac(6")-Ib" genes performed in pGEM-T vector after PCR amplification. Arrowheads represent primer positions and their orientations, and M and ∗ indicate the location of the start and the stop codon, respectively.
The aac(3)-Ib gene encoding a 3-N-aminoglycoside acetyltransferase was identical to that conferring gentamicin and fortimicin resistance previously described in a strain of P. aeruginosa (37), except for a silent substitution (C→A) at position 207 of the gene, and the absence of the 15 last nucleotides. The E. coli JM109(pA3) strain, which expressed the truncated aac(3)-Ib gene, exhibited a decreased susceptibility to gentamicin as demonstrated by MIC determination. Moreover, by the disk diffusion method, a small diameter of 10 mm around the fortimicin disk was observed with E. coli JM109(pA3) instead of a diameter of 30 mm as observed with the host strain. Thus, despite the lack of the four last amino acids the aac(3)-Ib gene conferred resistance to fortimicin and a low-level resistance to gentamicin, similar to results described by Schwocho et al. (37).
The aac(6")-Ib" sequence of the fused cassette encoded a 6"-N-aminoglycoside acetyltransferase that was characterized by a Leu-119→Ser substitution [numbering of the reference aac(6")-Ib sequence (42)], and differed by three amino acid substitutions—Val-18→Leu, Leu-42→Val, and Ser-100→Gly—from the already-described aac(6")-Ib" genes (5, 17, 31, 32, 41). The amino acid at position 119 has been found to be critical functionally in that a Leu-to-Ser switch at this position was responsible for the loss of amikacin resistance conferred by the aac(6")-Ib gene and the acquisition of gentamicin resistance, conveyed by the aac(6")-Ib" gene (17). These genes encoded a protein with an AAC(6")-II specificity, i.e., kanamycin, tobramycin, netilmicin, and gentamicin resistance and amikacin susceptibility. However, E. coli JM109(pA6), which harbored the aac(6")-Ib" gene, exhibited an unusual aminoglycoside resistance pattern, with kanamycin and tobramycin resistance, reduced netilmicin and amikacin susceptibility, and gentamicin susceptibility (Table 4). The natural plasmid pTK1 from K. pneumoniae ORI-1 also contains an aac(6")-Ib" gene cassette beside the blaGES-1 gene cassette in In52 (32). In order to evaluate whether the mutations influenced the enzyme specificity, the recombinant plasmid pA6In52 was constructed by cloning the aac(6")-Ib" gene of In52 in the pGEM-T vector and expressed in E. coli JM109. The MIC determination revealed slight differences between the two clones, E. coli JM109(pA6) and E. coli JM109(pA6In52) (Table 4). The recombinant plasmid pA6In52 conferred the expected resistance profile, i.e., reduced susceptibility to gentamicin and full susceptibility to amikacin, with MICs similar to those previously reported (5, 35). In contrast, the recombinant plasmid pA6 conferred a marked resistance to tobramycin, a reduced susceptibility to amikacin, and an increased susceptibility to gentamicin, suggesting a role of the mutations in the resistance pattern conferred by this enzyme. As shown in Fig. 2, several potential start codons have been proposed for the aac(6")-Ib genes (5, 10). However, in our construction the compared genes necessarily started from the initiation codon introduced in the primer, validating their comparison.
E. coli JM109(pA3A6), which expressed the product of the aac(3)-Ib/aac(6")-Ib" gene fusion, had a resistance profile identical to E. coli JM109(pI18), indicating that this protein was functional and expressed in the same manner as in the integron. The MIC determination showed that the AAC(3)-I/AAC(6") protein fusion had a broad activity, combining the effect of the protein encoded by the aac(6")-Ib" gene, with a decreased gentamicin susceptibility. However, it did not yield fortimicin resistance, indicating the activity of a unique protein rather than the additive effect of two enzymes. Northern blot experiments should allow the demonstration of whether one or two RNAs are transcribed from the fused gene. A single enzyme frequently modifies several antibiotics, via the same modification mechanism of each substrate. The sole example of a bifunctional aminoglycoside resistance enzyme described at present is the AAC(6")-APH(2"), found in strains of streptococci and staphylococci (9, 43). In the case of the fused gene product AAC(3)-I/AAC(6"), it is difficult to establish whether the enzyme is really bifunctional or the presence of the AAC(3)-I increases the activity of the AAC(6") protein.
Although the AAC(6") of type I is common in Enterobacteriaceae, this enzyme, along with AAC(3)-I, is infrequent in P. aeruginosa (23). The existence of a fused product of both genes in Pa695 raises the hypothesis, as does the finding of blaGES-1, that the integron studied here may have originated from enterobacteria. Moreover, the 5" sequences flanking the aac(6")-Ib cassette junctions display considerable genetic plasticity (5), and some studies have reported translational fusion with the aac(6")-Ib gene (3, 26, 40), suggesting that these sequences are favorable for expression of fused genes. On the other hand, the integron described here was bounded at the 5" end by a 25-bp inverted repeat (IRi) (TGTCGTTTTCAGAAGACGGCTGCAC) identical to the IRi sequence identified at the boundary of several integrons (13, 18, 29), and the nucleotide sequence upstream of the IRi was from Tn501, suggesting that the integron was inserted into Tn501 or a close relative transposon, which would itself be inserted in the chromosome.
In conclusion, we report here the characterization of a new class 1 integron found in P. aeruginosa. This integron contains the blaGES-1 gene previously reported in Enterobacteriaceae, showing the interspecies diffusion of this ESBL-encoding gene, as recently described for VEB-1 (12). Further molecular analysis led to the discovery of a functional fused gene encoding an AAC(3)-I/AAC(6") protein. This enzyme conferred a specific resistance pattern, combining the activity of an unusual AAC(6") with an increased effect on gentamicin. This work confirms the major role of integrons in the spread of resistance genes and gives an insight into the multiple and complex recombinations occurring in these genetic elements.
Acknowledgments
We thank Thierry Lambert for precious advice and Catherine André and Cécile Frigo for technical assistance.
This work was supported by grants from the French Network on β-lactamase study and from the Ministère de l'Education Nationale et de la Recherche (EA-525), Université de Bordeaux 2, Bordeaux, France.
REFERENCES
- 1.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, P. A. 1999. Automated thermal cycling is superior to traditional methods for nucleotide sequencing of blaSHV genes. Antimicrob. Agents Chemother. 43:2960-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunny, K. L., R. M. Hall, and H. W. Stokes. 1995. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob. Agents Chemother. 39:686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casin, I., F. Bordon, P. Bertin, A. Coutrot, I. Podglajen, R. Brasseur, and E. Collatz. 1998. Aminoglycoside 6"-N-acetyltransferase variants of the Ib type with altered substrate profile in clinical isolates of Enterobacter cloacae and Citrobacter freundii. Antimicrob. Agents Chemother. 42:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H. Y., M. Yuan, and D. M. Livermore. 1995. Mechanisms of resistance to beta-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J. Med. Microbiol. 43:300-309. [DOI] [PubMed] [Google Scholar]
- 7.Collis, C. M., G. Grammaticopoulos, J. Briton, H. W. Stokes, and R. M. Hall. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41-52. [DOI] [PubMed] [Google Scholar]
- 8.Collis, C. M., and R. M. Hall. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferretti, J. J., K. S. Gilmore, and P. Courvalin. 1986. Nucleotide sequence analysis of the gene specifying the bifunctional 6"-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J. Bacteriol. 167:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galimand, M., T. Lambert, G. Gerbaud, and P. Courvalin. 1993. Characterization of the aac(6")-Ib gene encoding an aminoglycoside 6"-N-acetyltransferase in Pseudomonas aeruginosa BM2656. Antimicrob. Agents Chemother. 37:1456-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giakkoupi, P., L. S. Tzouvelekis, A. Tsakris, V. Loukova, D. Sofianou, and E. Tzelepi. 2000. IBC-1, a novel integron-associated class A beta-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob. Agents Chemother. 44:2247-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum beta-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5" ends but variable 3" ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 15.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiska, D. L., and P. H. Gilligan. 1999. Pseudomonas, p. 517-525. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 17.Lambert, T., M. C. Ploy, and P. Courvalin. 1994. A spontaneous point mutation in the aac(6")-Ib" gene results in altered substrate specificity of aminoglycoside 6"-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol. Lett. 115:297-304. [DOI] [PubMed] [Google Scholar]
- 18.Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J. M. Frere, and G. M. Rossolini. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 21.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 23.Miller, G. H., F. J. Sabatelli, L. Naples, R. S. Hare, and K. J. Shaw. 1995. The most frequently occurring aminoglycoside resistance mechanisms—combined results of surveys in eight regions of the world. J. Chemother. 7:17-30. [PubMed] [Google Scholar]
- 24.Mugnier, P., I. Podglajen, F. W. Goldstein, and E. Collatz. 1998. Carbapenems as inhibitors of OXA-13, a novel, integron-encoded beta-lactamase in Pseudomonas aeruginosa. Microbiology 144:1021-1031. [DOI] [PubMed] [Google Scholar]
- 25.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 26.Nobuta, K., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1988. Sequencing and expression of the 6"-N-acetyltransferase gene of transposon Tn 1331 from Klebsiella pneumoniae. J. Bacteriol. 170:3769-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordmann, P., and M. Guibert. 1998. Extended-spectrum beta-lactamases in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 42:128-131. [DOI] [PubMed] [Google Scholar]
- 28.Nordmann, P., and T. Naas. 1994. Sequence analysis of PER-1 extended-spectrum beta-lactamase from Pseudomonas aeruginosa and comparison with class A beta-lactamases. Antimicrob. Agents Chemother. 38:104-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippon, L. N., T. Naas, A. T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirel, L., D. Girlich, T. Naas, and P. Nordmann. 2001. OXA-28, an extended-spectrum variant of OXA-10 beta-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 35.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-beta-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schwocho, L. R., C. P. Schaffner, G. H. Miller, R. S. Hare, and K. J. Shaw. 1995. Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 39.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolmasky, M. E., and J. H. Crosa. 1993. Genetic organization of antibiotic resistance genes (aac[6"]-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid 29:31-40. [DOI] [PubMed] [Google Scholar]
- 41.Toriya, M., M. Sakakibara, K. Matsushita, and T. Morohoshi. 1992. Nucleotide sequence of aminoglycoside 6"-N-acetyltransferase [AAC(6")] determinant from Serratia sp. 45. Chem. Pharm. Bull. 40:2473-2477. [DOI] [PubMed] [Google Scholar]
- 42.Tran Van Nhieu, G., and E. Collatz. 1987. Primary structure of an aminoglycoside 6"-N-acetyltransferase, AAC(6")-4, fused in vivo with the signal peptide of the Tn3-encoded beta-lactamase. J. Bacteriol. 169:5708-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ubukata, K., N. Yamashita, A. Gotoh, and M. Konno. 1984. Purification and characterization of aminoglycoside-modifying enzymes from Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 25:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vahaboglu, H., R. Ozturk, H. Akbal, S. Saribas, O. Tansel, and F. Coskunkan. 1998. Practical approach for detection and identification of OXA-10-derived ceftazidime-hydrolyzing extended-spectrum beta-lactamases. J. Clin. Microbiol. 36:827-829. [DOI] [PMC free article] [PubMed] [Google Scholar]