Abstract
Previous data have indicated that the development of resistance to amprenavir, an inhibitor of the human immunodeficiency virus type 1 protease, is associated with the substitution of valine for isoleucine at residue 50 (I50V) in the viral protease. We present further findings from retrospective genotypic and phenotypic analyses of plasma samples from protease inhibitor-naïve and nucleoside reverse transcriptase inhibitor (NRTI)-experienced patients who experienced virological failure while participating in a clinical trial where they had been randomized to receive either amprenavir or indinavir in combination with NRTIs. Paired baseline and on-therapy isolates from 31 of 48 (65%) amprenavir-treated patients analyzed demonstrated the selection of protease mutations. These mutations fell into four distinct categories, characterized by the presence of either I50V, I54L/I54M, I84V, or V32I+I47V and often included accessory mutations, commonly M46I/L. The I50V and I84V genotypes displayed the greatest reductions in susceptibility to amprenavir, although each of the amprenavir-selected genotypes conferred little or no cross-resistance to other protease inhibitors. There was a significant association, for both amprenavir and indinavir, between preexisting baseline resistance to NRTIs subsequently received during the study and development of protease mutations (P = 0.014 and P = 0.031, respectively). Our data provide a comprehensive analysis of the mechanisms by which amprenavir resistance develops during clinical use and present evidence that resistance to concomitant agents in the treatment regimen predisposes to the development of mutations associated with protease inhibitor resistance and treatment failure.
There are currently six protease inhibitors (PIs) approved for the treatment of human immunodeficiency virus type 1 (HIV-1) infection: saquinavir (SQV), ritonavir (RTV), indinavir (IDV), nelfinavir (NFV), amprenavir (APV), and lopinavir (LPV). Reduced susceptibility to each of these antiretroviral agents can arise, both in vitro and in vivo, following selection and outgrowth of viral mutant strains and is associated with specific amino acid substitutions in the viral protease (1, 3). Additional compensatory mutations may also be selected in the protease substrate Gag cleavage sites (8, 35).
APV, a novel hydroxyethylamine sulfonamide, is a potent and selective inhibitor of HIV-1 and HIV-2 proteases, with Ki of 0.6 and 19 nM, respectively (13). In vitro selection experiments in which virus was passaged in increasing concentrations of APV, identified an isoleucine-to-valine substitution at protease position 50 (I50V) as a key marker of resistance development to this protease inhibitor (21, 23, 31). The I50V mutation alone confers a two- to three-fold decrease in susceptibility compared to the wild-type virus (21, 23, 31). In the presence of other protease mutations, especially M46I/L and I47V, reduction in susceptibility to APV can increase to greater than 10-fold (21, 31). Protease substitutions L10F and I84V have been observed much more rarely in vitro (21). The eventual replacement of I84V by I50V during continued in vitro selection suggests that the latter genotype is more viable in the presence of the inhibitor at concentrations achieved during these experiments. Limited clinical data derived predominantly from patients receiving APV monotherapy (6, 20) have confirmed the role of the I50V protease mutation in the evolution of reduced viral susceptibility to this agent.
Data from some earlier studies have indicated that the resistance and cross-resistance profiles of APV appear to be distinct from those seen with other protease inhibitors (21, 31, 32). For example, the viral genotypes selected by APV in vitro confer minimal cross-resistance to other protease inhibitors, but cross-resistance, when it does occur, is confined to RTV. Indeed, the induction of increased sensitivity to SQV and IDV has been observed in some viral variants selected by APV in vitro (31). In other studies, clinical isolates which have been selected in vivo by protease inhibitors other than APV and that are resistant to one or more drugs in this class, frequently retain susceptibility to APV (24, 29). Increased sensitivity to APV induced by the protease substitution N88S, which is associated with prior IDV or NFV therapy, has also been observed in clinical isolates, and this effect has been confirmed by site-directed mutagenesis studies (36).
The primary objective of the current study was to explore and describe the evolution of viral genotypes and phenotypes in patients who have experienced virological failure on an APV-containing antiretroviral regimen. In order to accomplish this, a retrospective virological analysis was performed on plasma samples obtained from previously PI-naïve patients who experienced virological failure while receiving APV and nucleoside reverse transcriptase inhibitor (NRTI) combination therapy during a phase III clinical trial, PROAB3006. This open label study, comparing IDV to APV in PI-naïve, NRTI-experienced patients has generated the largest body of data thus far relating to the development of resistance to APV in the clinical setting. In vivo evolution of viral resistance to indinavir has been explored extensively and reported in previous studies (4, 5, 35) and is therefore not discussed in detail here.
The present study describes the distinct viral genotypes that evolved in response to APV exposure during clinical use, the impact of these genotypes on susceptibility to APV, and their impact on susceptibility to other PIs. In addition, it highlights the influence of baseline viral susceptibility to concomitantly administered NRTIs on subsequent viral protease evolution. These findings have important general implications for combination antiretroviral therapy that includes a PI.
MATERIALS AND METHODS
Study population.
A total of 504 patients were enrolled into study PROAB3006. All were NRTI experienced and PI naïve at entry and were randomized to receive either APV (1,200 mg twice daily) or IDV (800 mg three times daily) plus background NRTI therapy for 48 weeks in an open-label study design.
The study population comprised patients who were selected on the basis of experiencing virological failure by week 48 of the study. Patients identified as experiencing virological failure by week 48 either had plasma HIV-1 RNA levels of ≥400 copies/ml (at two consecutive time points at week 8 or beyond or at the last time point within the randomized phase) or had prematurely discontinued the randomized phase prior to week 48 due to virological failure. Patients who discontinued the randomized phase for reasons other than virological failure (e.g., adverse experience, lost to follow-up, etc.) were not included. Paired baseline and postvirological failure samples from 48 APV-treated and 28 IDV-treated patients were analyzed.
Genotypic and phenotypic analysis for protease (PRO), 3" gag terminus (CS), and reverse transcriptase (RT) was attempted for samples taken from this population at baseline and, where possible, postbaseline (weeks 8, 12, 16, 24, 32, and 48). However, where plasma viral RNA was insufficiently high (particularly <1,000 copies/ml), analysis was not always possible.
In addition to the samples from the patients who experienced virological failure, pretreatment samples from all patients who participated in the study were subjected to phenotypic analysis, wherever possible.
Viral RNA extraction and nested RT-PCR.
Plasma viral RNA was extracted using reagents provided with the Amplicor HIV-1 Monitor test kit (19) (Roche Diagnostics). Two nested RT-PCRs were performed to reverse transcribe and amplify protease (PRO)/3" gag terminus (CS) and RT (amino acids 15 to 418). SuperScript II (Life Technologies) and Taq polymerase (PE Applied Biosystems) were used for reverse transcription and amplification, respectively, by a method outlined previously (15).
Population sequencing.
The nucleotide sequence of amplified cDNA products was determined using the Applied Biosystems Inc. PRISM BigDye kit, and reaction products were resolved by electrophoresis on a 4.5% polyacrylamide gel (30% [wt/vol] acrylamide/Bis solution, 29:1; Gibco BRL) with the Applied Biosystems 377 sequencer. Sequences processed by FACTURA were then aligned, analyzed, and compared to consensus B sequences (Los Alamos database) by using the Sequence Navigator program.
Clonal sequencing.
The TOPO cloning kit (Invitrogen Inc.) was used to insert the viral protease/3" gag into the plasmid vector pCRII. Escherichia coli TOP10F" cells were transformed with the resulting ligation products. Following overnight growth on agar plates overlaid with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma), 10 white colonies from each transformation were sequenced as described above.
Drug susceptibility phenotype.
Drug susceptibility phenotypes for a panel of 12 anti-HIV drugs (zidovudine, lamivudine, abacavir, stavudine, didanosine, zalcitabine, nevirapine, APV, IDV, RTV, NFV, and SQV) were determined for baseline isolates by Virco NV (Mechelen, Belgium) using the recombinant virus assay (RVA) (11). Drug susceptibilities for the chimeric viruses produced by homologous recombination were determined using a colorimetric cytopathic effect protection assay (22). Susceptibility to all the test drugs of a wild-type IIIB virus was determined in parallel. The 50% inhibitory concentrations (IC50s) of the test virus and IIIB control virus were determined for each drug. A ratio, the fold reduction (FR) in susceptibility relative to the control (IC50 of test virus/IC50 of IIIB control), was calculated for each of the 12 drugs for each viral isolate. For the purposes of this study, a value greater than 10-fold was considered significant for nevirapine and lamivudine, while greater than 4-fold was considered significant for all other drugs. LPV susceptibility for viral isolates derived from patients experiencing APV therapy failure during the study was also evaluated (LPV was unavailable at study commencement; therefore, baseline isolate LPV susceptibility data were not available).
Site-directed mutagenesis.
A panel of 28 mutant viruses was generated by site-directed mutagenesis of an HXB2 clone using the QuikChange kit (Stratagene) in conjunction with custom-synthesized oligonucleotides. Mutant virus APV susceptibility was evaluated by an RVA. The method and construct used have been detailed previously (25).
Statistical analysis.
Fisher's Exact tests were performed. All data manipulation, tabulations, and calculations were performed using the SAS 6.12 system (SAS Institute, Cary, N.C.) under UNIX. A test was interpreted to be statistically significant if the P value was <0.05.
RESULTS
Phenotypic susceptibility of isolates from PI-naïve patients to APV and other PIs.
A large volume of PI susceptibility data was generated for pretreatment (i.e., PI-naïve) isolates from patients participating in the PROAB3006 study, and although some of the data were not required for the purposes of the specific analyses which are the focus of this report, these data are summarized here for completeness. Table 1 shows the mean, standard deviation, median, minimum, and maximum IC50s of all pretreatment isolates (n = 334) from patients with available data for APV, IDV, NFV, RTV, and SQV.
TABLE 1.
PI susceptibility of isolates from PI-naïve patients
| Parameter | Drug IC50 (μM)
|
||||
|---|---|---|---|---|---|
| APV (n = 334) | IDV (n = 332) | NFV (n = 334) | RTV (n = 324) | SQV (n = 334) | |
| Mean (± SD) | 0.029 ± 0.025 | 0.043 ± 0.037 | 0.061 ± 0.069 | 0.067 ± 0.055 | 0.0065 ± 0.004 |
| Median | 0.020 | 0.038 | 0.045 | 0.060 | 0.005 |
| Maximum | 0.204 | 0.374 | 0.574 | 0.478 | 0.032 |
| Minimum | 0.002 | 0.0014 | 0.0012 | 0.0025 | 0.0006 |
Association between baseline NRTI susceptibility and development of protease mutations.
Patients and investigators had been encouraged to change background NRTIs at the start of the study, but retrospective analysis revealed that many patients had either not switched or had switched to an alternative NRTI to which they had previously developed resistance.
Paired baseline and postvirological failure protease genotypic data were available for 48 APV-treated and 28 IDV-treated patients, of whom 31 out of 48 (65%) and 15 out of 28 (54%) harbored virus that developed protease mutations during treatment with APV or IDV, respectively. Evidence of baseline genotypic mutations associated with resistance to one or more of the NRTIs subsequently received in the study was present in 44 out of 48 (92%) and 25 out of 28 (89%) APV- and IDV-treated patients, respectively (data not shown).
Evidence of decreased susceptibility (>4 FR) at baseline to NRTIs administered subsequently during the study was shown to be significantly associated with an increased likelihood of the development of viral protease mutations in both treatment groups. Among the viruses from 41 out of 48 patients in the APV-treated group with available baseline NRTI phenotypic data, 21 of 28 (75%) that developed a protease mutation had evidence of resistance to an NRTI received during the study, compared to 4 of 13 (31%) that did not develop a protease mutation (Fisher's P value = 0.014). Similarly, for the 20 out of 28 IDV-treated patients with available baseline NRTI susceptibility data, 12 of 13 (92%) that developed a protease mutation had evidence of resistance to an NRTI subsequently received during the study, compared with 3 of 7 (43%) that did not develop a protease mutation (Fisher's P value = 0.031).
Pathways of viral protease evolution in response to APV therapy.
Four distinct genetic pathways of viral protease evolution producing a reduction in susceptibility to APV were identified among the isolates from 25 of the 48 patients who experienced APV/NRTI combination therapy failure and from whom paired baseline and postvirological failure genotypic and phenotypic data were obtained. These data are presented in Table 2, in which patients are grouped according to which of the four APV-selected viral genotypes were detected. Three of the four pathways were characterized by the selection of single protease amino acid changes at positions 50 (I50V), 54 (I54L or I54M), or, more rarely, 84 (I84V), respectively. The fourth pathway was characterized by an APV-selected viral genotype which harbored a double protease substitution at residues 32 and 47 (V32I+I47V). Additional concomitant amino acid changes, especially at M46 in protease and at p7/p1 (A431V) and p1/p6 (L449F) in Gag, were also observed. Sequential isolates from 2 of the 25 patients displayed one of these genotypes at a particular time point and a different one at a subsequent time point (Table 2, asterisks).
TABLE 2.
Summary of amino acid changes in viral protease and Gag amino acid positions A431 and L449 (p7/p1 and p1/p6 cleavage sites, respectively) selected by APV therapy and the impact of these genotypes on susceptibility to a panel of six PIsa
| Patient no. | Visit/week | APV-selected mutations in protease
|
Gag A431/L449 | Phenotypic susceptibility (FR)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Key | Other | APV | IDV | NFV | RTV | SQV | LPV | |||
| 1 | 16 | I50I/V | ND | ND | 16.00 | 0.40 | 1.40 | 5.00 | 0.90 | 2.40 |
| 2 | 48 | I50V | M46I, A71A/V | ND | NA | NA | NA | NA | NA | NA |
| 3 | 48 | I50V | M46I, V82I | L449F | NA | NA | NA | NA | NA | NA |
| 4 | 36 | I50V | C67W | L449F | 5.90 | 1.00 | 3.30 | 3.80 | 1.00 | 2.90 |
| 5 | 48 | I50V | V82I | L449F | 9.60 | 0.50 | 1.40 | 8.30 | 0.80 | 2.00 |
| 6∗ | 36 | I50V | M46I, L63R | L449F | 5.90 | 1.00 | 2.10 | 1.30 | 0.30 | 3.40 |
| 7 | 48 | I50V | M46I | L449F | 5.00 | 2.00 | 3.70 | 3.60 | 0.50 | 5.70 |
| 8 | 48 | I50V | M46I, A71A/V | ND | 5.30 | 1.30 | 1.00 | 2.50 | 1.30 | 4.00 |
| 9 | 48 | I50V | M46L | ND | 3.30 | 0.30 | 0.70 | 1.50 | 0.20 | 0.90 |
| 6∗ | 24 | V32I, I47V | ND | ND | 2.60 | 1.00 | 1.10 | 0.70 | 0.60 | 2.90 |
| 10 | 24 | V32I, I47V | ND | ND | 5.40 | 1.50 | 2.30 | 1.80 | 0.50 | 2.80 |
| 11 | 48 | V32I, I47V | L10I, M46I | ND | NA | 1.40 | 1.10 | 0.50 | 0.10 | 0.40 |
| 12 | 24 | V32I, I47V | ND | ND | 7.00 | 1.20 | 2.00 | 2.00 | 0.40 | 4.50 |
| 13 | 48 | V32I, I47V | ND | ND | NA | NA | NA | NA | NA | NA |
| 14 | 48 | V32V/I, I47V | L10F, M46M/L/I | A431A/V | 3.60 | 3.10 | 2.10 | 4.80 | 0.40 | 6.60 |
| 15∗∗ | 24 | V32I, I47V | ND | ND | 5.90 | 0.30 | 3.20 | 4.40 | 0.60 | 0.80 |
| 15∗∗ | 16 | I54L | ND | ND | NA | NA | NA | NA | NA | NA |
| 16 | 24 | I54M | ND | ND | 4.90 | 0.60 | 2.10 | 0.80 | 0.60 | 0.80 |
| 17 | 48 | I54L | L33F | ND | 7.60 | 2.10 | 5.90 | 1.80 | 0.70 | 1.80 |
| 18 | 48 | I54L | M46M/L | ND | 0.70 | 0.60 | 0.50 | 0.70 | 1.20 | 0.70 |
| 19 | 48 | I54M | ND | ND | 4.00 | 0.50 | 2.40 | 0.90 | 0.70 | 1.20 |
| 20 | 48 | I54L | M46L | ND | 2.00 | 1.70 | 6.20 | 2.00 | 0.90 | 1.80 |
| 21 | 48 | I54M | ND | ND | 3.40 | 0.80 | 2.30 | 2.60 | 0.80 | 1.40 |
| 22 | 48 | I54I/M | M46I | ND | 2.00 | 1.70 | 1.50 | 3.90 | 0.30 | 0.90 |
| 23 | 48 | I54L | M46L | ND | NA | NA | NA | NA | NA | NA |
| 24 | 48 | I84V | M46L, L63A | A431V | 4.50 | 1.00 | 0.80 | 1.60 | 0.90 | 4.50 |
| 25 | 48 | I84V | K55R | A431V | 10.30 | 1.10 | 1.30 | 4.60 | 0.60 | 3.20 |
FR, fold reduction in susceptibility relative to reference virus. ∗, V32I+I47V was subsequently displaced by I50V -containing genotype; ∗∗, I54L genotype subsequently displaced by V32I+I47V-containing genotype; ND, none detected; NA, not available.
Isolates from the remaining 23 of the 48 patients displayed either no genotypic changes in protease (n = 17) or developed M46I/L protease mutations in the absence of amino acid changes at positions characteristic of any of the four pathways listed above (n = 6). Since these isolates displayed no reduction in phenotypic susceptibility, they will not be discussed further.
The I50V protease substitution developed in nine isolates. The most frequent protease mutation observed in the presence of the I50V was M46I/L (detected in six isolates harboring I50V). There was an association between the emergence of the I50V mutation and the concurrent/subsequent emergence of the p1/p6 L449F mutation in Gag; five of nine viral isolates that developed the I50V developed this mutation.
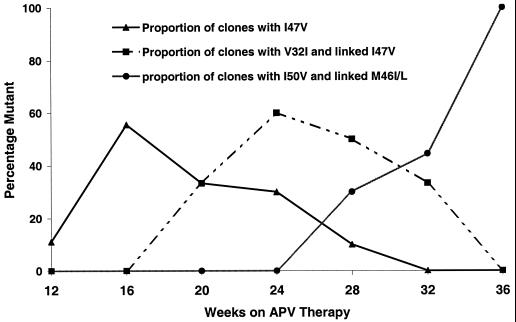
In seven viral isolates, a V32I+I47V double protease amino acid substitution was selected in response to APV. An additional M46I mutation was present in two isolates (patients 11 and 14). Two isolates bearing V32I+I47V (patients 6 and 15) also displayed evidence of evolution of other APV-associated genotypes during the course of APV exposure. In one isolate (patient 15), an I54L that had emerged in the week 16 sample, was displaced by a V32I+I47V viral genotype at week 24. Isolates from patient 6 displayed a distinctive pattern of viral evolution in which successive displacement of viral genotypes was observed. Clonal sequencing (10 clones per PCR product) of the viral protease RT-PCR products from patient 6 (Fig. 1) revealed the early emergence of the I47V, and its subsequent decline as the V32I+I47V genotype emerged to become the dominant genotype in the week 24 isolate. However, following continued APV therapy, the V32I+I47V genotype was displaced by an I50V-containing viral variant and was undetectable in the week 36 sample. Despite extensive clonal analysis of multiple samples from patient 6, the coexistence of V32I+I47V and I50V on the same genome was not observed.
FIG. 1.
Viral protease evolution in patient 6 in response to APV therapy examined by clonal sequencing (10 clones per isolate). Also shown is the displacement of V32I+I47V genotypes by I50V-containing genotypes.
Changes at viral protease amino acid position 54 involving either I54L or I54M substitutions were detected in nine viral isolates. The I54V substitution was not detected in any of the viral isolates derived from the 48 APV-treated patients described in this study. The consecutive appearance and displacement of I54L by V32I+I47V protease genotypes observed in patient 15 was described above. Four of the nine viral isolates that developed I54L/I54M protease mutations also developed M46L/I substitutions and a fifth concomitantly developed an L33F substitution, while the remaining four viral isolates developed I54L or I54/M without concomitant changes at other positions in protease.
Two patients harbored virus that developed the I84V protease substitution during the course of APV treatment. One isolate also carried an M46L substitution in the protease, and both carried p7/p1 A431V substitutions in Gag.
Impact of APV-selected genotypes on susceptibility to APV.
All four genotypes selected by APV in this study were also associated with decreased susceptibility to APV (Table 2). However, the degree of reduction in susceptibility conferred by each of the four genotypes varied (I50V: FR range, 3.3 to 16, and mean, 7.3; V32I+I47V: FR range, 2.6 to 7.0, and mean, 4.9; I54L and I54M: FR range, 0.7 to 7.6, and mean, 3.5). The two I84V-containing isolates exhibited reductions in APV susceptibility, of 4.5- and 10.3-fold, respectively. One of the I50V genotypes displayed a greater reduction in susceptibility to APV than may have been expected from the genotype (patient 1, 16 FR), which appeared to have no other changes in either PRO or CS. This observation cannot be easily explained, although it is possible that a minority population carrying additional changes that were not detected in the genotyping was selected in the generation of the recombinant virus from which the phenotype data were produced.
Impact of APV-selected genotypes on susceptibility to other PIs.
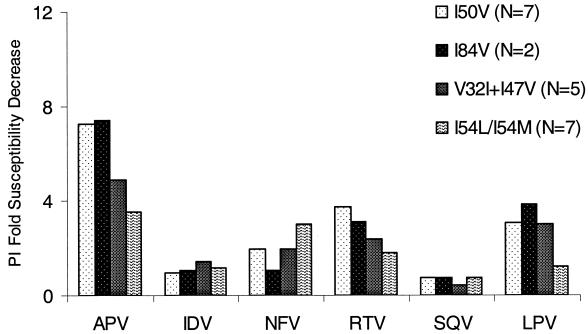
The APV-associated genotypes described above were associated with minimal cross-resistance to other PIs. The mean fold susceptibility decreases to IDV, NFV, RTV, SQV, and LPV were calculated for each of the APV-resistance genotypes and are shown in Fig. 2. All tested isolates exhibited <4-fold susceptibility shifts for IDV and SQV. Just two isolates displayed decreases in susceptibility to NFV (patients 17 and 20, 5.9- and 6.2-fold, respectively), and both of these harbored I54L mutations. Evidence of low-level cross-resistance to RTV was seen in 5 out of 22 isolates for which data were available, with a maximum reduction in susceptibility of 8.3-fold. Low-level (maximum 6.6-fold [patient 14]) cross-resistance to LPV was also observed in 5 out of 22 isolates.
FIG. 2.
Impact on PI susceptibility engendered by each of the four protease genotypic pathways selected by APV. Values given are the mean fold susceptibility decreases relative to that of HXB2 control for isolates for which data were available.
Impact of specific protease mutations on APV susceptibility.
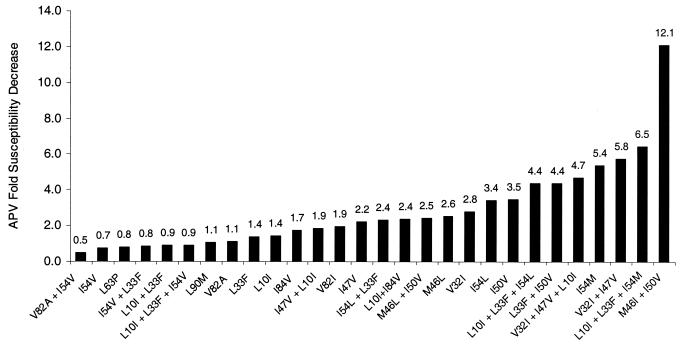
To further elucidate the specific contribution of individual mutations and combinations of mutations, including those selected by APV or other PIs in clinical use, a panel of 28 mutant viruses was generated by site-directed mutagenesis and the impact on susceptibility to APV was determined. APV susceptibilities for each of the 28 viruses varied widely (Fig. 3), but a pattern of increasing APV resistance was associated with the presence of protease mutations selected by APV therapy. All seven viruses displaying a >4-fold APV susceptibility decrease harbored either I54L, I54M, V32I+I47V, or I50V. Accessory mutations clearly play a role in augmenting APV resistance when present with these key mutations, as could be seen by the effect of adding the M46I mutation into an I50V-containing genetic background where the I50V+M46I virus displayed a more than four times greater reduction in APV susceptibility than the virus bearing I50V alone. Viruses harboring protease mutations associated with reduced susceptibility to other PIs, such as those with V82A, I54V, and L90M, remained fully susceptible to APV.
FIG. 3.
APV fold susceptibility change (relative to HXB2 control) for the panel of site-directed mutants of HIV-1 strain HXB2.
The V82I protease substitution emerged in two patient isolates (patients 3 and 5), both of which also developed I50V substitutions. Site-directed mutagenesis converting the isoleucine codon back to valine at position 82 and subsequent APV susceptibility determination for the patient 3 viral isolate failed to demonstrate an effect on APV susceptibility when compared to the same virus carrying V82I in an identical genetic background.
DISCUSSION
Amprenavir/NRTI combination therapy can result in the selection of one of four alternative viral protease genotypes characterized by the presence of either I50V, I54L or I54M, V32I+I47V, or rarely, I84V. Viral susceptibility data presented here for both clinical isolates and site-directed mutant viruses have confirmed the role of these mutations in reduction of APV susceptibility. However, clinical isolates resistant to APV exhibited infrequent cross-resistance to other PIs. A noteworthy finding of this study was the observed role of resistance to coadministered NRTIs in the subsequent development of protease mutations.
A higher frequency of protease mutation development has been detected in this study of PI-naïve/NRTI-experienced patients when compared to antiretroviral therapy-naïve patients initiating combination regimens including APV (study PROA3001; unpublished data) or IDV (7, 10, 17) where RT mutations, especially the lamivudine-associated M184V mutation, predominated in patients experiencing virological failure. The variable baseline susceptibility to NRTIs subsequently received during the study described here may have led to differential regimen potency and the possibility of a more rapid viral evolution in patients where the NRTIs were exerting a suboptimal antiviral effect. A statistically significant correlation between evidence of reduced baseline susceptibility to NRTIs subsequently coadministered with the PIs during the study and an increased likelihood of development of key PI mutations were identified for both the APV- and IDV-treated patient groups. This finding emphasizes the importance of ensuring that the virus is susceptible to as many components of the regimen as possible in order to achieve maximal suppression of viral replication and minimize the opportunity for the accumulation of additional mutations.
A previous study has reported a correlation between the rate of evolution of protease mutations and plasma trough concentrations in patients receiving RTV where higher RTV levels in plasma were associated with slower viral evolution (18). Selection of a maximally suppressive regimen may be aided by the use of resistance testing at the time of therapy switch and by including, in subsequent regimens, agents from a class to which the patient has not been previously exposed or for which the development of resistance or cross-resistance is distinguishable from that of other agents (12). There have been major advances in the standard of care for HIV-1-infected patients since the initiation of this clinical trial. A greater understanding of viral resistance and cross-resistance within the NRTI class of antiretroviral agents and recent widespread adoption of coadministration of low-dose RTV to enhance plasma APV levels are two factors which could be expected to improve virological response in this patient population.
The profile of mutations selected by APV is different from that observed for other PIs. Four PRO genotypes, characterized by the presence of substitutions I50V, I54L or I54M, V32I+I47V, or I84V, developed in APV/NRTI-treated patients who experienced virological failure in this study. Each of these key mutations were usually accompanied by one or more accessory mutations, most commonly M46I/L. Although I50V-bearing viral variants were generally more resistant to APV than variants harboring any one of the other three genotypes, all four groups of amino acid substitutions were found to be associated with reduced susceptibility to this agent.
Owing to recommended treatment strategies, evidence of virological failure resulted in a change in the antiretroviral regimen received by the patients described in this study. In most cases, discontinuation of APV occurred soon after virological failure, thereby precluding any assessment of further viral evolution in the presence of APV. However, in a small number of patients (n = 14), the APV-containing regimen was not changed for some time (range, 4 to 56 weeks) after virological failure, and further viral evolution was monitored. In 10 of 14 cases, the key APV-selected mutations initially identified (I50V, n = 4; I54L/I54M, n = 3; V32I+I47V, n = 1; I84V, n = 2) were retained and additional accessory mutations (M46I/L, L33F), if not already present, were acquired. More marked evolution of viral protease genotype occurred in the remaining 4 of 14 cases: in three cases, one each with an initial V32I+I47V, I54L/I54M, or I84V mutation at failure, these initial key APV mutations were lost and replaced with the I50V mutation. In the fourth case, the initial V32I+I47V mutation was lost and replaced with I84V (data not shown). Therefore, even though genotypic evolution was observed during continued APV therapy, the mutation that prevailed, I50V or I84V, was consistent with those previously described as characteristic of APV resistance. Furthermore, I50V appeared to be particularly stable: unlike the other genotypes, once selected, I50V was not lost or replaced while APV therapy continued. Key mutations characteristic of selection by other PIs, for example, L90M and V82A/F/T, were not observed during continued APV therapy in any of these patients.
The cross-resistance to other PIs engendered by each of the four resistance genotypes selected by APV was minimal, with low-level cross-resistance to RTV observed most frequently. Low-level cross-resistance to NFV was detected in a small number of isolates. However, it is known that NFV cross-resistance is frequently observed, even in isolates derived from patients who are NFV naïve (24).
Three of the four patterns of protease amino acid substitutions associated with APV resistance observed in this study have been identified previously by in vitro passage studies. Earlier studies identified I50V as a signature mutation of APV resistance development (21, 31) while I84V appeared transiently (21). More recently, in vitro passage of the RF HIV-1 viral strain in the presence of increasing APV concentrations in MT-2 cells selected for a viral genotype bearing V32I+I47V in addition to M46I and V82I (9). However, neither leucine nor methionine substitutions at protease residue I54 following in vitro APV selection experiments have been reported previously. The emergence of a particular genotype in response to APV selective pressure, either in vitro or in vivo, may be influenced by a number of factors. Viral protease polymorphisms present at baseline could predispose the virus to a particular pathway of resistance development. Although the possible correlation between the presence of individual baseline polymorphisms and the acquisition of specific protease mutations was not explored here, a previous study has highlighted the importance of such polymorphisms with regard to viral fitness. It was shown that the introduction of a single L10I mutation was sufficient to rescue the impaired growth of a virus harboring three protease mutations (G48V, A71T, and V82A) (26). In vitro selection studies with the investigational PI BMS-232632 demonstrated that the evolution of specific resistance genotypes was dependent on the starting viral strain (9). The quasispecies nature of HIV-1 infection, in addition to extensive inter- and intraclade sequence heterogeneity, ensure that the in vivo dynamics of viral resistance evolution are potentially much more complex than those prevailing in in vitro selection studies.
Lower drug exposure in vivo, due to either poor adherence or host physiological factors, may favor the selection of less resistant genotypes. APV susceptibility data presented here from site-directed mutants and clinical isolates demonstrate that there is a wide spectrum of APV susceptibility reduction associated with particular genotypes and that those with I50V in addition to M46I consistently exhibit the highest APV resistance. Lower APV concentrations in plasma have been shown to be associated with the preferential selection of less-APV-resistant I54L- or I54M-containing genotypes while higher APV levels in plasma were associated with selection of more resistant I50V-containing genotypes (R. Elston, S. Randall, R. Myers, M. Maguire, A. Rakik, M. Ait-Khaled, D. Stein, and W. Snowden, 8th Conf. Retrovir. Opportunistic Infect., abstr. 465, 2001). A relationship between PI concentrations in plasma and the differential selection of protease mutants has been demonstrated for SQV. These studies have shown that the relative frequency of the selection of G48V and V82A mutations by this agent in vivo was positively correlated with higher SQV dosing (28, 34). The V32I+I47V genotype evolved in two patients for whom it was known that there were periods of nonadherence or partial adherence. One patient (patient 6) discontinued APV for 17 days and then resumed it at a lower dose for 21 days prior to resumption of the standard dose, following which the previously evolved V32I+I47V genotype was displaced by a more resistant I50V-containing viral variant. Clonal analysis of multiple sequential samples revealed that the I50V and V32I+I47V viral variants were not genomically linked. They, therefore, appear to represent two mutually exclusive pathways of APV resistance development. In other patients, however, the V32I+I47V genotype persisted and a pattern of acquisition of additional protease mutations, usually M46I, was followed. Biochemical studies suggest that the I47V mutation is compensatory for V32I (27). These two residues form the S2 active site pocket of the protease enzyme, and the double V32I+I47V mutation yields a pocket whose volume is unchanged from that of the wild type.
Amino acid changes at protease residue 54 selected by IDV (4, 5, 35) and SQV (33, 34) are always isoleucine to valine (I54V). RTV also selects I54V variants (18, 30), but only rarely selects I54L variants (18). In this respect, APV appears quite distinct from the other PIs with regard to the selective pressure that it exerts on viral protease residue 54, resulting in substitutions I54L or I54M rather than I54V. APV susceptibility data for site-directed viral mutants substantiate this assertion that the I54V mutation, either alone or in the presence of other protease mutations (V82A, L33F, and L10I), was not associated with reduced APV susceptibility.
A strong association between the emergence of the I50V-containing genotype and coselection of the leucine-to-phenylalanine mutation in the Gag p1/p6 cleavage site was observed. This mutation has been detected in viral isolates that had acquired multiple protease mutations following in vitro passage with LPV (2) and BILA 1906 BS and BILA 2185 BS (8). The previously observed p7/p1 A431V Gag alanine-to-valine mutation (35) was also seen in this study but was not detected in any I50V-containing isolates.
The role in APV resistance of the V82I mutation, which occurs as a natural polymorphism in about 5% of untreated isolates (14, 16), is unclear. In this study, it emerged in two isolates, both of which also developed the I50V. However, susceptibility studies of recombinant viruses from one of these isolates, in which a valine codon was substituted for an isoleucine at residue 82, revealed that the V82I mutation does not enhance APV resistance. Consistent with this, site-directed mutant viruses harboring V82I remained susceptible to APV.
In summary, the role of the I50V mutation in conferring resistance to APV has been confirmed in a large phase III study. Three additional viral protease genotypes characterized by the development of substitutions I54L, I54M, V32I+I47V, and I84V, which may occur with concomitant accessory mutations (e.g., M46I/L, L33F, L10F), evolved in response to APV and generally conferred lower levels of APV resistance. Each of these four genotypes conferred little or no cross-resistance to other PIs. Finally, the significant association between preexisting viral resistance to NRTIs subsequently administered in the PI/NRTI combination regimen and the emergence of protease mutations, emphasizes the importance of optimising treatment regimens to ensure that the virus is susceptible to as many components as possible.
Acknowledgments
We thank all the clinical investigators and patients who participated in study PROAB3006. Thanks also to Terry Paul for assistance with preparation of the manuscript and to Marcus Oxer for his bioinformatics expertise which greatly aided our sequence analyses.
REFERENCES
- 1.Boden, D., and M. Markowitz. 1998. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 42:2775-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrillo, A., K. D. Stewart, H. L. Sham, D. W. Norbeck, W. E. Kohlbrenner, J. M. Leonard, D. J. Kempf, and A. Molla. 1998. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condra, J. H. 1998. Resistance to HIV protease inhibitors. Haemophilia 4:610-615. [DOI] [PubMed] [Google Scholar]
- 4.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Tang, H. Teppler, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:259-575. [DOI] [PubMed] [Google Scholar]
- 6.De Pasquale, M. P., R. Murphy, D. Kuritzkes, J. Martinez-Picado, J. P. Sommadosi, R. Gulick, L. Smeaton, V. DeGruttola, A. Caliendo, L. Sutton, A. V. Savara, and R. T. D'Aquila. 1998. Resistance during early virological rebound on amprenavir plus zidovudine plus lamivudine triple therapy or amprenavir monotherapy in ACTG protocol 347. Antivir. Ther. 3(Suppl.):50-51. [Google Scholar]
- 7.Descamps, D., P. Flandre, V. Calvez, G. Peytavin, V. Meiffredy, G. Collin, C. Delaugerre, S. Robert-Delmas, B. Bazin, J-P. Aboulker, G. Pialoux, F. Raffi, and F. Brun-Vezinet. 2000. Mechanisms of virologic failure in previously untreated. HIV-infected patients from a trial of induction-maintenance therapy. JAMA 283:205-211. [DOI] [PubMed] [Google Scholar]
- 8.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong, Y-F., B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno, and P-F. Lin. 2000. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrob. Agents Chemother. 44:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havlir, D. V., N. S. Hellmann, C. J. Petropoulos, J. M. Whitcomb, A. C. Collier, M. S. Hirsch, P. Tebas, J. P. Sommadossi, and D. D. Richman. 2000. Drug susceptibility in HIV infection after viral rebound in patients receiving indinavir-containing regimens. JAMA 283:229-234. [DOI] [PubMed] [Google Scholar]
- 11.Hertogs, K., M. P. de Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch, M. S., F. Brun-Vézinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection. Recommendations of an international AIDS society-USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 13.Kim, E. E., C. T. Baker, M. D. Dwyer, M. A. Murcko, B. G. Rao, R. D. Tung, and M. A. Navia. 1995. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J. Am. Chem. Soc. 117:1181-1182. [Google Scholar]
- 14.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 15.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 16.Lech, W. J., G. Wang, Y. L. Yang, Y. Chee, K. Dorman, D. McCrae, L. C. Lazzeroni, J. W. Erickson, J. S. Sinsheimer, and A. H. Kaplan. 1996. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J. Virol. 70:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maguire, M., M. Gartland, S. Moore, A. Hill, M. Tisdale, R. Harrigan, J-P. Kleim, and the AVANTI Study Group. 2000. Absence of zidovudine resistance in antiretroviral-naïve patients following zidovudine/lamivudine/protease inhibitor combination therapy: Virological evaluation of the AVANTI 2 and AVANTI 3 studies. AIDS 14:1195-1201. [DOI] [PubMed] [Google Scholar]
- 18.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 19.Mulder, J., N. McKinney, C. Christopherson, J. Sninsky, L. Greenfield, and S. Kwok. 1994. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J. Clin. Microbiol. 32:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers, R. E., W. Snowden, S. Randall, and M. Tisdale. 1998. Unique resistance profile of the protease inhibitor amprenavir (141W94) observed in vitro and in the clinic. Antivir. Ther. 3(Suppl.):59-60.10726056 [Google Scholar]
- 21.Partaledis, J. A., K. Yamaguchi, M. Tisdale, E. E. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, C. M. Stuver, R. A. Byrn, and D. K. Livingston. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauwels, R., J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter, and E. De Clercq. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20:309-321. [DOI] [PubMed] [Google Scholar]
- 23.Pazhanisamy, S., J. A. Partaledis, B. G. Rao, and D. J. Livingston. 1998. In vitro selection and characterization of VX-478 resistant HIV-1 variants. Adv. Exp. Med. Biol. 436:75-83. [DOI] [PubMed] [Google Scholar]
- 24.Race, E., E. Dam, V. Obry, S. Paulous, and F. Clavel. 1999. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS 13:2061-2068. [DOI] [PubMed] [Google Scholar]
- 25.Robinson, L. H., R. E. Myers, B. W. Snowden, M. Tisdale, and E. D. Blair. 2000. HIV type 1 protease cleavage site mutations and viral fitness: implications for drug susceptibility phenotyping assays. AIDS Res. Hum. Retrovir. 16:1149-1156. [DOI] [PubMed] [Google Scholar]
- 26.Rose, R. E., Y. F. Gong, J. A. Greytok, C. M. Bechtold, B. J. Terry, B. S. Robinson, M. Alam, R. J. Colonno, and P. F. Lin. 1996. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc. Natl. Acad. Sci. USA 93:1648-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sardana, V. V., A. J. Schlabach, P. Graham, B. L. Bush, J. H. Condra, J. C. Culberson, L. Gotlib, D. J. Graham, N. E. Kohl, R. L. LaFemina, C. L. Schneider, B. S. Wolanski, J. A. Wolfgang, and E. A. Emini. 1994. Human immunodeficiency virus type 1 protease inhibitors: evaluation of resistance engendered by amino acid substitutions in the enzyme's substrate binding site. Biochemistry 33:2004-2010. [DOI] [PubMed] [Google Scholar]
- 28.Schapiro, J. M., M. A. Winters, J. Lawrence, and T. C. Merigan. 1995. Clinical cross-resistance between the HIV-1 protease inhibitors saquinavir and indinavir and correlations with genotypic mutations. AIDS 13:359-365. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, B., K. Korn, B. Moschik, C. Paatz, K. Überla, and H. Walter. 2000. Low level of cross-resistance to amprenavir (141W94) in samples from patients pre-treated with other protease inhibitors. Antimicrob. Agents Chemother. 44:3213-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmit, J. C., L. Ruiz, B. Clotet, A. Raventos, A. Tor, J. Leonard, J. Desmyter, E. De Clercq, and A. M. Vandamme. 1996. Resistance-related mutations in the HIV-1 protease gene of patients treated for 1 year with the protease inhibitor ritonavir (ABT-538). AIDS 10:995-999. [DOI] [PubMed] [Google Scholar]
- 31.Tisdale, M., R. E. Myers, B. Maschera, N. R. Parry, N. M. Oliver, and E. D. Blair. 1995. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob. Agents Chemother. 39:1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tisdale, M., R. Myers, S. Randall, M. Maguire, M. Ait-Khaled, R. Elston, and W. Snowden. 2000. Evolution of resistance to the HIV protease inhibitor amprenavir in vitro and in clinical studies. Clin. Drug Investig. 20:267-285. [Google Scholar]
- 33.Vaillancourt, M., D. Irlbeck, T. Smith, R. W. Coombs, and R. Swanstrom. 1999. The HIV type 1 protease inhibitor saquinavir can select for multiple mutations that confer increasing resistance. AIDS Res. Hum. Retrovir. 15:355-363. [DOI] [PubMed] [Google Scholar]
- 34.Winters, M. A., J. M. Schapiro, J. Lawrence, and T. C. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J. Virol. 72:5303-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Y-M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. B. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziermann, R., K. Limol, K. Das, E. Arnold, C. J. Petropoulos, and N. T. Parkin. 2000. A mutation in human immunodeficiency virus type 1 protease, N88S, that causes in vitro hypersensitivity to APV. J. Virol. 74:4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]