Abstract
Extended-spectrum β-lactamases (ESBLs) are active against oxyimino cephalosporins and monobactams. Twenty-one Klebsiella pneumoniae isolates obtained between 1991 and 1995 at the Princess Alexandra Hospital in Brisbane, Australia, were subject to amplification and sequencing of the SHV β-lactamase-encoding genes. Thirteen strains were phenotypically ESBL positive. Of these, six strains carried the blaSHV-2a gene and seven strains carried the blaSHV-12 gene. Eight strains were phenotypically ESBL negative. Of these, seven strains carried the non-ESBL blaSHV-11 gene and one strain carried the non-ESBL blaSHV-1 gene. There was complete correspondence between the ESBL phenotype and the presence or absence of an ESBL-encoding gene(s). In addition, it was determined that of the 13 ESBL-positive strains, at least 4 carried copies of a non-ESBL-encoding gene in addition to the blaSHV-2a or blaSHV12 gene. A minisequencing-based assay was developed to discriminate the different SHV classes. This technique, termed “first-nucleotide change,” involves the identification of the base added to a primer in a single-nucleotide extension reaction. The assay targeted polymorphisms at the first bases of codons 238 and 240 and reliably discriminated ESBL-positive strains from ESBL-negative strains and also distinguished strains carrying blaSHV-2a from strains carrying blaSHV-12. In addition, this method was used to demonstrate an association between the relative copy numbers of blaSHV genes in individual strains and the levels of antibiotic resistance.
In recent years the use of oxyimino cephalosporins and monobactams such as cefotaxime, ceftriaxone, ceftazidime, and aztreonam has resulted in the selection of β-lactamases that recognize them as substrates. These enzymes are referred to as extended-spectrum β-lactamases (ESBLs), and most of these are derived from the β-lactamases TEM-1 or SHV-1 by one or more amino acid substitutions (13, 19, 31, 32, 39) and are encoded on large conjugative plasmids (12, 18, 34).
Klebsiella pneumoniae is a significant cause of hospital-acquired infections. Many K. pneumoniae isolates express ESBLs of the SHV family. Known SHV variants are listed at http://www.lahey.org/studies/webt.htm#SHV20. Not all are fully characterized, but it appears that the majority may be classed as ESBLs. Almost all SHV-derived ESBLs have a G-to-A mutation which specifies a glycine-to-serine substitution at amino acid 238 (numbering according to that of Ambler et al. [1]), although it has recently been found that substitutions with alanine or aspartate at that site can also give rise to ESBL activity (8, 44). An additional G-to-A mutation specifying a glutamate-to-lysine substitution at amino acid 240 is seen in a subset of ESBLs. In general, the substitution at position 238 confers a large increase in resistance to cefotaxime and a small increase in resistance to cetazidime, while the presence of substitutions at both positions confers a small additional increase in resistance to cefotaxime and a large additional increase in resistance to ceftazidime (4, 7, 31).
Although SHV ESBLs have been documented worldwide, there are only two published reports of outbreaks in Australian hospitals. An SHV-5-related ESBL was characterized in a Western Australian hospital (24) and SHV ESBL-producing K. pneumoniae was isolated at Princess Alexandra Hospital in Brisbane, Australia (38). In both cases the SHV ESBLs were identified and partially characterized on the basis of MICs, the double-disk synergy test (DDST), and isoelectric focusing (IEF) and in the former case also by enzyme kinetic analysis.
Accurate identification of ESBLs requires DNA-based methods. Previously used methods include DNA hybridization (5, 11, 29), oligotyping (20), restriction fragment length polymorphism analysis (27), an immunoassay system (6), PCR-single stranded conformation polymorphism (2, 25, 26), and the ligase chain reaction (17). The clinically significant effect on phenotype conferred by single-nucleotide changes in SHV ESBLs makes them ideal candidates for the development of a minisequencing protocol. The minisequencing procedure, or first-nucleotide change (FNC) method, has previously proved effective for the detection of polymorphic sites in humans (3, 9, 14, 22, 33). The method interrogates polymorphic sites through primer annealing immediately upstream of the polymorphic site followed by determination of the identity of a single base incorporated. The procedure is performed in microwells and requires no electrophoresis. It is therefore easy to automate and, since it produces quantitative results, requires neither skill nor subjectivity in interpretation.
In this study we completely characterized by nucleotide sequencing the SHV β-lactamase-encoding genes found in a collection of K. pneumoniae isolates from Princess Alexandra Hospital in Brisbane. These characterized samples were then used to evaluate the FNC method as a predictor of the resistance phenotype.
MATERIALS AND METHODS
Bacterial strains.
Twenty-one strains of K. pneumoniae isolated at Princess Alexandra Hospital between December 1991 and June 1995 have been previously described by Schooneveldt and colleagues (38). Thirteen of these were found to express ESBLs as defined by the DDST (15), and the MICs of cefotaxime, ceftazidime, and aztreonam supported this finding.
In addition, Escherichia coli JC411, which carries the blaSHV gene that encodes the partially characterized ESBL described by Mulgrave and Attwood (24), was included in some aspects of this study. This strain was supplied by Leigh Mulgrave at the WA Centre for Pathology and Medical Research, Perth, Western Australia, Australia.
All strains were cultured in Luria-Bertani broth and stored in cryovials with 12% glycerol at −80°C.
DNA extraction and PCR amplification of blaSHV genes.
Plasmid DNA was extracted from 1.5-ml cultures grown overnight in L-broth using alkaline lysis (21).
Overlapping regions of the genes were amplified by PCR using primers which hybridize either to the coding regions (SHV-F, nucleotide position 311 to 328, TCAGCGAAAAACACCTTG; SHV-R, nucleotide position 782 to 764, TCCCGCAGATAAATCACCA) (25) or sites adjacent to the coding sequence (BIG-F, nucleotide position −72 to −52 CGCCGGGTTATTCTTATTTG; BIG-R, nucleotide position +78 to +54 TCTTTCCGATGCCGCCGCCAGTCA. (The − and + refer to nucleotides upstream of transcription initiation and downstream of transcription termination, respectively.) PCRs (50 μl) were carried out in a solution containing 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3; Boehringer Mannheim), a 400 μM concentration of each deoxynucleoside triphosphate (Boehringer Mannheim), 20 pmol of each primer (Gibco-Life Technologies), 100 ng of plasmid DNA template, and 1 U of Taq polymerase (Boehringer Mannheim). After a 5-min denaturation at 96°C, 35 cycles of temperatures of 96, 55, and 72°C and a final extension step of 72°C for 3 min were performed. The length of the temperature steps for the 35 cycles were 30 s each for the coding sequence primers and 1 min each for the primers adjacent to the coding sequence. Dimethoxyl sulfoxide was added to a final concentration of 5% when the BIG-F primer was used.
Cloning of blaSHV encoding gene fragments.
Amplicons generated from the clinical isolates A1, F2, J1, J2, and L1 using primers SHV-F and SHV-R were cloned using the pGEMt plasmid kit (Promega) and JM109 High Efficiency competent Escherichia coli cells (Promega). Ten to twenty clones were selected for each isolate, and secondary PCR products were generated from these clones for sequencing and FNC analysis.
Sequencing.
PCR products were purified using Wizard mini-columns (Promega). Amplicon (50 to 100 ng [10 ng/100 bases]) was sequenced using 3.2 pmol of the amplification primer. The products were analyzed on an ABI 373A DNA automated sequencer. Sequence results were aligned using the Clustal W program (10), which was accessed through the Australian National Genome Information Service at http://www.angis.su.oz.au/.
FNC.
FNC is a microtiter plate-based minisequencing (single-nucleotide extension) assay. A biotinylated primer is hybridized with a PCR-generated target at a sequence 1 base upstream from a polymorphic site. This complex is captured by streptavidin-coated microtiter wells. The primer is extended by DNA polymerase in the presence of a single fluorescein-labeled dideoxynucleoside triphosphate (ddNTP) and unlabeled ddNTPs. Bound fluorescein is detected by enzyme-linked immunosorbent assay. The assay is carried out in four wells, each with a different fluorescein-labeled ddNTP.
A modification of the FNC protocol described by Pecheniuk et al. (33) was used. Briefly, in a 50-μl reaction mixture containing Sequenase buffer (40 mM Tris HCl [pH 7.51], 20 mM MgCl2, 50 mM NaCl) plus 5% Nonidet P-40 and 5% Tween 20, 1 pmol of biotinylated primer was bound to 0.6 pmol of amplified 471-bp DNA product by heating at 96°C for 3 min and then rapidly chilling on dry ice. Three hundred and sixty microliters of a cold solution containing phosphate-buffered saline (PBS), 0.1% Tween 20, and 10 mM EDTA was added to the thawed reaction mix, and 100 μl of the resulting mixture was added to each of four streptavidin-coated microwells. The primer-amplimer complex was allowed to bind to the microplate for 30 min at room temperature.
Unbound material was removed by six washes with PBS-0.1% Tween 20 and three washes with Sequenase buffer (Amersham Life Science). Extensions were carried out for 30 min at room temperature in Sequenase buffer using 0.5 U of Sequenase (Amersham Life Science). Each of the four wells contained a 0.024 μM concentration of one of the four fluorescein-labeled ddNTPs plus a 0.3 μM concentration of the remaining three unlabeled ddNTPs. The extension reactions were carried out in Sequenase buffer, which contained 0.4 mM dithiothreitol, 0.4 mM MnCl2, 0.7 mM Na isocitrate, 0.015% Tween 20, and 0.015% Nonidet.
Incorporation of fluorescein into the captured complex was quantified after washing six times with PBS-0.1% Tween 20 and incubating for 30 min with 0.03 U of antifluorescein-alkaline phosphatase conjugate (Boehringer Mannheim). p-Nitrophenyl phosphate (0.1 mg in 100 μl of 0.2 M Tris-HCl) was used to detect the incorporated fluorescein-labeled ddNTP, and the color development was monitored by absorbance reading at 405 nm with an enzyme-linked immunosorbent assay plate reader (Biomek Plate Reader; Beckman Coulter).
Two FNC primers were utilized. MSP-238, 5" biotin-GTTTATCGCCGATAAGACCGGAGCT, was designed to detect the G-to-A change at position 1 of codon 238 indicative of ESBLs. MSP-240, 5" biotin-TATCGCCGATAAGACCGGAGCTAGC, was designed to detect the G-to-A change at position 1 of codon 240 indicative SHV-12 and similar enzymes, such as SHV-5.
IEF.
Cell extracts were focused and bands with β-lactamase activity were visualized with the chromogenic substrate as described by Matthew et al. (23) with modifications as described by Rasheed et al. (36).
RESULTS
Nucleotide sequencing of the SHV ESBL-encoding genes.
The amplified blaSHV genes from all 21 K. pneumoniae isolates were sequenced. The results are summarized in Table 1. Genes encoding SHV variants 1, 2a, 11, and 12 were detected. SHV-1 and SHV-11 are non-ESBLs, and SHV-2a and SHV-12 are ESBLs. The MICs and DDST results obtained by Schooneveldt et al. (38) and included in Table 1 are consistent with these DNA sequence data. It is clear that the mutation at codon 238 confers an increase in MICs over that conferred by the wild-type precursor, while the mutation in codon 240 causes a considerable further increase in the MICs.
TABLE 1.
Characteristics of K. pneumoniae isolates in this study
| Isolate | DDSTa | MIC (μg/ml) ofb:
|
Polymorphic codon at position:
|
Genotype | ||||
|---|---|---|---|---|---|---|---|---|
| ATM | CAZ | CTX | 35 | 238 | 240 | |||
| A1 | + | 0.5 | 1 | 1 | CAA | (G/A)GC | GAG | SHV-11/2a |
| B1 | + | 1.0 | 1 | 1 | CAA | AGC | GAG | SHV-2a |
| B2 | − | <0.03 | 0.125 | <0.03 | CAA | GGC | GAA | SHV-11 |
| C1 | + | 64 | 64 | 4 | CAA | AGC | AAG | SHV-12 |
| D1 | + | 2 | 4 | 4 | CAA | AGC | GAG | SHV-2a |
| E1 | + | 0.5 | 1 | 1 | CAA | AGC | GAG | SHV-2a |
| F1 | + | 1 | 1 | 1 | CAA | AGC | GAG | SHV-2a |
| F2 | + | 0.25 | 0.5 | 0.5 | CAA | AGC | GAG | SHV-2a |
| G1 | + | >128 | >128 | 16 | CAA | AGC | AAG | SHV-12 |
| H1 | + | >128 | >128 | >128 | CAA | AGC | AAG | SHV-12 |
| I1 | + | >128 | >128 | 16 | CAA | AGC | AAG | SHV-12 |
| J1 | + | 64 | 32 | 4 | CAA | (G/A)GC | (G/A)AG | SHV-11/12 |
| J2 | + | 64 | 32 | 2 | CAA | (G/A)GC | (G/A)AG | SHV-11/12 |
| J3 | − | 0.125 | 0.5 | 0.125 | CAA | GGC | GAG | SHV-11 |
| J4 | − | 0.25 | 2 | 0.125 | CAA | GGC | GAG | SHV-11 |
| J5 | − | 0.06 | 0.5 | 0.125 | CAA | GGC | GAG | SHV-11 |
| K1 | − | 0.06 | 0.5 | 0.06 | CTA | GGC | GAA | SHV-1 |
| K2 | − | >0.03 | 0.125 | <0.03 | CAA | GGC | GAA | SHV-11 |
| L1 | + | 32 | 32 | 1 | CAA | (G/A)GC | (G/A)AG | SHV-11/12 |
| L2 | − | 0.125 | 0.25 | 0.25 | CAA | GGC | GAG | SHV-11 |
| M1 | − | <0.03 | 0.125 | <0.03 | CAA | GGC | GAA | SHV-11 |
These data have been previously reported by Schooneveldt et al. (38).
Abbreviations: ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime.
In addition, the blaSHV gene encoding the SHV-5-like enzyme described by Mulgrave and Attwood (24) was sequenced. It was found to encode SHV-12.
Amplicons from strains A1, F2, J1, J2, and L1 yielded sequences with clearly visible double G+A peaks at position 1 of codon 238 and/or codon 240. Several clones were constructed from PCR products derived from these strains, and sequence analysis revealed that they all possess copies of non-ESBL SHV-encoding genes in addition to the SHV-2a gene (strains A1 and F2) or the SHV-12 gene (strains L1, J1, and J2). Although this sequencing did not encompass codon 35, which discriminates SHV-2/5 from SHV-2a/12, sequence determination of the primary PCR products never yielded any sign of double peaks at that or any other polymorphic sites apart from at codons 238 and 240, thus indicating that the non-ESBL SHV blaSHV genes in these strains encode SHV-11.
Limited attempts were made to make use of the clones to determine the relative copy numbers of the different genes. Ten clones from strains J1 and J2 were analyzed by FNC at position 1 of codons 238 and 240. Strain J2 yielded one SHV-12 sequence, eight SHV-11 sequences, and one failed assay, while strain J1 yielded two SHV-12 sequences and eight SHV-11 sequences. This suggests that blaSHV-11 is present at a higher copy number than blaSHV-12 in these strains. Seven clones from strain F2 were sequenced across the codon 238 and 240 region, and this yielded four SHV-2a sequences and three SHV-11 sequences, indicating approximately equal copy numbers for the two genes. For strains A1 and L1, two clones only were sequenced across the codon 238 and 240 region. Strain A1 yielded one each of SHV-11 and SHV-2a, and strain L1 yielded one each of SHV-11 and SHV-12, thus indicating that the relative copy numbers are unlikely to be very different.
FNC results are concordant with β-lactamase class and MICs.
A procedure for interrogating the polymorphic nucleotides at the first positions of codons 238 and 240 was developed. At both positions, the presence of a G is indicative of the ancestral form while A is indicative of the extended spectrum of activity. The mutation in codon 238 converts a non-ESBL to an ESBL, while a second mutation in codon 240 confers very high activity against the extended-spectrum substrates. Subsequent to optimization of the procedure, the reliability of the method was assessed by carrying out the amplifications and FNC assays in triplicate.
The results of the FNC assays are shown in Table 2. In order to fully illustrate the results, the unprocessed absorbance readings (i.e., the absorbance reading using undeveloped color reagents as a blank) from one replicate are shown, together with the mean and standard deviation of the log(A/G) incorporation using all three replicate experiments. The FNC results are concordant with the genotyping, DDST, and MIC data. This indicates it may be feasible to predict MICs from the results of mutation analyses even when there are complications arising from the coexistence of different blaSHV variants within individual strains. However, this will only be confirmed through analysis of additional strains expressing a wider variety of SHV ESBL variants.
TABLE 2.
Results of FNC assaysd
| Isolate | Genotypea | Base at 238a | FNC result forb:
|
Log(A/G) at position 238c | Base at 240 | FNC result for:
|
Log(A/G) at position 240 | ||
|---|---|---|---|---|---|---|---|---|---|
| A 238 | G 238 | A 240 | G 240 | ||||||
| A1 | 2a/11 | G/A | 0.888 | 0.78 | 0.065 (0.014) | G | 0.059 | 1.362 | −1.192 (0.141) |
| B1 | 2a | A | 0.726 | 0.648 | 0.026 (0.031) | G | 0.189 | 2.138 | −1.011 (0.023) |
| B2 | 11 | G | 0.214 | 0.527 | −0.355 (0.026) | G | 0.101 | 0.58 | −0.583 (0.191) |
| C1 | 12 | A | 1.27 | 0.114 | 0.902 (0.156) | A | 1.632 | 0.102 | 1.17 (0.086) |
| D1 | 2a | A | 0.789 | 0.298 | 0.395 (0.025) | G | 0.147 | 1.344 | −0.962 (0.100) |
| E1 | 2a | A | 0.53 | 0.62 | −0.055 (0.013) | G | 0.134 | 1.571 | −0.936 (0.159) |
| F1 | 2a | A | 0.88 | 0.672 | 0.124 (0.013) | G | 0.181 | 0.848 | −0.762 (0.154) |
| F2 | 2a | A | 0.934 | 0.618 | 0.157 (0.029) | G | 0.083 | 1.632 | −1.034 (0.299) |
| G1 | 12 | A | 1.179 | 0.187 | 0.691 (0.111) | A | 2.252 | 0.183 | 0.989 (0.109) |
| H1 | 12 | A | 1.372 | 0.191 | 0.780 (0.092) | A | 1.632 | 0.132 | 0.972 (0.122) |
| I1 | 12 | A | 1.568 | 0.141 | 0.849 (0.200) | A | 2.685 | 0.127 | 1.201 (0.131) |
| J1 | 11/12 | G/A | 0.48 | 0.963 | −0.299 (0.012) | G/A | 0.688 | 0.832 | 0.195 (0.202) |
| J2 | 11/12 | G/A | 0.485 | 0.741 | −0.293 (0.162) | G/A | 0.707 | 0.565 | 0.084 (0.010) |
| J3 | 11 | G | 0.061 | 0.793 | −0.925 (0.155) | G | 0.054 | 0.692 | −0.852 (0.242) |
| J4 | 11 | G | 0.093 | 1.172 | −0.934 (0.167) | G | 0.073 | 0.681 | −0.734 (0.267) |
| J5 | 11 | G | 0.213 | 0.409 | −0.395 (0.163) | G | 0.12 | 0.475 | −0.520 (0.080) |
| K1 | 1 | G | 0.045 | 0.329 | −0.431 (0.314) | G | 0.117 | 0.43 | −0.327 (0.175) |
| K2 | 11 | G | 0.162 | 0.919 | −0.692 (0.059) | G | 0.133 | 0.691 | −0.657 (0.057) |
| L1 | 11/12 | G/A | 0.856 | 0.702 | 0.062 (0.044) | G/A | 1.328 | 0.468 | 0.489 (0.068) |
| L2 | 11 | G | 0.061 | 1.001 | −1.033 (0.195) | G | 0.054 | 0.61 | −0.821 (0.214) |
| M1 | 11 | G | 0.049 | 0.751 | −0.488 (0.543) | G | 0.112 | 1.032 | −0.804 (0.129) |
Genotypes as determined by PCR, cloning, and sequencing.
The unprocessed absorbance readings from one replicate of the FNC assays.
The mean of three replicates with the standard deviations in parentheses. The ratios were calculated from unprocessed absorbance readings.
The signals for the incorporation of the other two bases were also determined (data not shown). These absorbance readings did not exceed 0.23 and in the great majority of cases were <0.1.
IEF.
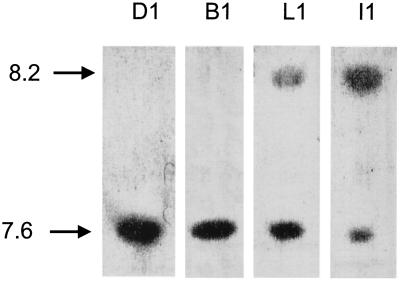
There is a clear difference in MICs between the strains that have been found to have both blaSHV-12 and blaSHV-11 and those in which blaSHV-11 was not detected. It may be hypothesized that these groups of strains have different levels of expression of SHV-12, with expression in the proven heterozygotic strains being lower. To test this, cell extracts were subjected to IEF and then stained for β-lactamase activity staining using nitrocefin. IEF would be expected to resolve SHV-11 and -2a (pI 7.6) from SHV-12 (pI 8.2). The results were concordant with the DNA-based assays, and representative strains are shown in Fig. 1. Strains shown to possess genes encoding SHV-1/11 and/or SHV-2a yielded bands at pI 7.6, while strains previously shown to possess genes encoding SHV-12 yielded bands at pI 8.2 in addition to bands at pI 7.6. Interestingly, all strains yielded bands at pI 7.6. This indicates that all strains carrying blaSHV-12 also carry blaSHV-11 and/or blaSHV-1. Consistent with this, the pI 7.6 bands were more intense than the pI 8.2 bands in strains L1, J1, and J2, which gave genetic evidence for the coexpression of SHV-11 and SHV-12, while the relative band intensities were reversed for strains I1, G1, and H1, in which SHV-11-encoding genes had not been detected. The only exceptions to this were strains K1 and M1, which yielded bands that were at pI 7.6 (as expected) but considerably fainter than the bands from other strains, and the presence of β-lactamases with a pI of 5.35 in extracts from strains B2, C1, K1, and M1. This may be a TEM β-lactamase. Its expression by strains with sensitive (B2, K1, and M1) and very resistant (C1) phenotypes suggests that it does not contribute to resistance to these antibiotics.
FIG. 1.
IEF of extracts from four representative K. pneumoniae strains. The strain designations are shown above each lane. pIs are shown at left.
DISCUSSION
This study constitutes the first full characterization of SHV-ESBLs of Australian origin. Sequence analysis revealed that in this collection of K. pneumoniae isolates from Princess Alexandra Hospital in Brisbane, Queensland, the ESBLs were exclusively of classes 2a and 12, while, with one exception, the non-ESBL enzymes were SHV-11. These classes are all characterized by a Leu-Gln substitution at amino acid 35 with respect to SHV-1 and differ from each other only at position 1 of codons 238 and 240. While the existence of SHV-2a was first reported in 1991 (35) the existence of SHV-12 has only been reported more recently in a study of strains of Swiss origin (28). Although there have been few subsequent reports of these enzymes, it may be incorrect to conclude that these classes are rare. In the study carried out by Nuesch-Inderbinen et al. (28) SHV-2a was found in a high percentage of strains, while in a recent studies in Eastern Asia they were the most abundant classes in K. pneumoniae (16, 30, 43). It may be that in certain parts of the world, SHV-2a and SHV-12 are very common. Consistent with this, the strain from Western Australia reported to express an SHV-5-like enzyme (24) has now been shown to possess an SHV-12-encoding gene.
The 21 K. pneuoniae isolates had previously been reported to possess ESBLs with pI values different from those reported here (38). It appears that in those experiments, ESBLs with pIs of 8.2 were not reliably detected. The IEF analyses were repeated for this report, and it is now clear that IEF results are consistent with the genotypes.
The finding that all ESBLs in this study possessed the Leu-Gln substitution at amino acid 35 is consistent with the model advanced by Du Bois et al. (7), which states that SHV ESBLs evolve repeatedly in parallel through mutations at amino acids 238 and 240 and that this can occur in a number of ancestral non-ESBL enzymes that differ as a result of genetic drift. The non-ESBL phenotype conferred by SHV-11 shows that the Leu-Gln substitution has little or no significance with respect to hydrolysis of expanded-spectrum cephalosporins, and therefore its appearance is likely to be due to drift rather than antibiotic selection.
These data strongly indicate that the ESBLs we have characterized arose from an SHV-11 ancestor. The presence of SHV-11 in a number of ESBL-negative strains suggests either that SHV-2a and SHV-12 appeared as result of antibiotic-mediated selection pressure at the Princess Alexandra Hospital in Brisbane or that K. pneumoniae strains expressing these enzymes are ubiquitous in southeast Queensland. The fact that these strains were isolated in a hospital (38) lends support to the first alternative.
During the course of this study, it was demonstrated that a number of strains carried both ESBL and non-ESBL encoding genes. Elucidation of the precise molecular basis for this phenomenon awaits further analysis, although it is clear that at least a subset of these strains harbor multiple ESBL-encoding genes. Indeed, the results of the IEF would indicate that all the strains harboring ESBL-encoding genes also carry the non-ESBL-encoding precursor. This result is consistent with the observations of Xiang et al. (42), who reported that very high levels of ESBL-mediated resistance are due to the gene amplification on low-copy-number plasmids. The failure to detect non-ESBL-encoding clones derived from all amplimers from ESBL-positive strains may be explained by the variation in the relative copy numbers of the ESBL- and non-ESBL-encoding genes and the limited number of amplimer-derived clones that were subject to sequence analysis.
It is currently unknown whether the genes encoding non-ESBLs in ESBL-positive strains are located on the chromosome or on plasmids. All genetic analyses in the present study were carried out using alkaline lysis plasmid preparations which would be unlikely to contain significant chromosomal DNA. However, there are reports of chromosomally located blaSHV-1 genes in K. pneumoniae (37), and it now seems as if the great majority of K. pneumoniae isolates carry the blaSHV-1 gene (G. S. Babini and D. M. Livermore, Letter, Antimicrob. Agents Chemother. 44:2230, 2000). We cannot rule out the possibility that the plasmid preparations were contaminated with small quantities of chromosomal DNA and that some of the genes we have detected are chromosomally located.
It is of interest that in the case of the isolates carrying blaSHV-2a, there were a significant (up to eightfold) range of MICs without any corresponding differences in copy numbers as evidenced by the FNC results. This may be due to, e.g., differences in the porin content of the isolates. This has been demonstrated to modulate resistance levels (41).
In this study we demonstrated the potential of the FNC minisequencing technique in diagnostic microbiology. FNC and similar methods have previously been used in human genetics (4, 9, 14, 22), but this approach has not been previously applied to the genotyping of bacteria. ESBL detection was considered a suitable application for FNC analysis because a very small number of known polymorphisms result in phenotypic changes of clinical significance. Since non-DNA-based methods for ESBL detection are relatively easy and inexpensive, it is possible that DNA-based methods will not prove to be competitive in the clinical laboratory environment. Nevertheless, the FNC method is carried out in microplates, is quantitative, and does not involve electrophoresis. It is therefore suitable for automation and may prove effective in situations requiring the rapid screening of large numbers of samples. Additionally, the reaction could be incorporated into a DNA SNP array format as described by Tonisson et al. (40).
We have demonstrated the potential of the FNC method to accommodate the presence of multiple blaSHV variants within single strains and variations in the relative copy numbers of these different variants. Of most significance is the correlation of MICs, gene identities, and relative copy numbers. It is remarkable that with these strains, the blaSHV-11 genes can serve as intracellular standards for measuring the copy number of ESBL-encoding genes, thus providing a simple strategy for predicting resistance using a DNA-based assay.
Acknowledgments
Particular thanks go to James Biddle and Kamile Rasheed for carrying out the IEF analyses. Thanks also go to Leigh Mulgrave, Ross Barnard, and Adele Millis for constructive comments on the manuscript and to Natalie Pecheniuk for advice and assistance with the FNC procedure.
This work was supported by the Australian Federal Government Program for Cooperative Research Centres.
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for class A β-lactamases. Biochem. J. 256:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanawong, A., F. H. M'Zali, J. Heritage, A. Lulitanond, and P. M. Hawkey. 2000. Characterisation of extended-spectrum β-lactamases of the SHV family using a combination of PCR-single strand conformational polymorphism (PCR-SSCP) and PCR-restriction fragment length polymorphism (PCR-RFLP). FEMS Microbiol. Lett. 184:85-89. [DOI] [PubMed] [Google Scholar]
- 3.Chiang, F. T., K. L. Hsu, C. D. Tseng, W. H. Hsiao, T. H. Chern, and Y. Z. Tseng. 1997. Molecular variant M235T of the angiotensinogen gene is associated with essential hypertension in Taiwanese. J. Hypertens. 15:607-611. [DOI] [PubMed] [Google Scholar]
- 4.Collatz, E., R. Labia, and L. Gutmann. 1990. Molecular evolution of ubiquitous β-lactamases towards extended-spectrum enzymes active against newer β-lactam antibiotics. Mol. Microbiol. 4:1615-1620. [DOI] [PubMed] [Google Scholar]
- 5.Cooksey, R. C., N. C. Clark, and C. Thornsberry. 1985. A gene probe for TEM type β-lactamases. Antimicrob. Agents Chemother. 28:154-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curran, R., D. C. S. Talbot, and K. J. Towner. 1996. A rapid immunoassay method for the direct detection of PCR products: applications to detection of TEMβ-lactamase genes. J. Med. Microbiol. 45:76-78. [DOI] [PubMed] [Google Scholar]
- 7.DuBois, S. K., M. S. Marriott, and S. G. B. Amyes. 1995. TEM- and SHV-derived extended-spectrum β-lactamases: relationship between selection, structure and function. J. Antimicrob. Chemother. 35:7-22. [DOI] [PubMed] [Google Scholar]
- 8.Essack, S. Y., L. M. C. Hall, D. G. Pillay, M. L. McFadyen, and D. M. Livermore. 2001. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum β-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hietala, M., P. Aula, A. C. Syvanen, A. Isoniemi, L. Peltonen, and A. Palotie. 1996. DNA-based carrier screening in primary healthcare: screening for the aspartylglucosaminuria mutations in maternity health offices. Clin. Chem. 42:1398-1404. [PubMed] [Google Scholar]
- 10.Higgins, D. J., J. D. Thompson, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huovinen, S., G. Huovinen, and G. A. Jacoby. 1988. Detection of plasmid mediated β-lactamases with DNA probes. Antimicrob. Agents Chemother. 32:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby, G. A. 1994. Genetics of extended-spectrum beta-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 13(Suppl. 1):2-11. [DOI] [PubMed] [Google Scholar]
- 13.Jacoby, G. A. 1997. Extended-spectrum beta-lactamases and other enzymes providing resistance to oxyimino-beta-lactams. Infect. Dis. Clin. N. Am. 11:875-887. [DOI] [PubMed] [Google Scholar]
- 14.Jalanko, A., J. Kere, E. Savilahti, M. Schwartz, A. C. Syvanen, M. Ranki, and H. Soderlund. 1992. Screening for defined cystic fibrosis mutations by solid-phase minisequencing. Clin. Chem. 38:39-43. [PubMed] [Google Scholar]
- 15.Jarlier, V., M.-H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J., Y. Kwon, H. Pai, J. Kim, and D. Cho. 1998. Survey of Klebsiella pneumoniae strains producing extended-spectrum β-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J. Clin. Microbiol. 36:1446-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J., and H.-J. Lee. 2000. Rapid discriminatory detection of genes coding for SHV β-lactamases by ligase chain reaction. Antimicrob. Agents Chemother. 44:1860-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, P., J. Tung, S. Ke, and S. Chen. 1998. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a district hospital in Taiwan. J. Clin. Microbiol. 36:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mabilat, C., and P. Courvalin. 1990. Development of “oligotyping” for characterization and molecular epidemiology of TEM β-lactamases in members of the family Enterobacteriaceae. Antimicrob. Agents Chemother. 34:2210-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Mariotti, C., N. Saverse, A. Suomalainen, M. Rimoldi, G. Comi, A. Prelle, C. Antozzi, S. Servidei, L. Jarre, S. DiDonato, et al. 1995. Genotype to phenotype correlations in mitochondrial encephalomyopathies associated with the A3243G mutation of mitochondrial DNA. J. Neurol. 242:304-312. [DOI] [PubMed] [Google Scholar]
- 23.Mathew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 24.Mulgrave, L., and P. V. Attwood. 1993. Characterisation of an SHV-5 related extended broad-spectrum beta-lactamase in Enterobacteriaceae from Western Australia. Pathology 25:71-75. [DOI] [PubMed] [Google Scholar]
- 25.M'Zali, F. H., D. M. Gascoyne-Binzi, J. Heritage, and P. M. Hawkey. 1996. Detection of mutations conferring extended-spectrum activity on SHV β-lactamases using polymerase chain reaction single strand conformational polymorphism (PCR-SSCP). J. Antimicrob. Chemother. 37:797-802. [DOI] [PubMed] [Google Scholar]
- 26.M'Zali, F. H., J. Heritage, D. M. Gascoyne-Binzi, A. M. Snelling, and P. M. Hawkey. 1998. PCR single strand conformational polymorphism can be used to detect the gene encoding SHV-7 extended-spectrum β-lactamase and to identify different SHV genes within the same strain. J. Antimicrob. Chemother. 41:123-125. [DOI] [PubMed] [Google Scholar]
- 27.Nuesch-Inderbinen, M. T., H. Hachler, and F. H. Kayser. 1996. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 15:398-401. [DOI] [PubMed] [Google Scholar]
- 28.Nuesch-Inderbinen, M. T., F. Kayser, and H. Hachler. 1997. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob. Agents Chemother. 41:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouellette, M., G. C. Paul, A. M. Philippon, and P. H. Roy. 1988. Oligonucleotide probes (TEM-1, OXA-1) versus isoelectric focusing in β-lactamase characterization of 114 resistant strains. Antimicrob. Agents Chemother. 32:397-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pai, H. 1998. The characteristics of extended-spectrum β-lactamases in Korean isolates of enterobacteriaceae. Yonsei Med. J. 39:514-519. [DOI] [PubMed] [Google Scholar]
- 31.Palzkill, T. 1998. β-Lactamases are changing their activity spectrums. ASM News 64:90-95. [Google Scholar]
- 32.Payne, D. J., and S. G. B. Amyes. 1991. Transferable resistance to extended-spectrum β-lactams: a major threat or a minor inconvenience? J. Antimicrob. Chemother. 27:255-261. [DOI] [PubMed] [Google Scholar]
- 33.Pecheniuk, N. M., N. A. Marsh, T. P. Walsh, and J. L. Dale. 1997. Use of first nucleotide change technology to determine the frequency of factor V Leiden in a population of Australian blood donors. Blood Coagul. Fibrinolysis 8:491-495. [DOI] [PubMed] [Google Scholar]
- 34.Philippon, A., G. Arlet, and P. H. Lagrange. 1994. Origin and impact of plasmid-mediated extended-spectrum beta-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 13(Suppl. 1):517-529. [DOI] [PubMed] [Google Scholar]
- 35.Podbielski, A., J. Schonling, B. Melzer, K. Warnatz, and H.-G. Leusch. 1991. Molecular characterisation of a new plasmid-encoded SHV-type β-lactamase (SHV-2 variant) conferring high-level cefotaxime resistance upon Klebsiella pneumoniae. J. Gen. Microbiol. 137:569-578. [DOI] [PubMed] [Google Scholar]
- 36.Rasheed, J. K., G. J. Anderson, H. Yigit, A. M. Queenan, A. Doménech-Sánchez, J. M. Swenson, J. W. Biddle, W. J. Ferraro, G. A. Jacoby, and F. C. Tenover. 2000. Characterization of the extended-spectrum β-lactamase reference strain, Klebsiella pneumoniae K6 (ATCC 700603), which produces the novel enzyme SHV-18. Antimicrob. Agents Chemother. 44:2382-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice, L. B., L. L. Carias, A. M. Hujer, M. Binafede, R. Hutton, C. Hoyen, and R. A. Bonomo. 2000. High-level expression of chromosomally encoded SHV-1 β-lactamase and an outer membrane change confer resistance to ceftazidime and piperacillin-tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schooneveldt, J. M., G. R. Nimmo, and P. Giffard. 1998. Detection and characterisation of extended spectrum β-lactamases in Klebsiella pneumoniae causing nosocomial infection. Pathology 30:164-168. [DOI] [PubMed] [Google Scholar]
- 39.Sirot, D. 1995. Extended-spectrum plasmid mediated β-lactamases. J. Antimicrob. Chemother. 36(Suppl. A):19-34. [DOI] [PubMed] [Google Scholar]
- 40.Tonisson, N., A. Kurg, K. Kaasik, E. Lohmussaar, and E. Metspalu. 2000. Unravelling genetic data by arrayed primer extension. Clin. Chem. Lab. Med. 38:165-170. [DOI] [PubMed] [Google Scholar]
- 41.Wu, T. L., L. K. Sui, L. H. Su, T. L. Lauderdale, F. M. Lin, H. S. Leu, T. Y. Lin, and M. Ho. 2001. Outer membrane protein change combined with co-existing TEM and SHV-1 β-lactamases lead to false identification of ESBL-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 47:755-761. [DOI] [PubMed] [Google Scholar]
- 42.Xiang, X., K. Shannon, and G. French. 1997. Mechanism and stability of hyperproduction of the extended spectrum β-lactamase SHV-5 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 40:525-532. [DOI] [PubMed] [Google Scholar]
- 43.Yan, J.-J., S.-M. Wu, S.-H. Tsai, J.-J. Wu, and I.-J. Su. 2000. Prevalence of SHV-12 among clinical isolates producing extended-spectrum β-lactamases and identification of a novel AmpC enzyme (CMY-8) in southern Taiwan. Antimicrob. Agents Chemother. 44:1438-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan, M., L. M. C. Hall, J. Hoogkaamp-Korstanje, and D. M. Livermore. 2001. SHV-14, a novel β-lactamase variant in Klebsiella pneumoniae isolates from Nijmegen, The Netherlands. Antimicrob. Agents Chemother. 45:309-311. [DOI] [PMC free article] [PubMed] [Google Scholar]