Abstract
A new natural TEM derivative, named TEM-87, was identified in a Proteus mirabilis isolate from an Italian hospital. Compared to TEM-1, TEM-87 contains the following mutations: E104K, R164C, and M182T. Kinetic analysis of TEM-87 revealed extended-spectrum activity against oxyimino cephalosporins (preferentially ceftazidime) and aztreonam. Expression of blaTEM-87 in Escherichia coli decreased the host susceptibility to these drugs.
Since first being identified at the beginning of the 1980s (10), extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae have spread worldwide by nosocomial routes (2, 7, 9, 19, 21). ESBLs comprise a group of active-site serine enzymes that variably confer resistance to oxyimino cephalosporins and monobactams (8). Most ESBLs found in clinical isolates of Enterobacteriaceae are variants of the original TEM-1 and SHV-1 enzymes in which one or more amino acid substitutions extend the substrate specificities (3, 4, 8, 14, 15, 21).
In this paper, we report the characterization of a new natural TEM derivative with ESBL activity, named TEM-87, that was identified in a Proteus mirabilis isolate during a survey that was recently undertaken to evaluate the prevalence of ESBL-producing isolates of Enterobacteriaceae in Italian hospitals (G. Amicosante, M. Perilli, M. R. De Massis, E. Dell'Amico, B. Segatore, N. Franceschini, C. Bianchi, and G. M. Rossolini, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1992, 2000; G. Amicosante, M. Perilli, M. R. De Massis, F. Giuliani, D. Setacci, A. Zollo, and G. M. Rossolini, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1993, 2000).
(This work was presented in part at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, 17 to 20 September 2000.)
P. mirabilis NO-113 was isolated in 1999 from a wound of an inpatient in a surgical ward at Novara Hospital (Novara, Italy). MICs were determined by a macrodilution broth procedure with Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.) as recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (13). β-Lactam compounds were from Sigma Chemical Co. (St. Louis, Mo.), with the following exceptions. Clavulanic acid and ceftazidime were from GlaxoSmithKline (Verona, Italy), piperacillin and tazobactam were from Wyeth-Lederle (Catania, Italy), cefepime and aztreonam were from Bristol-Myers Squibb (Wallingford, Conn.), and nitrocefin was from Unipath (Milan, Italy).
Hybridizations with blaTEM and blaSHV probes were carried out essentially as described previously (17). Briefly, the blaTEM and blaSHV probes were PCR amplicons containing the complete blaTEM-1 (20) or blaSHV-1 (1) coding sequences, respectively, and were labeled with 32P by the random priming technique (17). Colony blot hybridization was carried out on bacteria grown directly on nitrocellulose filters (Schleicher & Schuell, Dassel, Germany) layered onto MacConkey (for P. mirabilis) or Mueller-Hinton (for Escherichia coli) agar plates (Difco). In situ lysis of bacterial colonies and the setting of hybridization conditions were carried out as described previously (17). Plasmid purifications were carried out by the alkaline lysis method (17). E. coli DH5α (Life Technologies, Milan, Italy) was used as a recipient for the ESBL-encoding plasmid carried by NO-113 in transformation experiments (6). PCR amplification of the blaTEM gene and flanking regions was carried out using primers Mab/F (5"-GGGGAGCTCATAAAATTCTTGAAGAC) and Mab/R (5"-GGGGGATCCTTACCAATGCTTAATCA) that had been modified from the versions described previously (12), the AmpliTaq Gold DNA polymerase (Perkin-Elmer, Milan, Italy), and the ESBL-encoding plasmid pPm87 purified from E. coli (100 ng) as a template. Cycling conditions were as follows: denaturation at 95°C for 30 s, annealing at 42°C for 1 min, and extension at 72°C for 1 min, with all three steps repeated for 30 cycles. Direct sequencing of PCR amplicons was performed with a dRhodamine Terminator Cycle Sequencing Ready kit (Perkin-Elmer), an ABI PRISM 377 automatic DNA sequencer (Perkin-Elmer), and custom sequencing primers. The sequences were determined for both strands of the entire coding sequence. Sequencing was performed on PCR products derived from two independent reactions with identical results.
TEM-87 was purified from E. coli DH5α(pPm87) grown overnight at 37°C under aerobic conditions in 6 liters of brain heart infusion broth containing ceftazidime at a concentration of 30 μg/ml. Cells were harvested by centrifugation and resuspended in 300 ml of 50 mM Tris-HCl buffer (pH 8.0). A crude extract was prepared by sonication (five times for 30 s each time at 60 W), and cell debris was removed by high-speed centrifugation (105,000 × g for 30 min at 4°C). The clarified extract was loaded (flow rate, 3 ml/min) onto a Q-Sepharose FF column (2 by 20 cm; Amersham-Pharmacia Biotech, Milan, Italy) and equilibrated with 50 mM Tris-HCl buffer (pH 8.0), and the β-lactamase was eluted with a linear gradient of NaCl (0 to 1 M) in the same buffer. The active fractions were pooled, dialyzed overnight at 4°C against 25 mM bis-Tris buffer (pH 6.5), loaded (flow rate, 1 ml/min) onto a Mono-Q HR 5/5 column (Amersham-Pharmacia Biotech), and equilibrated with the same buffer, and the enzyme was eluted with a linear gradient of NaCl (0.2 to 0.8 M) in the same buffer. The fractions containing β-lactamase activity were pooled, dialyzed overnight at 4°C against 25 mM bis-Tris buffer (pH 7.1), and loaded (flow rate, 0.5 ml/min) onto a Mono-P HR 5/20 column (Amersham-Pharmacia Biotech) equilibrated with the same buffer. The proteins were eluted with 25 ml of 10-fold-diluted Polybuffer 74 in the pH range of 7 to 4. During purification, the β-lactamase activity was monitored by spectrophotometrically by measuring the hydrolysis of 200 μM ceftazidime as a substrate at 30°C in 50 mM sodium phosphate buffer (pH 7.0).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analytical isoelectric focusing (IEF) for detection of β-lactamases in crude bacterial extracts were carried out as previously described (14).
Steady-state kinetic parameters (Km and kcat) were determined by measuring substrate hydrolysis under initial rate conditions and by using the Hanes linearization of the Michaelis-Menten equation (18). Substrate hydrolysis was measured with a lambda 2 spectrophotometer (Perkin-Elmer) at 30°C in 50 mM sodium phosphate buffer (pH 7.0) containing 0.2 M KCl to prevent enzyme instability. The low Km values for penicillins were determined both as Kis, as previously described (18), with 100 μM nitrocefin as a reporter substrate, and by analyzing complete hydrolysis time courses (5). Final values were calculated by averaging the results yielded by the two methods. Inhibition by clavulanic acid and tazobactam was monitored with 100 μM nitrocefin as the reporter substrate.
Identification of a new natural TEM-derived ESBL, TEM-87, in a P. mirabilis clinical isolate.
P. mirabilis NO-113 was resistant to ampicillin, piperacillin, cefazolin, ceftazidime, and aztreonam; intermediate to amoxicillin-clavulanate and cefepime; and susceptible to piperacillin-tazobactam and cefotaxime (Table 1). ESBL production was suspected on the basis of the susceptibility pattern and of a positive double-disk synergy test (9) between clavulanate and ceftazidime. Analytical IEF of a crude extract of NO-113 revealed the presence of three β-lactamase activities of pIs 5.8, 7.1, and 8.7 (data not shown). A colony blot hybridization, carried out by using blaTEM- and blaSHV-specific probes, revealed the presence of blaTEM- but not of blaSHV-related sequences in NO-113 (data not shown). Analysis of a plasmid preparation from this strain revealed the presence of an approximately 7-kb plasmid, named pPm87. Transformation of E. coli DH5α with this preparation yielded ceftazidime-resistant transformants that contained an apparently identical plasmid (data not shown). These transformants were recognized by the blaTEM probe, produced a single β-lactamase activity of pI 5.8 as shown by analytical IEF, and exhibited a β-lactam resistance phenotype compatible with the production of an ESBL (Table 1).
TABLE 1.
MICs of various β-lactams for P. mirabilis NO-113 and E. coli DH5α(pPm87), producing the TEM-87 enzymea
| Antibiotic(s) | MIC (μg/ml) for:
|
||
|---|---|---|---|
| P. mirabilis NO-113b | E. coli DH5α(pPm87)c | E. coli DH5αd | |
| Ampicillin | >64 | >64 | 4 |
| Amoxicillin-clavulanatee | 16 | 16 | 4 |
| Piperacillin | >256 | >256 | 1 |
| Piperacillin-tazobactamf | 8 | 0.5 | 0.5 |
| Cefazolin | >64 | 32 | 2 |
| Cefotaxime | 2 | 1 | ≤0.06 |
| Ceftazidime | 64 | >64 | 0.12 |
| Cefepime | 16 | 4 | ≤0.06 |
| Aztreonam | 32 | 64 | 0.12 |
The in vitro susceptibility of E. coli DH5α is also shown for comparison.
The enzymes detected in the crude extract of this strain by analytical IEF had pIs of 5.8, 7.1, and 8.7.
The enzyme detected in the crude extract of this strain by analytical IEF had a pI of 5.8.
No enzymes were detected in the crude extract of this strain.
Clavulanic acid was used at a 1:2 ratio.
Tazobactam was used at a fixed concentration of 4 μg/ml.
PCR for blaTEM alleles performed on plasmid pPm87 purified from one of the E. coli transformants yielded an amplicon of the expected size (1.04 kb). Direct sequencing of this amplicon revealed the presence of a blaTEM allelic variant encoding a new natural TEM derivative, named TEM-87, which carried the following substitutions compared to TEM-1: Q4K, E104K, R164C, and M182T. Apart from the substitution in the signal peptide, TEM-87 exhibits the same set of mutations shown by TEM-43 (21), except for the presence of a cysteine instead of a histidine at position 164. Assuming a cleavage of the signal peptide identical to that of TEM-1, the predicted molecular weight and pI of mature TEM-87 would be 28,845 and 5.84, respectively.
Purification and properties of TEM-87.
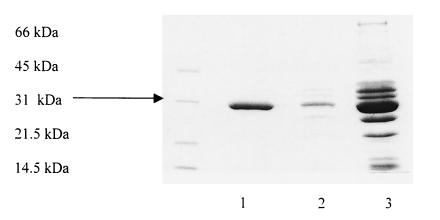
TEM-87 was purified from E. coli DH5α(pPm87) by three chromatographic steps, which yielded an enzyme that was more than 95% pure as evaluated by SDS-PAGE analysis (Fig. 1). IEF showed that the purified TEM-87 enzyme focused at pH 5.8. No other contaminating β-lactamase band was observed.
FIG. 1.
SDS-PAGE analysis of various purification steps of TEM-87 β-lactamase. Lane 1, final preparation after fast chromatofocusing; lane 2, enzyme-containing fraction after Mono-Q chromatography; lane 3, enzyme-containing fraction after Q-Sepharose FF chromatography. Protein size standards (in kilodaltons) are indicated to the left of the gel..
Kinetic parameters of TEM-87, determined with various β-lactams, revealed that the enzyme is able to efficiently hydrolyze penicillins, cephalosporins (including oxyimino cephalosporins), and aztreonam (Table 2). The highest hydrolytic efficiencies (kcat/Km ratio, >106 M−1 · s−1) were observed with penicillins. With cephalosporins, the hydrolytic efficiencies were 10- to 100-fold lower, being notably higher with ceftazidime, cefazolin, and cefepime than with cefotaxime. Aztreonam also behaved as a good substrate for TEM-87, with a kcat and a hydrolytic efficiency that were almost threefold higher than those of cefotaxime (Table 2). Both clavulanic acid and tazobactam inhibited TEM-87. The 50% inhibitory concentration of tazobactam (55 nM) was lower than that of clavulanic acid (130 nM). In short competition assays, these inhibitors provided a competitive model.
TABLE 2.
Kinetic parameters for TEM-87 β-lactamasea
| Antibiotic | Km (μM) | kcat (s−1) | kcat/Km (M−1 · s−1) |
|---|---|---|---|
| Benzylpenicillin | 6.2b | 39 | 6.3 × 106 |
| Ampicillin | 5.5b | 18 | 3.3 × 106 |
| Carbenicillin | 1.5b | 6.7 | 4.5 × 106 |
| Piperacillin | 14 | 70 | 5.0 × 106 |
| Cefazolin | 54 | 13 | 2.4 × 105 |
| Nitrocefin | 16 | 31 | 1.9 × 106 |
| Cefotaxime | 53 | 2.2 | 4.1 × 104 |
| Ceftazidime | 94 | 23 | 2.4 × 105 |
| Cefepime | 23 | 4.4 | 1.9 × 105 |
| Aztreonam | 50 | 5.3 | 1.1 × 105 |
Data for individual kinetic parameters are mean values of three measurements. Standard deviations were always lower than 10%.
The Km values for benzylpenicillin, ampicillin, and carbenicillin were calculated both as Kis and by analyzing complete hydrolysis time courses (5), as described in the text. Data represent averages of the results obtained with the two methods.
Compared with DH5α. E. coli DH5α(pPm87) showed a pattern of decreased susceptibility to β-lactams that was consistent overall with kinetic data. The MICs of cefotaxime and cefepime, although increased, remained below the breakpoint for susceptibility. When combined with amoxicillin and piperacillin, respectively, clavulanic acid and tazobactam were able to reduce their MICs, but only with the latter combination did the MIC fall below the breakpoint for susceptibility (Table 1).
Concluding remarks.
TEM-87 is a new natural TEM-type ESBL containing, in the mature protein, a set of mutations (E104K, R164C, and M182T) which resemble that of TEM-43 (21), except for the presence of a cysteine residue instead of a histidine at position 164. The roles of the E104K and M182T mutations in the evolution of TEM-type β-lactamases have already been described (16). The presence of a cysteine at position 164 likely contributes to the extension of substrate specificity by the same mechanism proposed for other mutations already found at this position. i.e., by increasing the Ω loop flexibility following a reduction of the hydrogen bonds at the neck of the loop (11). Compared to TEM-43 (21), TEM-87 exhibits an overall similar kinetic behavior but also some differences, including higher affinity and efficiency toward penicillins with bulky side chains (carbenicillin and piperacillin) and a more marked preference for ceftazidime than for cefotaxime and aztreonam. These findings suggest that the nature of the substitution at position 164 may significantly influence the kinetic properties of the enzyme.
Nucleotide sequence accession number.
The nucleotide sequence of the blaTEM-87 gene has been submitted to the EMBL-GenBank database and assigned the accession number AF250872.
Acknowledgments
This work was supported in part by a grant from MURST-PRIN 99 to G.A. and partially by a grant from Wyeth-Lederle.
REFERENCES
- 1.Barthélémy, M., J. Peduzzi, and R. Labia. 1988. Complete amino acid sequence of p453-plasmid-mediated PIT-2 β-lactamase (SHV-1). Biochem. J. 251:73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R., C. De Champs, D. Sirot, C. Chanal, R. Labia, and J. Sirot. 1999. Diversity of TEM mutants in Proteus mirabilis. Antimicrob. Agents Chemother. 43:2671-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, P. A., C. E. Cherubin, V. Idemyor, B. A. Rasmussen, and K. Bush. 1994. Multiply resistant Klebsiella pneumoniae strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing β-lactamases in a single isolate. Antimicrob. Agents Chemother. 38:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A., N. V. Jacobus, N. Bhachech, and K. Bush. 1996. TEM-28 from an Escherichia coli clinical isolate is a member of the His-164 family of TEM-1 extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 40:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Meester, F., B. Joris, G. Reckinger, C. Bellefroid-Bourguignon, J. M. Frère, and S. G. Waley. 1987. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and dd-peptidases. Biochem. Pharmacol. 36:2393-2403. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan, D., J. Jessee, and F. R. Bloom. 1995. Techniques for transformation of E. coli. In B. D. Hames and D. M. Glover (ed.), DNA cloning: a practical approach, 2nd ed., vol. 1. IRL Press, Oxford, England.
- 7.Jacoby, G. A., and A. A. Medeiros. 1991. More extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35:1697-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby, G. A. 1997. Extended-spectrum β-lactamases and other enzymes providing resistance to oxyimino-β-lactams. Infect. Dis. Clin. N. Am. 11:875-887. [DOI] [PubMed] [Google Scholar]
- 9.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 10.Knothe, H., P. Shah, V. Kremery, M. Anatal, and S. Mitsuhashi. 1983. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315-317. [DOI] [PubMed] [Google Scholar]
- 11.Knox, J. R. 1995. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob. Agents Chemother. 39:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mabilat, C., S. Goussard, W. Sougakoff, R. C. Spencer, and P. Courvalin. 1990. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum β-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid 23:27-34. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard. NCCLS document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Perilli, M., A. Felici, N. Franceschini, A. De Santis, L. Pagani, F. Luzzaro, A. Oratore, G. M. Rossolini, J. R. Knox, and G. Amicosante. 1997. Characterization of a new TEM-derived β-lactamase produced in a Serratia marcescens strain. Antimicrob. Agents Chemother. 41:2374-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perilli, M., B. Segatore, M. R. De Massis, M. L. Riccio, C. Bianchi, A. Zollo, G. M. Rossolini, and G. Amicosante. 2000. TEM-72, a new extended-spectrum β-lactamase detected in Proteus mirabilis and Morganella morganii in Italy. Antimicrob. Agents Chemother. 44:2537-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrosino, J., C. Cantu III, and T. Palzkill. 1998. β-Lactamases: protein evolution in real time. Trends Microbiol. 6:323-327. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Segel, I. H. 1976. Biochemical calculations, 2nd ed., p. 236-241. John Wiley & Sons, New York, N.Y.
- 19.Sirot, J., C. Chanal, A. Petit, D. Sirot, R. Labia, and G. Gerbaud. 1988. Klebsiella pneumoniae and other Enterobacteriaceae producing novel plasmid-mediated β-lactamases markedly active against third-generation cephalosporins: epidemiologic studies. Rev. Infect. Dis. 10:850-859. [DOI] [PubMed] [Google Scholar]
- 20.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, Y., N. Bhachech, P. A. Bradford, B. D. Jett, D. F. Sahm, and K. Bush. 1998. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli isolates producing TEM-10 and TEM-43 β-lactamases from St. Louis, Missouri. Antimicrob. Agents Chemother. 42:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]