Abstract
We studied the evolutionary relationships between the two protease inhibitor (PI) resistance mutations, D30N and L90M, of human immunodeficiency virus type 1 (HIV-1). The former is highly specific for nelfinavir resistance, while the latter is associated with resistance to several PIs, including nelfinavir. Among patients with nelfinavir treatment failure, we found that D30N acquisition was strongly suppressed when L90M preexisted. Thus, D30N/L90M double mutations not only were detected in a very limited number of patients but also accounted for a minor fraction within each patient. In the disease course, the D30N and L90M clones readily evolved independently of each other, and later the D30N/L90M double mutants emerged. The double mutants appeared to originate from the D30N lineage but not from the L90M lineage, or were strongly associated with the former. However, their evolutionary pathways appeared to be highly complex and to still have something in common, as they always contained several additional polymorphisms, including L63P and N88D, as common signatures. These results suggest that D30N and L90M are mutually exclusive during the evolutionary process. Supporting this notion, the D30N/L90M mutation was also quite rare in a large clinical database. Recombinant viruses with the relevant mutations were generated and compared for the ability to process p55gag and p160pol precursor proteins as well as for their infectivity. L90M caused little impairment of the cleavage activities, but D30N was detrimental, although significant residual activity was observed. In contrast, D30N/L90M demonstrated severe impairment. Thus, the concept of mutual antagonism of the two mutations was substantiated biochemically and functionally.
Protease is an essential enzyme for human immunodeficiency virus type 1 (HIV-1) replication (11, 26) and thus has been a target of anti-HIV-1 treatment (4, 11). One of the characteristic features of the protease is its high polymorphism and flexibility. Nearly 47% of the loci can be mutated naturally (12). However, natural mutations are not randomly scattered through the protease sequence, and variable regions and conserved regions have been identified. These conserved regions are located in the inner side of the protease homodimer and form subsites which are important conformations for substrate binding and expression of the enzyme activity (13).
Today, six protease inhibitors (PIs) are available clinically (10, 16, 18, 25, 28, 30), and all induce drug resistance mutations (2, 17, 23, 24, 27). Interestingly many of these PI resistance mutations are located within the subsites (6, 8), indicating that the acquisition of these mutations might affect protease activity. Indeed, several PI resistance mutations have been reported to demonstrate impaired enzyme activity (14) and reduced viral fitness (15, 19, 34). This reduced activity could be due to some conformational change in the protease, causing a reduced affinity to the natural substrates, p55gag and p160pol precursors (32), or instability of the protease homodimer (33). As flexible as the protease is, accumulation of mutations in the protease continues after the acquisition of so-called primary mutations responsible for drug resistance. The most fit virus will be selected gradually along with the acquisition of additional mutations, which may complement reduced protease activity, and will become the predominant population. A well-known example of such complementary mutations is L63P in protease, which recovers the viral fitness in a background of multiple combinations of other resistance mutations (22). The interactions of accumulated mutations are still not well understood, but there should be more patterns of complementary interaction among the mutations.
It is also plausible that there could be combinations of mutations that are incompatible and might enhance the level of protease activity impairment. However, this type of mutational combination would be difficult to find, as virus with such mutations would be a minor or underrepresented population in vivo. We have been interested in identifying such combinations, as they would provide important information not only on the structure-function relationships of the protease but possibly on the strategic use of drugs. Here we report a specific pair of mutations that greatly impair protease activity, with almost complete loss of viral infectivity and replication capacity. This combination comprises two major drug resistance mutations, a substitution of asparagine (N) for aspartic acid (D) at codon 30 (D30N) and a substitution of methionine (M) for leucine (L) at codon 90 (L90M). D30N is known to be a primary nelfinavir resistance mutation, which appears to be very specific to this inhibitor (24), whereas L90M is a primary mutation responsible for resistance to both nelfinavir and saquinavir (7, 27) and also appears to be associated with resistance to other PIs (31). We demonstrate an extremely low incidence of these two mutations in combination in the clinical setting, suggesting a highly exclusive relationship between the two mutations in the evolutionary process within each patient. Furthermore, we demonstrate their antagonism in vitro, including in protease activity itself as well as viral infectivity and replication capacity.
MATERIALS AND METHODS
Clinical specimens and database.
Two different patient groups were analyzed for their patterns of acquisition and frequencies of D30N, L90M, and other PI resistance mutations. The first group was selected from the patient samples sent to the National Institute of Infectious Diseases, Japan, for routine drug resistance genotyping. Patients in whom plasma viral RNA levels were not below 50 copies/ml even after 3 months of a nelfinavir-containing regimen were defined as nelfinavir resistant and selected for the study. A total 43 cases were selected according to this definition. Of these 43 patients, 19 were naive to PI treatment before nelfinavir administration (PI-naive patients), whereas 24 had previously received the other PIs and had then been given nelfinavir treatment (PI-experienced patients). For these 43 patients, genotypic analyses of HIV-1 protease genes were performed. The incidence of the D30N mutation before and after administration of nelfinavir and the subsequent occurrence of the mutation were examined. To evaluate statistical significance, the chi-square test was performed with the STAT View program (SAS Institute Inc., Cary, N.C.).
The other study group was derived from patient samples in a large database (n = 16,996). We used the data to define the frequencies of D30N, L90M, N88D, and V82A and their combinations. The samples were obtained from patients who had received antiretroviral therapy. These were submitted in the United States during 1998 and 1999 for routine assessment of drug susceptibility. Due to the nature of sample collection, we were unable to obtain therapeutic and clinical histories of the registered patients. The patients with viral loads of >1,000 HIV-1 RNA copies/ml were selected, and the frequencies of the mutations D30N, L90M, N88D, and V82A as well as their combinations were determined.
Extraction, amplification, and sequencing of viral RNA.
Viral RNA was extracted from 200 μl of patient plasma by using a commercially available kit (Roche Diagnostics, Mannheim, Germany). Extracted RNA was reverse transcribed using avian myeloblastosis virus reverse transcriptase (RT) (Takara, Otsu, Japan) and the specific primer DRPRO-2 (5"-ATTTTCAGGCCCATTTTTTGA). Subsequently, the protease region was amplified by nested PCR using Taq (Takara) and Pyrobest (Takara) DNA polymerases, and the specific outer and inner primers were used for outer and inner PCR, respectively. The outer primers used were DRPRO-1 (5"-CCAACAGCCCCACCAGA) and DRPRO-2 in the reverse transcription reaction, and the inner primer pairs used were DRPRO-3 (5"-AGCAGGAGACGATAGACAAGG) and DRPRO-4 (5"-CTGGCTTTAATTTTACTGGTA). Outer and inner PCRs were performed with the following program: a 2-min hold at 95°C and then 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. Nucleotide sequences of amplicons were analyzed by cycle sequencing methods using Big-dye terminator (PE Biosystems, Foster City, Calif.) and an ABI-377 autosequencer (PE Biosystems).
Clonal analysis for acquisition of PI resistance mutations.
Clonal analyses of the protease sequences were performed for eight cases, for which plasma samples could be obtained at multiple points, chosen from the PI-experienced cases described above. These patients also demonstrated virological failure with a nelfinavir-containing regimen. The protease genes were cloned, and the nucleotide sequences were determined. For two patients (JPR-1 and JPR-3), the sequence results were aligned by the Clustal-W program of the Phylip package and then analyzed by both the neighbor-joining and the maximum-likelihood methods.
Generation of recombinant virus clones with PI resistance mutations.
A recombinant virus was constructed using HXB2 and the NL4-3 clone as a backbone virus DNA. Initially, an NL4-3 DNA ApaI (restriction site at nucleotide 2011 of NL4-3)-KpnI (restriction site at nucleotide 3831) fragment, which contained the complete protease gene and the 5" half of the RT gene, was inserted into pGEM7zf(−) (Promega, Madison, Wis.). The construct was designated pGEM7/4-3ApaI-KpnI. Using pGEM7/4-3ApaI-KpnI as a template, two restriction sites, a NotI site at nucleotide 2275 and a SmaI site at nucleotide 2592, were introduced into the pol gene as synonymous mutations. Mutagenesis reactions were performed by employing the ExSite PCR-based site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The introduced NotI and SmaI sites corresponded to the 10th codon of protease and the 15th codon of RT, respectively. The following primer sets were used to introduce the restriction sites: NotI, 5"-GATCACTCTTTGGCAGCGGCCGCTGCTCGTCACAATAAAGATA and 5"-TATCTTTATTGTGACGAGCGGCCGCTGCCAAAGAGTGATC; SmaI,5"-AAAATTAAAGCCCGGGATGGATGGCCCAA and 5"-TTGGGCCATCCATCCCGGGCTTTAATTTT. The entire sequence was verified after the mutagenesis reaction, and a clone with introduced NotI and SmaI sites was designated pGEM7/4-3Not-SmaI. An ApaI (restriction site at nucleotide 2011)-to-AgeI (restriction site at nucleotide 3486) fragment, which contained the NotI-to-SmaI fragment, was excised from pGEM7/4-3Not-SmaI and cloned back into the wild-type molecular clone HXB2. This HXB2/NL4-3 recombinant virus clone with NotI and SmaI sites was designated HXB2cv and used as the host vector for the subsequent HIV-1 recombinant viruses with mutant proteases. The six protease mutants D25N, D30N, L90M, N88D, D30N plus L90M (D30N/L90M), and D30N plus N88D plus L90M (D30N/N88D/L90M) were constructed on the basis of the pGEM7/4-3ApaI-KpnI clone. The primer pairs used to construct each protease mutation were as follows: D25N, 5"-TAAAGGAAGCTATATTAAATACAGGAGCAGATG and 5"-CATCTGTCCTGTATTTAATAGAGCTTCCTTTA; D30N, 5"-GGAGCAGATAATACAGTA and 5"-TACTGTATTATCTGCTCC; L90M, 5"-AGAAATCTGATGACTCAG and 5"-CTGAGTCATCAGATTTCT; and N88D, 5"-ATAATTGGAAGAGATCTGTTGACTCAGATT and 5"-AATCTGAGTCAACAGATCTCTTCCAATTAT. After each mutagenesis reaction, the entire sequence was verified and cloned back to HXB2cv as a NotI-SmaI fragment. The full-length virus DNA was transfected into Cos7 cells by using Fugene6 (Roche Molecular Biochemicals), and culture supernatants were harvested 72 h after transfection and used as virus stocks.
Analysis of p55gag and p160pol processing of mutant viruses.
The recombinant DNA clones were transfected into 0.5 × 107 Cos7 cells by electroporation using a Gene Pulser apparatus (Bio-Rad, Hercules, Calif.) at 250 V and 250 μF. The transfected cells were transferred to Falcon T-150 (Becton Dickinson, Lincoln Park, N.J.) culture flasks containing prewarmed Dulbecco modified Eagle medium with 10% fetal calf serum, and the whole medium was changed 24 h after transfection. Seventy-two hours after transfection, the culture supernatant was harvested, and virus lysates were prepared according to a previously described method (20) with a minor modification. In brief, cell debris was removed from the collected culture supernatant by centrifugation at 3,000 rpm (Beckman Allegra 6KR centrifuge) for 15 min at 4°C. The cleared medium was filtered through a 0.2-μm-pore-size filter (Millipore, Bedford, Mass.), overlaid onto a 20% sucrose cushion, and centrifuged at 25,000 rpm (Beckman SW28 rotor) for 1.5 h at 4°C by Optima L-90K (Beckman Coulter, Fullerton, Calif.). The virus pellet was dissolved in an appropriate amount of radioimmunoprecipitation assay lysis buffer (0.05 M Tris-Cl [pH 7.2], 0.15 M NaCl, 0.1% sodium dodecyl sulfate, 1.0% TritonX-100, 1.0% sodium deoxycholate) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins in the gels were transferred to Immobilon P membranes (Millipore) by passive transfer. The membranes were incubated with an HIV-1-seropositive serum for 1 h at 37°C, followed by incubation with a biotinylated secondary antibody (Amersham Pharmacia Biotec, Uppsala, Sweden) and then with avidin-horseradish peroxidase (avidin-HRP) (Amersham Pharmacia Biotec). The HIV-1-positive control human serum used in this study was kindly supplied by T. Lee, Harvard School of Public Health. 3,3"-Diaminobenzidine (Sigma, St Louis, Mo.) was used as a substrate for HRP. For gp120 staining, sheep anti-gp120 C-terminal polyclonal antibody (International Enzymes, Inc., Fallbrook, Calif.) was used, followed by incubation with biotinylated anti-sheep immunoglobulin G secondary antibody (Amersham Pharmacia Biotec) and then avidin-HRP. 3,3"-Diaminobenzidine was used as a substrate for HRP.
Infectivity assay and replication kinetics of recombinant viruses with PI resistance mutations.
The infectivity of each protease mutant virus was assayed using the reporter cell line MAGIC5.B1-4. Details of this cell line are described elsewhere (21). Briefly, the cell line expresses CD4, CXCR4, and CCR5 on the cell surface and also contains the HIV-1 long terminal repeat-driven β-galactosidase gene. Thus, HIV-1-infected cells can be visualized as blue cells when stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The stocks of PI-resistant viruses were produced from transfected Cos7 cells as described above. The amount of virus in each stock was determined by RT activity assay as described below. MAGIC5.B1-4 cells were plated in six-well culture plates at a density of 105 cells per well the day before infection and inoculated with each virus adjusted to contain same amount of RT activity. Each infection was done in triplicate wells. Forty-eight hours after infection, the cells were fixed with 1% formaldehyde-0.5% glutaraldehyde in phosphate-buffered saline and stained with an X-Gal staining kit (Roche Diagnostics). The number of blue cells in each well was counted under a light microscope, and mean values from three wells were calculated.
RESULTS
Relationship between baseline incidence of L90M mutation and acquisition of D30N.
The relationship between baseline L90M mutation prior to nelfinavir administration and subsequent acquisition of D30N was studied with 43 nelfinavir-resistant cases, and the results are summarized in Table 1. There were 16 patients with the L90M mutation at baseline, and of these, only three (19%) acquired the D30N mutation after nelfinavir administration; this is significantly fewer than for the patients without L90M mutations (P = 0.032).
TABLE 1.
Baseline L90M incidence and acquisition of the D30N nelfinavir resistance mutation
| Existence of L90M prior to nelfinavir treatment | n | No. with acquisition of D30N mutation after nelfinavir treatmenta
|
|
|---|---|---|---|
| + | − | ||
| + | 16 | 3 | 13 |
| − | 27 | 13 (10) | 14 (9) |
Numbers in parentheses indicate numbers of PI-naive cases.
Sequence analysis was done by direct sequencing of the mixture of individual PCR products and hence did not rule out the possibility that the D30N detected might not always add to the preexisting L90M mutations at a clonal level but rather could represent populations that newly emerged independently of the preexisting L90M population. To assess this possibility, we cloned PCR amplicons from three D30N/L90M double-positive cases in direct sequencing, and the nucleotide sequences were determined for individual clones. No D30N/L90M double mutants were found in 15 clones derived from one of the three patients (JPR-2) (Table 2). These clones were positive for either D30N or L90M or were negative for both. Thus, this patient acquired no detectable intrinsic double mutations. In the other two patients (JPR-1 and JPR-3), minor fractions (2 of 30 clones and 10 of 35 clones, respectively) had the intrinsic double mutant (Table 2). Of 27 patients with baseline mutations other than L90M, 13 (48%) developed the D30N mutation (Table 1). The frequency of D30N acquisition was therefore significantly higher in the absence of L90M than in its presence. These results clearly demonstrated that the D30N mutation readily arose without the preexisting L90M mutation but was strongly restricted in its presence, not only among patients but also within a single patient. Thus, D30N and L90M appeared to be considerably incompatible with each other in clinical cases. Five cases were arbitrarily chosen from the patients who did not acquire the D30N mutation, and similar clonal analyses were performed. Neither D30N/L90M nor D30N was found in a total of 57 clones. They were either positive for only L90M or negative for both D30N and L90M (Table 2). Thus, D30N/L90M double mutants were quite rare in this group of patients.
TABLE 2.
Clonal analyses of D30N acquisition pattern in eight patients undergoing protease inhibitor treatment
| Genotype of clone | No. of clones with acquired D30N in:
|
Total no. of clones without D30N | ||
|---|---|---|---|---|
| JPR-1 | JPR-2 | JPR-3 | ||
| D30N− L90M− | 4 | 2 | 11 | 11 |
| D30N+ L90M− | 12 | 4 | 6 | 0 |
| D30N− L90M+ | 12 | 9 | 8 | 46 |
| D30N+ L90M+ | 2 | 0 | 10 | 0 |
| Total | 30 | 15 | 35 | 57 |
Phylogenetic analysis of D30N and L90M emergence within a single patient.
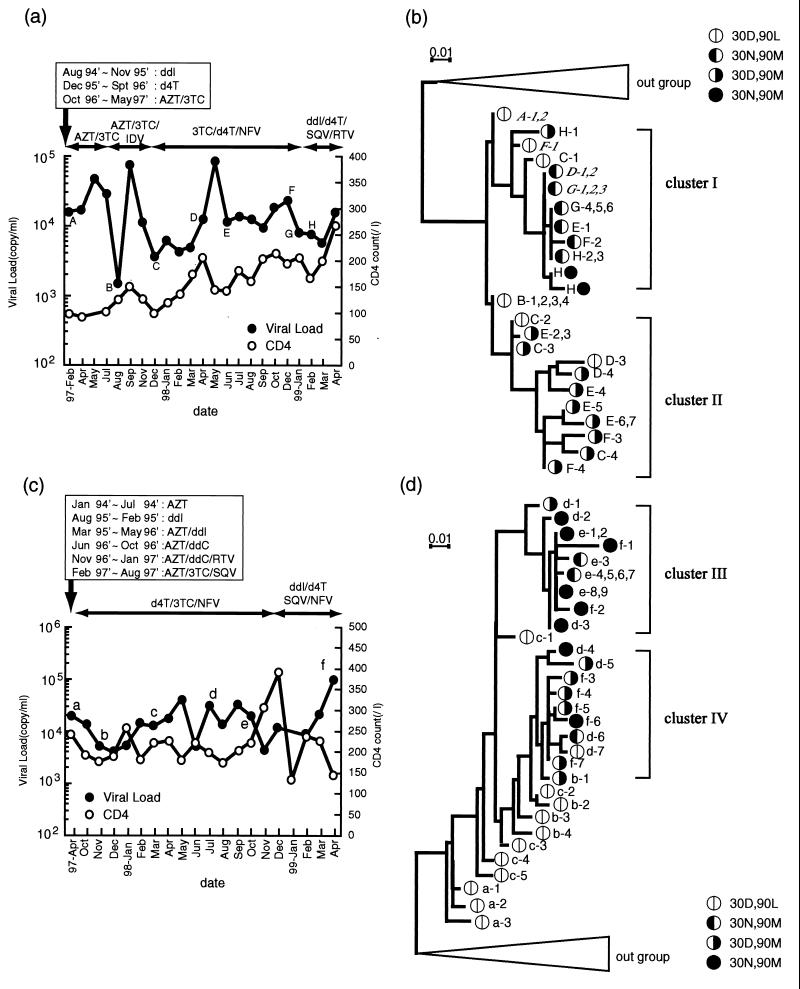
Patients JPR-1 and JPR-3, who acquired the intrinsic D30N/L90M double mutations, were further analyzed to determine the evolutionary pathways of D30N, L90M, and D30N/L90M. Their blood was collected systematically at multiple time points (marked with uppercase letters A to H in Fig. 1a and lowercase letters a to f in Fig. 1c). Subcloning and sequencing of the protease genes were performed for samples obtained at these time points. The sequence data were subjected to phylogenetic analyses. The trees in Fig. 1b and d were drawn by use of the maximum-likelihood method. The clones in the trees were named after their sampling time points and serial numbers at each point.
FIG. 1.
(a and c) Clinical courses of JPR-1 (a) and JPR-3 (c). Treatment protocols and durations are demonstrated with arrows. AZT, zidovudine; ddI, dideoxyinosine; IDV, idinavir; NFV, nelfinavir; SQV, saquinavir; RTV, ritonavir; ddC, dideoxycytosine. (b and d) Development of clones with various mutations, and their phylogenetic relationships, of JPR-1(b) and JPR-3(d). A to H (JPR-1) and a to f (JPR-3) indicate the sampling time points and are referred to the respective clone names with additional serial numbers. Codon 30 and 90 amino acid patterns of the clones are indicated.
Patient JPR-1 was a 56-year-old male hemophiliac patient infected with HIV-1 through contaminated blood products in early 1980s. Although lack of drug adherence had not been reported, the plasma viral load remained above 5,000 copies/ml during most of the observation period. The patient had been treated with several nucleoside RT inhibitors for nearly 8 years and then had switched to zidovudine-lamivudine (3TC)-indinavir triple therapy in May 1997. As this triple therapy turned out to be ineffective in November 1997, the therapy was then switched to 3TC plus stavudine (d4T) plus nelfinavir. At the time of the treatment switch (point C), two of four clones (C-3 and C-4) had the L90M mutation. The second triple therapy also did not lead to plasma viral RNA levels of less than 50 copies/ml, and treatment was further changed to dideoxyinosine plus d4T plus saquinavir plus nelfinavir. During this second treatment, a total of 21 clones from four time points (D to G) were sequenced, and it was found that 47% were D30N clones and 43% were L90M clones. Interestingly, the tree (Fig. 1b) clearly shows that L90M clones and D30N clones evolved from the 30D/90L wild type independently and clustered differently from each other (clusters I and II). It appeared highly unlikely during the whole period that L90M reverted to D30N or vice versa.
It was quite late in the clinical process, after nelfinavir therapy had been switched to saquinavir-ritonavir double-protease therapy, that two D30N/L90M clones (H-4 and H-5) emerged, suggesting a requirement of strong selection pressure for their evolution. Both of the clones appeared to have descended from the D30N lineage. However, their evolutionary process was complex, because they contained several additional substitutions. Interestingly, these substitutions included L10V, K14R, M64I, L63P, and N88D in common, suggesting similar, albeit complex, pathways for the evolution of these two mutants.
A similar phylogenetic analysis of case JPR-3 is shown in Fig. 1d, and the clinical and therapeutic courses are shown in Fig. 1c. The patient is a 24-year old male hemophiliac patient infected in 1980s who also experienced a long treatment history with RT inhibitors and PIs. Again, viral load suppression was not complete during the observation period. It also appeared that the D30N and L90M clones were derived from 30D/90L wild type clones and evolved independently of each other (clusters III and IV). It was also late in nelfinavir therapy (sampling point d and thereafter) that the D30N and L90M double mutants were detected. The evolutionary tree was more complex than that of JPR-1, and hence it was difficult to define the lineage from which the D30N/L90M double mutants emerged. However, at least 8 out of 10 D30N/L90M clones (d-2, d-3, e-1, e-2, e-8, e-9, f-1, and f-2) appeared to be associated with a D30N lineage (Fig. 1d). This is compatible with the view that D30N clones could acquire an additional L90M mutation much more easily than L90M clones could acquire a D30N mutation. Another feature shared with case JPR-1 was that all D30N/L90M mutants contained additional mutations, including N88D and L63P. Overall, the longitudinal clonal analyses of viral populations in the two clinical cases suggested that D30N and L90M mutations tended to evolve independently of each other. The D30N/L90M double mutants could evolve only under strong selection pressure, possibly via acquisition of particular substitutions such as N88D and L63P.
Frequency of PI resistance mutations in a large database.
The incidence of D30N, L90M, D30N/L90M, and other PI resistance mutations was estimated using genotype data derived from a large database (n = 16,996). As shown in Table 3, the frequency of the D30N/L90M double mutation was only 0.8%, whereas that of another double mutation, V82A/L90M, was 5.9%. The frequencies of D30N, L90M, N88D, and V82A single point mutations were found to be 6.3, 29.1, 5.2, and 12.3%, respectively. Given that two residues in either pair, D30N and L90M or V82 and L90M, can change independently of each other, one can predict a value of nearly 1.8% (6.3% × 12.3%) for the D30N/L90M double mutation, which is about half of that (3.6%) for V82A/L90M. The actual estimate (0.8%) for D30N/L90M, however, was less than this prediction. Thus, the results derived from the database also suggest mutual incompatibility between the D30N and L90M mutations. In addition, as these data were derived from population-based sequencing, the D30N/L90M double mutants observed may not all have been together on the same viral genome.
TABLE 3.
Frequencies of PI resistance mutations estimated from a large database (n = 16,996)
| Mutation pattern | No. | Frequency (%) |
|---|---|---|
| D30N | 1,065 | 6.3 |
| L90M | 4,952 | 29.1 |
| V82A | 2,085 | 12.3 |
| N88D | 883 | 5.2 |
| D30N/L90M | 140 | 0.8 |
| V82A/L90M | 999 | 5.9 |
| D30N/N88D | 759 | 4.5 |
| D30N/N88D/L90M | 139 | 0.8 |
Precursor protein processing by mutant proteases.
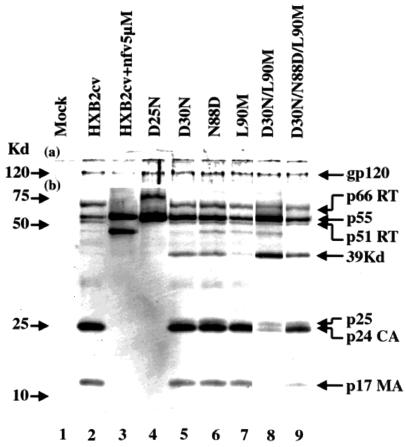
The processing of p55gag and p160pol precursors was analyzed by Western blotting for the recombinant viruses containing D30N, N88D, L90M, D30N/L90M, and D30N/N88D/L90M. The aspartic acid residue at codon 25 is critically involved in the catalytic activity, and its mutation was found to completely abolish the protease activity. We therefore further created a D25N virus and used it as a control of complete protease activity knockout (11). Another control was HXB2cv wild-type virions grown in the presence of 5 μM nelfinavir. The amount of viral protein loaded in each lane was adjusted to give equal intensities of the gp120 band.
As shown in Fig. 2, the products, CA (p24) and MA (p17), generated from p55gag precursor were clearly seen for the HXB2cv wild type (lane 2), whereas they were totally absent in nelfinavir-treated virions (lane 3) and in the D25N virions (lane 4). There was residual precursor p55gag in HXB2cv, but its intensity was much higher in nelfinavir-treated virions and D25N virions (lane 3). The mature Pol products, p66 RT and p51 RT, were seen in HXB2cv, whereas they were not detectable in nelfinavir-treated virions and D25N virions. With all three single point mutations, D30N, N88D, and L90M, both p24 and p17 were seen as clearly as they were in HXB2cv (Fig. 2, lanes 5, 6, and 7). At the same time, however, p55gag precursor was found in remarkable amounts in two (D30N and N88D) of the three mutants, whereas it was at a marginal level in the third mutant (L90M), as in HXB2cv. The 39-kDa band could represent an intermediate cleavage product of p55gag and was seen clearly in the first two mutants but only in a trace amount in the third and not at all in HXB2cv. The p160pol products, p66 RT and p51 RT, were detected in these three single point mutants as clearly as in the parental HXB2cv (Fig. 2). These results indicated that the protease could tolerate almost completely the L90M as well as the D30N or N88D mutation.
FIG. 2.
Analyses of Gag-Pol precursor protein processing of the wild type (HXB2cv) and various mutant viruses by Western blotting. Conditions and viruses used are indicated above each lane. The top part of the membrane (a) was probed with anti-gp120 C-terminal polyclonal antibody, and the bottom part (b) was probed with anti-HIV-1-positive human serum. Positions of molecular markers are indicated on the left, and positions of mature proteins, partially processed intermediate products, and unprocessed precursors are indicated on the right. nfv, nelfinavir.
In contrast, the D30N/L90M double mutation caused a severe impairment, as the mutant generated only a trace amount of p24 and no detectable p17 and reciprocally retained the precursor p55gag and the poorly processed intermediate 39-kDa product in large amounts (Fig. 2, lane 8). Thus, the two mutations in combination were found to be almost intolerable for protease function in relation to p55gag cleavage. However, p160pol processing to yield p66 RT and p51 RT appeared to be retained at least partially. When an additional N88D mutation was introduced into the double mutant, the processing capability was restored at least partially, as seen by the generation of p24 and p17 and a concomitant decrease of p55gag and the 39-kDa product (Fig. 2, lane 9).
Infectivities of recombinant viruses with protease mutations.
We next examined the infectivities of protease mutant viruses by using the MAGIC5.B.1-4 reporter cell assay. The virus inocula were adjusted in each case to contain the same level of RT activity. Each infection was performed in triplicate, and mean blue cell numbers were determined. The wild-type and D30N mutant viruses gave 350 ± 28 and 308 ± 15 positive cells, and L90M displayed 463 ± 13 positive cells, the highest among those tested (Table 4). The virus with D30N/L90M mutations gave only 35 ± 9 positive cells, and this was the lowest among those tested. This is consistent with the lowest cleavage capacity of the mutant protease (Fig. 2). The addition of N88D to D30/L90M (Fig. 2) to give the corresponding D30N/L90M/N88D virus resulted in significantly higher infectivity (191 ± 7 positive cells) than for D30N/L90M, which was also consistent with the protease cleavage activities. Thus, the greater the overall protease impairment, the lower the viral infectivity.
TABLE 4.
Infectivities of protease mutant viruses evaluated with MAGIC5B.1-4 reporter cells
| Virus | No. of blue cells (mean ± SD)a |
|---|---|
| Wild type | 350 ± 28 |
| D30N | 308 ± 15 |
| L90M | 463 ± 13 |
| N88D | 365 ± 41 |
| D30N/L90M | 35 ± 9 |
| D30N/L90M/N88D | 191 ± 7 |
| Mock | 0 |
All infections were performed in triplicate.
DISCUSSION
Previous reports have shown that some PI resistance mutations can cause a reduction in protease activity and function (15, 19, 34). These mutations are mostly located in substrate binding subsites of the protease, and conformational changes due to substitutions reduce the binding affinity between the substrate and the protease, which can cause reduced protease activity and impaired viral fitness (14, 15, 19, 34). Codon 30D is located in the S2 subsite of the protease (24), and nelfinavir resistance mutation D30N has been reported to demonstrate reduced protease activity and viral growth compared to the wild type (19). Codon 90L is not located in this subsite, which is different from the case for most other primary PI resistance mutations. The level of impairment conferred by the L90M mutation has been reported to be less significant than that conferred by D30N (19). These different impacts of D30N and L90M on virus fitness were also confirmed in our analyses of p55gag processing and study of virus growth. Interactions of multiple mutations sometimes causes dramatic effects, and we found that the combination of D30N and L90M strongly impaired the ability of the protease to process p55gag precursor into mature proteins, with the RT generating capacity being somewhat retained. In a previous study, Martinez-Picado et al. (19) described that the D30N/L90M double mutant clone emerged in proviral DNA during a competition culture between a D30N clone and a L90M clone; however, they could not detect the D30N/L90M double mutant virus in the culture supernatant. This suggests that the D30N/L90M clone was less fit in their study, agreeing with our findings.
Combinations of V32I plus I84V/A (3, 5) and G48V plus L90M (9) also have been reported to demonstrate low protease activity and reduced viral fitness; however, the levels of the impairment seem to be less than those we observed with the D30N/L90M pair. Thus, to our knowledge, D30N/L90M is the most incompatible mutation pair described. The double mutation caused almost complete elimination of infectivity and replication capacity and can be regarded as being a virtually mutually exclusive combination. On the other hand, the same series of assays for processing, infectivity, and replication suggested that either D30N or L90M did not compromise protease activity to any significant degree. These results explain why either mutation is selected so frequently but the combination of the two is so infrequent (both in the large database and in the smaller study group). We previously reported that the frequency of D30N acquisition was higher in patients who received nelfinavir as an initial PI than in those who received nelfinavir as an alternative (29). Here we demonstrated that the incidence of D30N acquisition was strikingly low when the L90M mutation already existed as a background mutation.
The longitudinal phylogenetic analysis of virus clones in a nelfinavir treatment failure case, JPR-1, strongly suggested that clones with D30N and those with L90M emerged directly from a 30D/90L wild-type clone and formed two independent clusters during almost the entire observation period. No clear evidence which suggested conversion of L90M to D30N or vice versa was obtained. These data strongly suggested mutual exclusion of D30N and L90M in the process of evolution of PI resistance mutants within a body. However, two clones with D30N/L90M (H-4 and H-5) eventually clearly appeared from the D30N lineage but not from the L90M lineage. Both of the clones possessed additional mutations, many of which were commonly shared. These included L10V, K14R, M64I, L63P, and N88D. Indeed, one such additional change, N88D, was found to restore, at least in part, the impaired processing activity and infectivity. In another nelfinavir treatment failure case, JPR-3, D30N/L90M clones also appeared late in the clinical course. In this case, their ascendants could not be defined well. However, most of them were associated with the D30N lineage. In addition, the L63P and N88D mutations were again commonly found in all D30N/L90M clones from JPR-3. Interestingly, correlation of the N88D mutation with D30N/L90M was also observed in the large database study (data not shown). These additional changes might have compensated sufficiently to allow the enzyme to tolerate the D30N/L90M mutational combination.
In the present study, we did not study drug susceptibility or replication of the viruses under drug selective pressure in vitro. However, in case JPR-1, the predominance of L90M clones (D-3, D-4, E-2 to -7, F-1, F-3, and F-4) during 3TC-d4T-nelfinavir treatment indicates an advantage of L90M for viral replication in vivo. Clones with D30N (G-1 to -6, H-2, and H-3) appeared after the treatment was switched to dideoxyinosine-d4T-saquinavir-ritonavir. It seems that D30N virus has enhanced fitness and higher drug resistance than clones with L90M.
The D30N/L90M protease processed p160pol precursor into p51 and p66 RT proteins, although a little less efficiently than HXB2cv, but it was hardly able to produce the final products of p55gag, MA and CA,. Thus, D30N/L90M protease behaved differently from the completely inactivated D25N protease and the HXB2cv protease whose function was inhibited by 5 μM nelfinavir. These results suggested that a certain level of catalytic activity was retained by D30N/L90M protease, but the substrate-enzyme interaction, particularly interaction with p55gag, might be significantly disturbed or altered.
The exact mechanism of the incompatibility between D30N and L90M is not understood. The three-dimensional structure of the HIV-1 protease dimer shows that the 30D residue is located in the S2 subsite (24), while the 90L residue is rather distal to the catalytic site. Thus, these two loci are not juxtaposed to each other. Therefore, direct steric hindrance or interaction between these residues is unlikely to occur. Interestingly, 88N is located between 30D and 90L, and the N88D mutation was compensatory to some extent. Thus, some surface area including 88N and 90L and the S2 subsite may be involved in or important for anchoring the p55gag substrate on the enzyme. A subtle change of the nonpolar side chain due to L90M and loss of negative charge due to D30N may somehow disturb the enzyme-substrate interaction. Acquisition of a negative charge due to N88D may compensate for the loss of a negative charge at codon 30 due to the D-to-N change.
Here we have identified an antagonistic relationship between the D30N and L90M mutations in the HIV-1 protease in terms of protease function and viral infectivity. We have also suggested that since the protease is highly flexible in structure, it can accommodate additional mutations to overcome incompatible mutations. Theoretically, evolutionary forces drive an impaired protease towards the recovery of its activity (1). Accumulation and selection of such mutations in vivo appeared to have taken a long time period in our clinical cases. The development of mutations that reduce viral fitness might cause a slowing of disease progression and possibly enable the partial restoration of host immune systems. As our analyses were performed mainly by using artificially constructed cloned viruses with limited mutation combinations, further investigations using clinically isolated viruses to identify interactions of drug resistance mutations are required. Accumulation of clinical data may provide guidance for a more rational multiple-drug protocol to help control HIV-1 replication and HIV-1 disease progression.
Acknowledgments
This study was supported by a grant from the Organization of Pharmaceutical Safety and Research (OPSR) of Japan.
We thank the clinicians of the Research Committee on Prevention of Developing Illnesses and Therapy for HIV-Infected Patients of Japan for their collaboration and support.
REFERENCES
- 1.Berkhout, B. 1999. HIV-1 evolution under pressure of protease inhibitors: climbing the stairs of viral fitness. J. Biomed. Sci. 6:298-305. [DOI] [PubMed] [Google Scholar]
- 2.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Teppler, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 3.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debouck, C. 1992. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res. Hum. Retroviruses 8:153-164. [DOI] [PubMed] [Google Scholar]
- 5.Gulnik, S. V., L. I. Suvorov, B. Liu, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 34:9282-9287. [DOI] [PubMed] [Google Scholar]
- 6.Hertogs, K., M. VanHoutte, B. Larder, and V. Miller. 1999. Testing for HIV-1 drug resistance: new developments and clinical implications. Recent Res. Dev. Antimicrob. Agents Chemother. 3:83-104. [Google Scholar]
- 7.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society--USA Panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch, M. S., B. Conway, R. T. D'Aquila, V. A. Johnson, F. Brun-Vezinet, B. Clotet, L. M. Demeter, S. M. Hammer, D. M. Jacobsen, D. R. Kuritzkes, C. Loveday, J. W. Mellors, S. Vella, and D. D. Richman. 1998. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. International AIDS Society--USA Panel. JAMA 279:1984-1991. [DOI] [PubMed] [Google Scholar]
- 9.Jacobsen, H., M. Hanggi, M. Ott, I. B. Duncan, S. Owen, M. Andreoni, S. Vella, and J. Mous. 1996. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J. Infect. Dis. 173:1379-1387. [DOI] [PubMed] [Google Scholar]
- 10.Kim, E. E., C. T. Baker, M. D. Dwyer, B. G. Mureko, B. G. Rao, R. D. Tung, and M. A. Navia. 1995. Crystal structure of HIV-1 protease in complex with VX478, a potent and orally bioavailable inhibitor of the enzyme. J. Am. Chem. Soc. 117:1181-1182. [Google Scholar]
- 11.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 13.Loeb, D. D., R. Swanstrom, L. Everitt, M. Manchester, S. E. Stamper, and C. A. D. Hutchison. 1989. Complete mutagenesis of the HIV-1 protease. Nature 340:397-400. [DOI] [PubMed] [Google Scholar]
- 14.Mahalingam, B., J. M. Louis, C. C. Reed, J. M. Adomat, J. Krouse, Y. F. Wang, R. W. Harrison, and I. T. Weber. 1999. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 263:238-245. [DOI] [PubMed] [Google Scholar]
- 15.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and Gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markowitz, M., M. Conant, A. Hurley, R. Schluger, M. Duran, J. Peterkin, S. Chapman, A. Patick, A. Hendricks, G. J. Yuen, W. Hoskins, N. Clendeninn, and D. D. Ho. 1998. A preliminary evaluation of nelfinavir mesylate, an inhibitor of human immunodeficiency virus (HIV)-1 protease, to treat HIV infection. J. Infect. Dis. 177:1533-1540. [DOI] [PubMed] [Google Scholar]
- 17.Markowitz, M., H. Mo, D. J. Kempf, D. W. Norbeck, T. N. Bhat, J. W. Erickson, and D. D. Ho. 1995. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J. Virol. 69:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz, M., M. Saag, W. G. Powderly, A. M. Hurley, A. Hsu, J. M. Valdes, D. Henry, F. Sattler, A. La Marca, J. M. Leonard, and D. D. Ho. 1995. A preliminary study of ritonavir, an inhibitor of HIV-1 protease, to treat HIV-1 infection. N. Engl. J. Med. 333:1534-1539. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuda, Z., X. Yu, Q. C. Yu, T. H. Lee, and M. Essex. 1993. A virion-specific inhibitory molecule with therapeutic potential for human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 90:3544-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mochizuki, N., N. Otsuka, K. Matsuo, T. Shiino, A. Kojima, T. Kurata, K. Sakai, N. Yamamoto, S. Isomura, T. N. Dhole, Y. Takebe, M. Matsuda, and M. Tatsumi. 1999. An infectious DNA clone of HIV type 1 subtype C. AIDS Res. Hum. Retroviruses 15:1321-1324. [DOI] [PubMed] [Google Scholar]
- 22.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 23.Partaledis, J. A., K. Yamaguchi, M. Tisdale, E. E. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, C. M. Stuver, R. A. Byrn, and D. J. Livingston. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patick, A. K., H. Mo, M. Markowitz, K. Appelt, B. Wu, L. Musick, V. Kalish, S. Kaldor, S. Reich, D. Ho, and S. Webber. 1996. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob Agents Chemother 40:292-297. (Erratum, 40:1575.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng, C., B. K. Ho, T. W. Chang, and N. T. Chang. 1989. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J. Virol. 63:2550-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts, N. A. 1995. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS 9(Suppl. 2):27-32. [PubMed] [Google Scholar]
- 28.Roberts, N. A., J. A. Martin, D. Kinchington, A. V. Broadhurst, J. C. Craig, I. B. Duncan, S. A. Galpin, B. K. Handa, J. Kay, A. Krohn, R. W. Lambert, J. H. Merrett, J. S. Mills, K. E. B. Parkes, S. Redshaw, A. J. Ritchie, D. L. Taylor, G. J. Thomas, and P. J. Machin. 1990. Rational design of peptide-based HIV proteinase inhibitors. Science 248:358-361. [DOI] [PubMed] [Google Scholar]
- 29.Sugiura, W., T. Oishi, A. Okano, M. Matsuda, H. Abum, K. Yamada, M. Koike, M. Taki, M. Ishikawa, T. Miura, K. Fukutake, K. Gouchi, A. Ajisawa, A. Iwamoto, H. Hanabusa, J. Mimaya, J. Takamatsu, N. Takata, E. Kakishita, S. Higasa, S. Kashiwagi, A. Shirahata, and Y. Nagai. 1999. Two possible pathways for acquisition of mutations related to nelfinavir resistance. Jpn. J. Infect. Dis. 52:175-176. [PubMed] [Google Scholar]
- 30.Vacca, J. P., B. D. Dorsey, W. A. Schleif, R. B. Levin, S. L. McDaniel, P. L. Darke, J. Zugay, J. C. Quintero, O. M. Blahy, E. Roth, V. V. Sardana, A. J. Schlabach, P. I. Graham, J. H. Condra, L. Gotlib, M. K. Holloway, J. Lin, I.-W. Chen, K. Vastag, D. Ostovic, P. S. Anderson, E. A. Emini, and J. R. Huff. 1994. L-735,524: an orally bioavailable human immunodeficiency virus type 1 protease inhibitor. Proc. Natl. Acad. Sci. USA 91:4096-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaillancourt, M., D. Irlbeck, T. Smith, R. W. Coombs, and R. Swanstrom. 1999. The HIV type 1 protease inhibitor saquinavir can select for multiple mutations that confer increasing resistance. AIDS Res. Hum. Retroviruses 15:355-363. [DOI] [PubMed] [Google Scholar]
- 32.Weber, I. T., and R. W. Harrison. 1999. Molecular mechanics analysis of drug-resistant mutants of HIV protease. Protein Eng. 12:469-474. [DOI] [PubMed] [Google Scholar]
- 33.Xie, D., S. Gulnik, E. Gustchina, B. Yu, W. Shao, W. Qoronfleh, A. Nathan, and J. W. Erickson. 1999. Drug resistance mutations can affect dimer stability of HIV-1 protease at neutral pH. Protein Sci. 8:1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]