Abstract
The phenomenon of cross-resistance to antiretroviral agents used to treat human immunodeficiency virus type 1 infection is well known but so far has been only qualitatively described. Here, we quantitate the degree of cross-resistance among all commonly prescribed antiretroviral agents in almost 5,000 clinically derived recombinant isolates collected in the United States since January 2000.
Highly active antiretroviral therapy for the treatment of human immunodeficiency virus type 1 (HIV-1) infection has produced substantial decreases in morbidity and mortality (8, 9; for a review, see reference 4). Despite these successes, resistance and cross-resistance within all three classes of antiretroviral agents have been described (for reviews, see references 1, 5, and 9). Although it is known that the degree of cross-resistance varies with the number and type of mutations, descriptions of cross-resistance have been largely qualitative (2, 6), with a growing awareness of the cross-resistance that occurs among the nucleoside analogues. Here, we quantify the degree of cross-resistance observed in a large number of clinically derived isolates by using a linear regression of the logfold change in susceptibility among different antiretrovirals. The results are expressed in terms of the slope and r2.
Phenotypic susceptibility, expressed as logfold resistance to commonly used antiretrovirals, was determined by using the Virco Antivirogram recombinant virus assay (3) for 4,995 consecutive clinical samples collected in the United States and submitted to Virco for routine drug resistance testing in 2000. The highest value tested was used in cases where the level of resistance was too high to be measured accurately. Although these samples showed a wide range of susceptibilities to antiretroviral agents, details about each sample (including antiretroviral treatment history) are in most cases unknown. Furthermore, it is worth noting that these samples may not be representative of the general HIV-positive population in the United States, as they were derived only from those patients for whom drug resistance testing had been ordered, often because of treatment failure.
The most commonly observed substitutions in the HIV reverse transcriptase (RT) associated with resistance in this data set corresponded to mutations at codons 215 (occurring in 44% of cases), 184 (39%), 41 (33%), 67 (30%), 103 (25%) 210 (23%), 219 (21%), 70 (19%), 69 (19%), 181 (16%), 118 (15%), 190 (12%), 74 (11%), 98 (10%), and 44 (10%). The most commonly observed substitutions associated with protease inhibitor (PI) resistance corresponded to mutations at codons 63 (83%), 10 (37%), 71 (34%), 77 (33%), 90 (28%), 82 (19%), 20 (18%), 46 (17%), 54 (16%), 84 (11%), and 73 (11%). These mutations occurred in a great variety of permutations, and other well-described mutations appeared at prevalences below 10%. In descending order, decreases in phenotypic susceptibility of greater than fourfold were observed for lamivudine (3TC) (47%), delavirdine (44%), nevirapine (43%), nelfinavir (37%), zidovudine (34%), efavirenz (33%), ritonavir (33%), indinavir (17%), abacavir (19%), amprenavir (13%), didanosine (9%), stavudine (7%), and zalcitabine (7%).
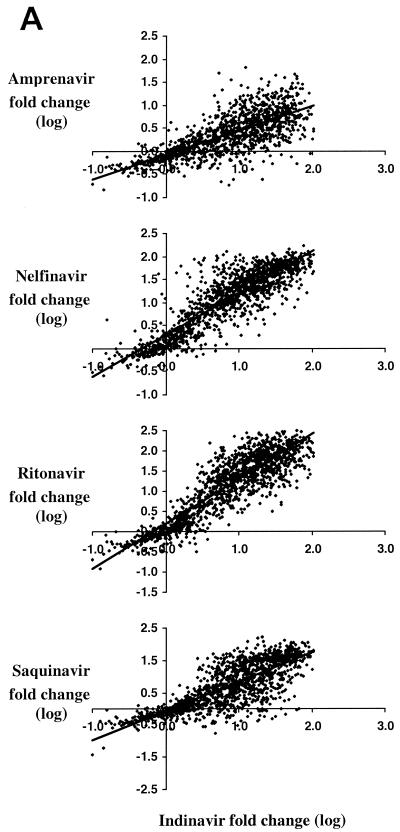
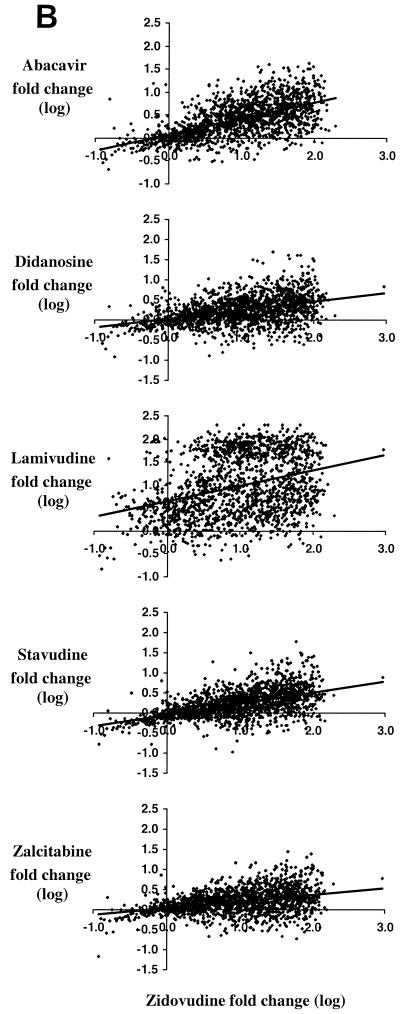
Results of linear regression (log-log) analysis of the susceptibility of HIV-1 to drugs within each of the three drug classes are shown (Fig. 1). For conciseness, indinavir, zidovudine, and efavirenz were arbitrarily chosen as comparators for the PI, nucleoside analogue RT inhibitor (NRTI), and nonnucleoside RT inhibitor (NNRTI) drug classes, respectively. In general, linear regression provided a reasonable fit to the data, except for the data for 3TC and the NNRTIs, likely because a single mutation can confer high-level resistance to these drugs.
FIG. 1.
Relationships among antiretroviral drug susceptibilities within available drug classes in recent clinical isolates. The observed fold changes in susceptibility for over 4,900 clinically derived recombinant HIV-1 isolates are shown on a log-log scale for each of three drug classes, PI (A), NRTI (B), and NNRTI (C), in comparison to indinavir, zidovudine, and efavirenz susceptibility, respectively, used as representative examples.
As expected, the highest level of cross-resistance was observed within the PI and NNRTI classes. In all cases except for those involving amprenavir, comparisons of susceptibilities (log-log) between PIs yielded slopes between 0.75 and 1.3, confirming considerable cross-resistance within this class. Indinavir susceptibility correlated most highly with ritonavir susceptibility (r2 = 0.79, compared with values of 0.70 for nelfinavir, 0.69 for saquinavir, and 0.55 for amprenavir) (Fig. 1). Values of r2 for any PI phenotype pair (except those involving amprenavir) ranged from 0.63 to 0.79. Amprenavir cross-resistance was generally lower; the greatest correlations were with ritonavir (r2 = 0.58) and indinavir (0.55), followed by those with saquinavir (0.49) and nelfinavir (0.45). This may reflect the effects of both inherent differences among the molecules and the patterns of exposure to these PIs in the population.
Almost all samples with >10-fold-decreased efavirenz susceptibility also displayed >10-fold-decreased nevirapine susceptibility, though this was not the case for delavirdine (Fig. 1C). Even with relatively poor fits, r2 values between inhibitors within the NNRTI class were at least 0.62 (Table 1).
TABLE 1.
Correlations among antiretroviral susceptibilitiesa
| Drug | Degree of correlation (r2) of indicated drug
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rit | Nel | Saq | Amp | ABC | 3TC | AZT | d4T | ddC | ddI | EFV | Nev | Del | |
| PIs | |||||||||||||
| Ind | 0.79 | 0.70 | 0.69 | 0.55 | 0.24 | 0.14 | 0.26 | 0.11 | 0.10 | 0.15 | 0.11 | 0.14 | 0.09 |
| Rit | 0.69 | 0.67 | 0.58 | 0.25 | 0.15 | 0.27 | 0.11 | 0.09 | 0.15 | 0.10 | 0.13 | 0.08 | |
| Nel | 0.63 | 0.45 | 0.25 | 0.18 | 0.26 | 0.10 | 0.10 | 0.14 | 0.10 | 0.13 | 0.08 | ||
| Saq | 0.49 | 0.21 | 0.10 | 0.24 | 0.11 | 0.08 | 0.14 | 0.10 | 0.12 | 0.08 | |||
| Amp | 0.21 | 0.08 | 0.22 | 0.12 | 0.10 | 0.15 | 0.08 | 0.09 | 0.06 | ||||
| NRTIs | |||||||||||||
| ABC | 0.42 | 0.35 | 0.24 | 0.36 | 0.38 | 0.08 | 0.10 | 0.04 | |||||
| 3TC | 0.09 | 0.03 | 0.25 | 0.21 | 0.04 | 0.06 | 0.02 | ||||||
| AZT | 0.29 | 0.14 | 0.19 | 0.06 | 0.08 | 0.03 | |||||||
| d4T | 0.19 | 0.23 | 0.03 | 0.03 | 0.02 | ||||||||
| ddC | 0.33 | 0.02 | 0.03 | 0.01 | |||||||||
| ddI | 0.06 | 0.07 | 0.05 | ||||||||||
| NNRTIs | |||||||||||||
| EFV | 0.70 | 0.62 | |||||||||||
| Nev | 0.68 | ||||||||||||
| Del | |||||||||||||
The degree of correlation in fold change in 50% inhibitory concentration compared to that of a reference strain (log scale) among over 4,900 clinically derived recombinant isolates is expressed as r2. Even with low values of r2, most correlations are highly statistically significant due to the large number of observations. Ind, indinavir; Rit, ritonavir; Nel, nelfinavir; Saq, saquinavir; Amp, amprenavir; ABC, abacavir; 3TC, lamivudine; AZT, zidovudine; d4T, stavudine; ddC, zalcitabine; ddI, didanosine; EFV, efavirenz; Nev, nevirapine; Del, delavirdine.
Importantly, both the slope and r2 within the NRTI class were much lower than those of the NNRTIs or PIs (Fig. 1, Table 1). Thus, although there is considerable cross-resistance between stavudine and zidovudine, for example, the correlation is much lower than it is between the PIs. Thus, only approximately one-third of the observed susceptibility of stavudine can be rationalized on the basis of observed zidovudine susceptibility. It is also important to note that the magnitudes of the decreases in susceptibility to didanosine, stavudine, and zalcitabine were all relatively small. Within the NRTI class, the highest correlations observed between susceptibilities were with abacavir (Table 1); these correlations were higher than that observed between zidovudine and stavudine, for example. There was essentially no relationship between 3TC susceptibility and either zidovudine or stavudine susceptibility. It is important to note that a large number of samples actually had modestly decreased levels of 3TC susceptibility rather than simply very high or very low levels associated with the presence or absence, respectively, of a mutation at codon 184.
Most of the correlations shown are highly statistically significant (P < 0.001), but this is in part driven by the very large number of samples, and these correlations may not always reflect clinical significance or even cause-and-effect relationships. The correlations observed are not necessarily all due to true cross-resistance (resistance to multiple compounds of similar inhibitory mechanisms as a consequence of exposure to one or more compounds of this type) but are in some cases due to concurrent resistance (coresistance resulting from past exposure to two or more drugs). The effect of concurrent resistance is most strongly demonstrated in the positive correlation observed between susceptibility to the RT inhibitors and susceptibility to the PI drug classes, for which little or no common genetic basis for cross-resistance exists. The significant correlation between zidovudine (NRTI) resistance and PI resistance, for example, is therefore most likely due to concurrent resistance, where individuals have most likely had past exposure to drugs of both classes.
Despite this information, we argue that the data are most consistent with a high degree of true cross-resistance within drug classes, as within-class resistance for the PI and NNRTI classes was much more strongly correlated (minimum r2 = 0.45 and 0.62 for the PI and NNRTI classes, respectively) than any correlation observed for resistance across drug classes (maximum r2 = 0.27). Furthermore, the slope and r2 of the regression lines were close to unity across all four PIs, despite the improbability of large numbers of patients having experienced all four PIs. The observation that strong correlations in susceptibility exist between members of the nucleoside drug class is consistent with previously reported results (J. Whitcomb, M. Maranta, N. Parkin, N. Hellman, and C. Petropoulos, Abstr. 1st IAS Conference on HIV Pathogenesis and Treatment, abstr. 592, 2001).
Finally, the data point to a common mechanism of phenotypic resistance: the fact that only a fairly small number of resistance mutations are able to confer the observed phenotypic susceptibility changes implies that the majority of mutations have broader, incremental effects that alter resistance within the entire class. While some mutations confer resistance almost uniquely to a single compound (such as the mutation coding for the D30N change in the protease or the M184V change in the RT), these appear to be the exception rather than the rule. It is also important to note that the recombinant phenotype assay used essentially rules out mechanisms of phenotypic resistance other than those resulting from mutations in the protease and/or RT gene (and a small portion of gag).
The observation that significant levels of phenotypic cross-resistance exist within drug classes may have important clinical implications in the treatment of drug-experienced individuals failing their current regimens and highlights the need for the development of new therapies. The quantitative analyses of prevalent mutations, reduced sensitivity, and phenotypic cross-resistance from a large cross section of clinical isolates presented here may provide insight for the development of newer antiretrovirals.
REFERENCES
- 1.Deeks, S. G. 2001. Nonnucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S25-S33. [DOI] [PubMed] [Google Scholar]
- 2.Hanna, G. J., and R. T. D'Aquila. 1999. Antiretroviral drug resistance in HIV-1. Curr. Infect. Dis. Rep. 1:289-297. [DOI] [PubMed] [Google Scholar]
- 3.Hertogs, K., M.-P. de Béthune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan, J. E., D. Hanson, M. S. Dworkin, T. Frederick, J. Bertolli, M. L. Lindegren, S. Holmberg, and J. L. Jones. 2000. Epidemiology of human immunodeficiency virus-associated opportunistic infections in the United States in the era of highly active antiretroviral therapy. Clin. Infect. Dis. 30(Suppl. 1):S5-S14. [DOI] [PubMed] [Google Scholar]
- 5.Loveday, C. 2001. Nucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S10-S24. [DOI] [PubMed] [Google Scholar]
- 6.Miller, V. 2001. Resistance to protease inhibitors. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S34-S50. [DOI] [PubMed] [Google Scholar]
- 7.Moore, R. D., and R. E. Chaisson. 1999. Natural history of HIV infection in the era of combination antiretroviral therapy. AIDS 13:1933-1942. [DOI] [PubMed] [Google Scholar]
- 8.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg, et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 9.Swanstrom, R., and J. Eron. 2000. Human immunodeficiency virus type-1 protease inhibitors: therapeutic successes and failures, suppression and resistance. Pharmacol. Ther. 86:145-170. [DOI] [PubMed] [Google Scholar]