Abstract
The gene encoding the plasma membrane proton pump (H+-ATPase) of Aspergillus fumigatus, PMA1, was characterized from A. fumigatus strain NIH 5233 and clinical isolate H11-20. An open reading frame of 3,109 nucleotides with two introns near the N terminus predicts a protein consisting of 989 amino acids with a molecular mass of approximately 108 kDa. The predicted A. fumigatus enzyme is 89 and 51% identical to H+- ATPases of Aspergillus nidulans and Saccharomyces cerevisiae, respectively. The A. fumigatus PMA1 is a typical member of the P-type ATPase family that contains 10 predicted transmembrane segments and conserved sequence motifs TGES, CSDKTGT, MLTGD, and GDGVN within the catalytic region. The enzyme represents 2% of the total plasma membrane protein, and it is characteristically inhibited by orthovanadate, with a 50% inhibitory concentration of ∼1.8 μM. H+-ATPases from Aspergillus spp. contain a highly acidic insertion region of 60 amino acids between transmembrane segments 2 and 3, which was confirmed for the membrane-assembled enzyme with a peptide-derived antibody. An increasing A. fumigatus PMA1 copy number confers enhanced growth in low-pH medium, consistent with its role as a proton pump. These results provide support for the development of the A. fumigatus H+-ATPase as a potential drug discovery target.
Opportunistic fungal infections have increased markedly in recent years, and Aspergillus fumigatus, an ubiquitous filamentous fungus, has become a prevalent cause of invasive fungal infections in immunocompromised patients (8, 9). After inhalation of its airborne conidia, A. fumigatus causes a variety of diseases including allergic bronchopulmonary aspergillosis in asthma patients and invasive pulmonary aspergillosis (IPA) in immunocompromised patients (3, 9, 18). IPA is rare in immunocompetent individuals; but its prevalence has increased due to transplantation procedures, aggressive chemotherapy for cancer, immunosuppressive regimens for patients with autoimmune disease, and the emergence of AIDS (9). A. fumigatus is estimated to be responsible for 30% of fungal infections among cancer patients and 10 to 25% of fungal infections among leukemia patients (9), and the mortality rate associated with A. fumigatus infections is high (18). Early diagnosis of invasive fungal infections is critical for a successful therapeutic outcome, although such diagnoses are frequently difficult to achieve (2, 8, 19). Despite the availability of azoles with broader reactivities against filamentous fungi, such as itraconazole (11) or the recently introduced echinocandins, amphotericin B remains the first choice for the treatment of severe or refractory mycoses, especially IPA (4, 10). However, the clinical efficacy of amphotericin B is limited by its well-known nephrotoxicity (36). Lipid formulations of amphotericin B improve the therapeutic index of this drug (9, 27), but cost and toxicity remain problems (34). In order to improve therapeutic outcomes, additional drug targets are needed to develop new antifungal drugs.
One such target is the fungal plasma membrane H+-ATPase, which is an ATP-dependent proton pump. It plays a critical role in fungal cell physiology by regulating intracellular pH, maintaining ionic balance, and generating the electrochemical proton gradient necessary for nutrient uptake (37). The H+-ATPase from Saccharomyces cerevisiae has been shown to be essential by gene disruption experiments (38), and it displays a number of biochemical and genetic properties that make it attractive as a drug discovery target (25, 31). It is a member of the P-type ATPase family that mediates ATP-dependent cation transport and is closely related to ion-translocating enzymes from plants (H+-ATPase), bacteria (K+-ATPase and Mg2+-ATPase) and animals (Na+, K+-ATPase, Ca2+-ATPase, and H+, K+-ATPase) (20, 22). The fungal pump has been extensively characterized from model systems such as S. cerevisiae (30). It comprises a single subunit of about 100 kDa that consists of a membrane-bound domain with 10 transmembrane segments, a large cytoplasmic ATP hydrolysis domain, and a narrow stalk domain that links the two larger domains (20). These enzymes couple ATP hydrolysis in the cytoplasmic domain to ion transport in the membrane-embedded domain, forming an acyl-phosphate intermediate during catalysis (20).
The PMA genes encoding H+-ATPases from numerous fungi and plants have been characterized and have been shown to be highly similar, with similarity from 45 to 95% at the amino acid level (13), although they show only about 25% similarity with those from members of the higher eukaryotes (41). The P-type cardiac and gastric ATPases are well-known targets for therapeutics, and there is a high degree of target specificity for these ATPases (29). The highly conserved functional properties of the fungal H+-ATPases suggest that a specific antagonist could present broad reactivity to fungal species. In addition, prominent acid efflux induced by the H+- ATPases may be an important pathogenicity determinant for tissue infiltration (25).
In this report, we describe the cloning and biochemical characterization of the H+-ATPase from the pathogenic fungus A. fumigatus, which could serve as a target for novel antifungal drug development.
MATERIALS AND METHODS
Strains and cell culture.
A. fumigatus strains NIH 5233 (ATCC 13073) and H11-20 were used in the study. Escherichia coli strain TOP10 (Invitrogen) was used for plasmid propagation, while Epicurian XL1-Blue MRF" (Stratagene) was used for titration and propagation of the genomic library. A. fumigatus mycelia were grown at 37°C in YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 2% [wt/vol] dextrose).
Identification of PMA1 gene from A. fumigatus cDNA library.
The NIH 5233 (ATCC 13073) cDNA library constructed in phagemid pBluescript SK(±)was obtained from Stratagene. Conserved regions of PMA1 genes from S. cerevisiae and other fungi (DQSAITGESL, AMTGDGVNDAPSLKKAD, and LAGVEILCSCSDKTGTLTKNKL) were used to identify P-type ATPases from a genomic sequence database of A. fumigatus (Astra-Zeneca). These regions were aligned to pmaA of Aspergillus nidulans (33). Primers (5"-GA GCC GAA GCA ACA ATG GCG GAG and 5"-GC GAG AGA TTC ACC AGT GAT GGC) designed from identical regions of the nucleotide sequence alignment between the identified A. fumigatus P-type ATPase sequences and the pmaA gene of A. nidulans were initially used to amplify a 0.9-kb fragment of A. fumigatus PMA1 cDNA of strain NIH 5233. The library was propagated and plated on 100-mm Luria-Bertani agar plates to yield 500 to 100 plaques per plate. The plates were divided into quadrants, and the plaques were pooled. cDNA from the phage pools was extracted with LambdaSorb phage adsorbent (Promega). The 0.9-kb fragment of A. fumigatus PMA1 was amplified from the cDNA extracted from these pools, and positive pools were replated and single plaques were evaluated. A. fumigatus PMA1 was amplified from a single enriched plaque by using primers T3 and T7 present in the pBluescript SK(±) phagemid. The T3-T7 fragment was cloned into vector pCR-XL-TOPO (Invitrogen) to create plasmid pHB AfPMA1.
Genomic DNA isolation and identification of A. fumigatus PMA1.
A. fumigatus strains were maintained on YPD agar plates at 37°C. Spores (2.7 × 106) were inoculated into 25 ml of YPD medium and were grown overnight at 37°C with shaking. The cells were filtered through a 0.2-μm-pore-size SFCA filter (Nalgene) and collected in a porcelain bowl, frozen with dry ice, and ground with a pestle. Total genomic DNA was extracted with a slightly modified version of the Wizard Genomic DNA Purification kit (Promega). After the addition of the nucleus lysis solution, the protocol was modified by incubating the sample in a 37°C water bath for 1 h. The sample was centrifuged for 20 min at top speed (13,000 rpm) in a microcentrifuge (Marathon 16 KM; Fisher) after the addition of the protein precipitation solution. A. fumigatus PMA1 was amplified from chromosomal DNA with primers derived from the cDNA sequence of A. fumigatus NIH 5233 PMA1 (5"-GCC ATT CCT GCC ATT TCT TGT TTG and 5"-GCC GAA GCC CAG CCC TAT GCA TGT ATG).
Transformation of Aspergillus.
Electroporation was performed in a 1.5-ml microcentrifuge tube by mixing 2 to 5 μg of DNA (in 10 μl) with 50 μl of conidia (∼108) along with 2 μg of pyrG DNA amplified by PCR from plasmid pCDA14 (6), as described by Weidner et al. (43). Spheroplast transformation was performed as follows. A suspension of 107 A. fumigatus spores in 50 ml of YG (0.5% yeast extract and 2% d-glucose supplemented with 5 mM uridine and 5 mM uracil) containing trace elements was incubated for 12 h at 28°C with gentle circular agitation (200 rpm) until the formation of germ tubes, as observed by light microscopy. Germ tubes were collected by filtration under vacuum or centrifugation at 5,000 × g for 10 min. The germ tubes were washed by centrifugation with sterile water and resuspended in 50 ml of a solution containing 0.32 M ammonium sulfate, 32 mM citric acid (pH 6), 0.4% yeast extract, 0.8% sucrose, 130 mM magnesium sulfate, 400 mg of bovine serum albumin, 0.1 ml of beta-glucuronidase (Sigma), and 300 mg Glucanex (InterSpex Products, San Mateo, Calif.). The cells were swirled (120 rpm) for 5 to 6 h at 30°C until protoplast formation. The cells were collected by centrifugation at 2,500 × g for 10 min and resuspended in 0.4 M ammonium sulfate-1% sucrose-50 mM citric acid (pH 6). The cells were centrifuged for 5 min at 4,000 × g, and the cell pellet was resuspended in 1 ml of 0.6 M KCl-50 mM CaCl2-10 mM morpholineethanesulfonic acid (pH 6) and placed at 4°C for 10 min. An aliquot (100 μl) of the protoplast suspension was added to 4 to 20 μg of DNA. A solution (50 μl) containing 25% (wt/vol) polyethylene glycol (PEG) 6000, 100 mm CaCl2, 0.6 M KCl, and 10 mM Tris-HCl (pH 7.5) was added to the DNA-cell suspension, which was kept at 4°C for 20 min. The suspension was then added to 1 ml of the PEG-containing solution and placed at 22°C for 20 min. The transformation mixture (100 to 200 μl) was plated on selective medium (YG without uracil supplementation) containing 0.6 M KCl.
Total RNA isolation, cDNA preparation, and identification of A. fumigatus PMA1.
Total RNA was extracted from H11-20 with an RNeasy Plant Mini kit (Qiagen). The protocol was modified by grinding the cells with dry ice and by washing with 500 μl of the RW1 buffer provided with the kit. cDNA was obtained by reverse transcriptase PCR with the Omniscript kit (Qiagen). A. fumigatus PMA1 was amplified from cDNA with the same primers used for amplification of the gene from genomic DNA.
Disruption and restriction enzyme marking of A. fumigatus PMA1.
The entire region between the StuI sites (1,299 bp) was replaced by the 4.0-kb Aspergillus niger pyrG gene (6) from plasmid pCDA14 digested with XbaI, creating pHBΔAfPMA1. The pyrG-disrupted A. fumigatus PMA1 gene was amplified with primers (5"-GCT TTG TGA GCC GAA GCA ACC CAT GGC GGA GCG GAGAT CTC C and 5"-CCA GAT GAG ATA TTA GTA GAG TAA TCT CCG GAT TAC TCA TCA TCC) and was used for transformation of strain CEA17 (6) by both electroporation and spheroplasting methods (7). Stable transformants were selected on minimal medium plates, and genomic DNA was isolated as described above. Primers (5"-GCC ATC ACT GGT GAA TCT CTC GC and 5"-CGT AGG CAA CAG CGA TGG TGG) were used to amplify part of the A. fumigatus PMA1 gene with or without disruption. PCR products were analyzed by agarose gel electrophoresis. Introduction of a marked copy of A. fumigatus PMA1 was accomplished by modifying the gene with a silent mutation encoding a new EcoRI restriction enzyme site at nucleotide position 1389 with a QuickChange Site-Directed Mutagenesis kit (Stratagene).
Purification of plasma membranes and H+-ATPase-mediated ATP hydrolysis measurements.
Plasma membranes were prepared from a 24-h culture of A. fumigatus strain H11-20 by centrifugation on a sucrose step gradient, as described previously (39). Cells were collected in cheesecloth and resuspended in homogenization buffer (50 mM Tris-HCl [pH 7.5], 0.3 M sucrose, 1% glucose, 1 mM EDTA, 2 mM dithiothreitol [DTT]) containing 1 mM phenylmethylsulfonyl fluoride and 2.5 μg of chymostatin per ml. The cells were broken in a French pressure cell at a pressure of 20,000 lb/in2. After cell disruption, the cells were centrifuged in a Sorval SS-34 rotor for 20 min at 5,000 rpm. A crude membrane fraction was pelleted from the supernatant by centrifugation at 49,000 rpm for 1 h in a Beckman 50.2 Ti rotor. The crude membranes were washed in membrane wash buffer (10 mM Tris-HCl [pH 7], 1 mM EGTA, 1 mM DTT, 20% glycerol) with 0.5 mM phenylmethylsulfonyl fluoride and were resuspended in membrane wash buffer. Purified plasma membranes were recovered at the 53.5%-43.5% (wt/wt) sucrose interface of a step gradient containing 1 mM EDTA, 1 mM DTT, and 10 mM Tris (pH 7.0) after centrifugation for 3 h at 39,000 rpm in a SW41 rotor. The membranes were washed in membrane wash buffer for 1 h at 49,000 rpm in a 50.2 Ti rotor and resuspended in membrane wash buffer.
ATPase assays.
ATP hydrolysis assays were performed in triplicate 96-well microplates as described previously (42). The amount of inorganic phosphate released was determined by measuring the absorbance at 660 nm in a microtiter plate reader (Tecan SLT Instruments) after a 15-min incubation at 37°C. The optimal pH for ATP hydrolysis was determined in a standard reaction medium with the pH adjusted to 5.0 to 7.5. Km and Vmax values were determined by measuring ATP hydrolysis with equimolar concentrations of ATP and MgSO4 from 0 to 10 mM containing 0.5 mM NaN3 to eliminate possible contributions from mitochondrial ATPase activities. Vanadate sensitivity was assayed by measuring ATP hydrolysis in the presence of 0 to 1,000 μM sodium vanadate. The protein concentration was determined with Coomassie Plus Reagent (Pierce) and was measured at 595 nm in a microplate reader (Tecan SLT Instruments) with bovine gamma globulin as the standard.
Antibodies and Western blot analysis.
Peptide-derived antibodies (Research Genetics, Inc.) were produced against an Aspergillus-specific 16-amino-acid stretch (KPEMFETYKEYLATAN) contained within a highly acidic insertion region of 39 amino acids between transmembrane segments 2 and 3. The antibodies were used to analyze Western blotted plasma membrane proteins on polyvinylidene difluoride membranes as described previously (24). Plasma membranes proteins were separated by sodium dodecyl sulfate (SDS)-gel electrophoresis on 10% precast minigels (Novex) and transferred to a polyvinylidene difluoride membrane in an Xcell II Mini-Cell blotting apparatus according to the manufacturer's instructions.
PCR amplification, quantitative real-time PCR, and sequence analysis.
PCR was carried out in a PTC-100 MiniCycler (MJ Research). Amplifications were carried out with Expand Long Template Enzyme mixture with salt buffer 2 provided with the enzyme (Boehringer Mannheim). PCR products were purified prior to sequence analysis with a QIAquick PCR Purification kit and a GEL Extraction kit (Qiagen). Quantitative real-time PCR with an A. fumigatus PMA1-specific molecular beacon to determine copy number was performed as described by Manganelli et al. (21). Both cDNA and genomic DNA sequences were analyzed at the New York University Medical Center DNA sequencing facility. The sequences of the PCR products were compared and aligned with the genomic sequence by using various Genetics Computer Group programs (University of Wisconsin). Hydropathy profiles were generated by the method of Kyte and Doolittle (17).
Nucleotide sequence accession numbers.
The nucleotide sequence of the cloned fragment containing A. fumigatus PMA1 is available from GenBank under accession numbers AY040608 and AY040609.
RESULTS
Cloning of A. fumigatus PMA1.
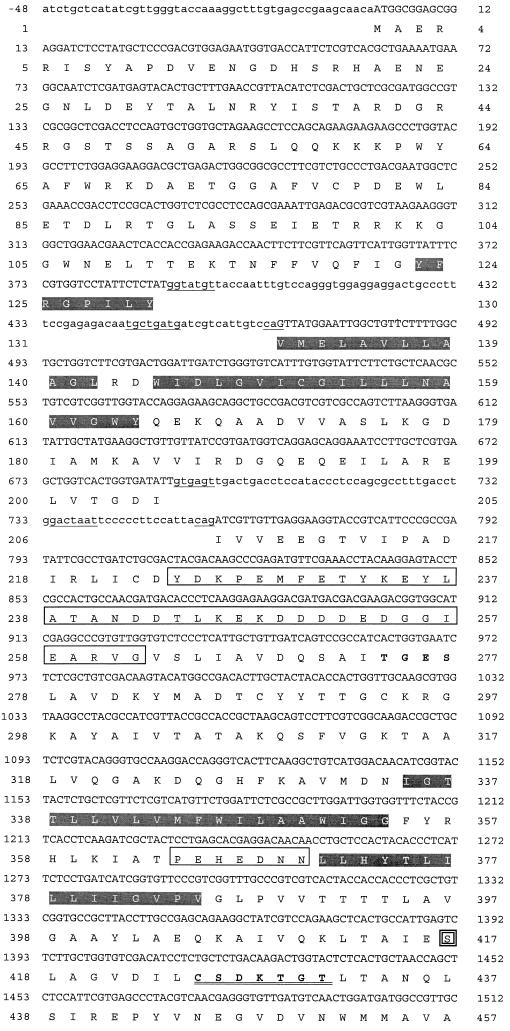
Conserved regions of PMA1 genes from S. cerevisiae and other fungi were used to identify P-type ATPases from an incomplete genomic sequence database of A. fumigatus (Astra-Zeneca). A clone with a 3.2-kb insert containing the entire coding region of A. fumigatus PMA1 was obtained from the screening of a commercial genomic library of A. fumigatus. Total genomic DNA was extracted from A. fumigatus strains NIH 5233, a standard reference strain, and H11-20, a clinical isolate from a cancer patient. The genomic DNA was used to amplify A. fumigatus PMA1 with primers derived from the cDNA sequence. A. fumigatus PMA1 cDNA and the genomic DNA sequences of both strains showed 100% identity to one another. The nucleotide sequence and predicted amino acid sequence for A. fumigatus PMA1 are shown in Fig. 1. The highly conserved motif CSDKTGT, which contains the site of phosphorylation, Asp427, and predicted transmembrane segments are also shown in Fig. 1. Comparison of the genomic DNA sequence and the cDNA sequence indicated that the A. fumigatus PMA1 gene contains two introns, both of which are located in the N-terminal half. Intron splice sequences and intron sizes were typical of those for filamentous fungi (Table 1) (32). The coding region initiates with an ATG codon and terminates with a TAA stop codon. The predicted protein consists of 989 amino acids and a predicted molecular mass of 108,862 Da. A. fumigatus PMA1 and A. nidulans pmaA were approximately 85% identical at the DNA level and 89% at the protein level, and A. fumigatus PMA1 had at least 40% similarity with all known fungal enzymes (Table 2).
FIG. 1.
Nucleotide and predicted amino acid sequences of the A. fumigatus PMA1 gene. Untranslated regions and intron sequences are shown in lowercase letters. The deduced amino acid sequence of the PMA1 gene is shown by the one-letter amino acid designations below the nucleotide sequence. Amino acid residues are numbered beginning with the first methionine, and the translation termination codon is denoted by an asterisk. The site of phosphorylation, D427KTGT, is double underlined. Ten putative hydrophobic transmembrane segments are shaded, consensus intron sequences are underlined, and the three extra insertion regions are boxed. Ser-417 near the site of phosphorylation has been boxed with a double line.
TABLE 1.
Intron splice sequences of A. fumigatus PMA1 gene
| Gene | In- tron | Length (bp) | Sequence (5" to 3")b |
|---|---|---|---|
| PMA1 | 1 | 76 | GTATGT . . . . . . . . TGCTGATG . . . . . . . CAG |
| PMA1 | 2 | 66 | GTGAGT . . . . . . . . GGACTAAT . . . . . . . CAG |
| Consensusa | 49-85 | GTPuNGT . . . . . . NPuCTPuACN . . . . PyAG |
Consensus data for introns in filamentous fungal genes (32).
Pu, purine; Py, pyrimidine.
TABLE 2.
Similarity between A. fumigatus PMA1 and related P-type ATPases
| % Similarity | % Identity | GenBank accession no. | |
|---|---|---|---|
| Fungal P-type ATPases | |||
| A fumigatus NIH 5233 PMA1 | 100 | 100 | This study (AY040608) |
| A. fumigatus H11-20 PMA1 | 100 | 100 | This study (AY040609) |
| A. nidulans PMAA | 93.3 | 89.8 | AF043332 |
| A. nidulans pmaA | 92.9 | 89.3 | AF036763 |
| Saccharomyces pombe pma2 | 63.6 | 53.4 | M60471 |
| Neurospora crassa PMA1 | 63.5 | 52.4 | J02602 |
| Pseudalle scheria angusta PMA1 | 63.4 | 51.9 | AAD19960 |
| C. albicans PMA1 | 63.2 | 52.1 | M74075 |
| Kluyveromyces lactis PMA1 | 62.97 | 51.9 | L37875 |
| Zygosaccharomyces rouxii PMA1 | 62.9 | 51.6 | D10764 |
| S. cerevisiae PMA1 | 62.6 | 51.4 | X03534 |
| S. pombe PMA1 | 62.8 | 51.7 | J03498 |
| Pneumocystis carinii PMA1 | 61.1 | 50.4 | U65004 |
| S. cerevisiae pma2 | 60.5 | 50.0 | J04421 |
| Uromyces fabae PMA1 | 49.9 | 40.3 | AJ003067 |
| C. neoformans PMA1 | 47.1 | 37.9 | AF077766 |
| Dicytostelium discoideum PMA1 | 40.3 | 29.5 | 1709664 |
| Animal P-type ATPases | |||
| Scyliorhinus caniculus caniculus Ca+ ATPase | 41.6 | 31.5 | 114307 |
| Rattus norvegicus Na+, K+-ATPase | 42.8 | 29.4 | 203037 |
| Rattus norvegicus Na+, K+-ATPase | 43.4 | 30.8 | 114376 |
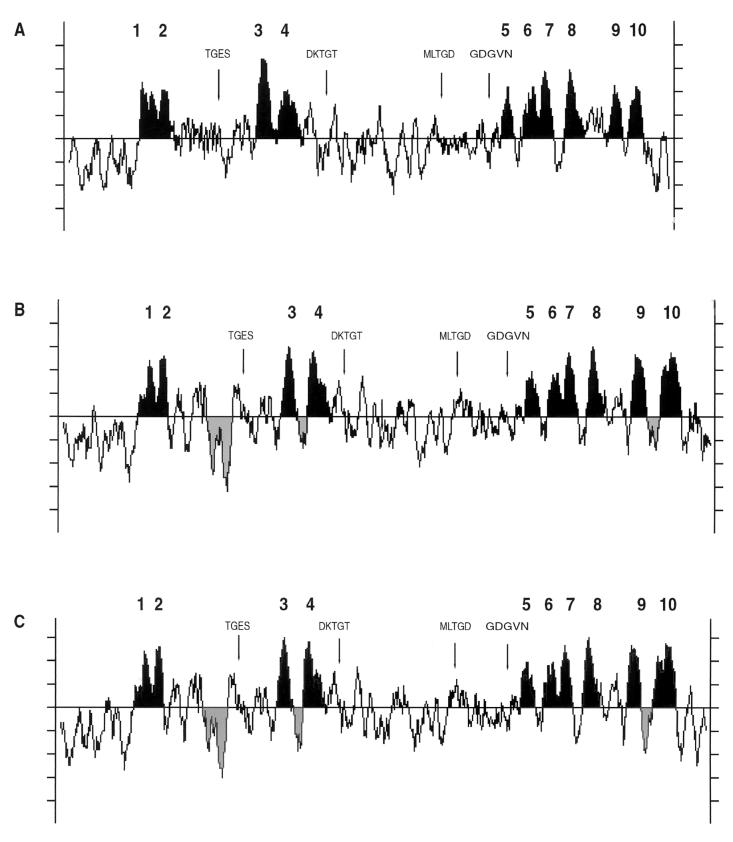
The A. fumigatus H+-ATPase contained three unique insertion sequences in the conserved central region of the protein consisting of 39, 7, and 16 amino acids at positions 224, 364, and 886, respectively (Fig. 2). These insertions are absent from all other fungal membrane H+-ATPases but were present in A. nidulans pmaA. Hydrophobicity and homology analyses of the deduced amino acid sequence of the encoded protein suggested the presence of 10 transmembrane (TM) domains with a long cytoplasmic loop between segments TM4 and TM5 (Fig. 3). By use of BLAST analysis (1) and the PILEUP alignment program (12), the A. fumigatus H+-ATPase was found to have amino acid sequence identities and similarities of 29 to 53% and 40 to 63%, respectively, relative to those of non-Aspergillus fungal enzymes.
FIG. 2.
Alignment of conserved regions of P-type ATPases. Amino acid sequence alignment of A. fumigatus PMA1 (AfPMA1) relative to other fungal PMA genes from A. nidulans (AnPMA1), S. cerevisiae (ScPMA1), and C. neoformans (CnPMA1). The highly conserved sequence TGES and transmembrane segments TM3, TM4, TM9, and TM10 are shaded. The Aspergillus amino acid insert regions are boxed.
FIG. 3.
Hydropathy profiles for the plasma membrane H+-ATPases from A. fumigatus, A. nidulans, and S. cerevisiae. Highly conserved regions of the H+-ATPases from S. cerevisiae (A), A. nidulans (B), and A. fumigatus (C) are noted, including sequence motifs involved with phosphorylation (CSD427KTGT), nucleotide binding (MLTGD and GDGVN), and dephosphorylation (TGES). Transmembrane segments TM1 to TM10 are designated, and the Aspergillus insertion regions are shaded.
Functional properties of A. fumigatus PMA1.
H+-ATPases play an important role in cell physiology in lower eukaryotes and have been shown to be essential to the cell (38). To assess gene essentiality in A. fumigatus, the intact gene was disrupted by deleting the region containing amino acids 297 to 730 and inserting the A. niger pyrG gene (6). The linearized construct was used to transform pyrG-deficient strain A. fumigatus CEA17 (6). More than 500 transformants were selected following electroporation or protoplast transformation on minimal medium in the absence of uracil or uridine. PCR analysis of the transformant genotype with A. fumigatus PMA1-specific primers indicated that the A. fumigatus PMA1 disruption allele had not replaced the wild-type homologue of A. fumigatus PMA1. In all transformants, the wild-type copy of A. fumigatus PMA1 remained and the disrupted copy of A. fumigatus PMA1297-730::pyrG was randomly distributed in the genome. Attempts to introduce a well-characterized mutation, S417F, which causes attenuation of catalytic activity in PMA1 genes from other organisms (30), was also unsuccessful. The effects of these mutations on A. fumigatus PMA1 are unknown but may have been more severe than in other fungi, resulting in apparent lethality.
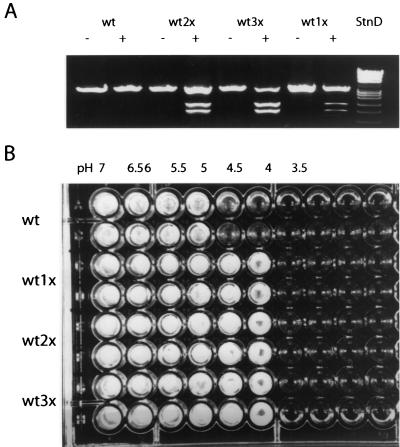
In the course of disrupting PMA1, we observed that cotransformation of cells with pyrG and an intact A. fumigatus PMA1 marked with a silent mutation that resulted in a novel EcoRI restriction site resulted in a significant number of transformants with multiple copies of A. fumigatus PMA1. The copy number could be increased by repeated 5-fluorootic acid treatment and retransformation with the marked wild-type gene. Figure 4A shows a restriction enzyme (EcoRI) digest of PCR-amplified wild-type and polyploid strains carrying the native and marked genes. Quantitative evaluation of the gene in wild-type strains and strains with the marked gene fragments indicated that the gene copy number varied from one to three.
FIG. 4.
Effect of gene dosage on pH-dependent growth. (A) The A. fumigatus PMA1 gene was amplified by PCR from wild-type (wt) cells and transformants containing marked copies of PMA1 with a unique EcoRI restriction enzyme site. The fragments were either untreated or digested with EcoRI. The amount of native PMA1 relative to the amount of marked enzyme was used to determine whether cells contained a single copy (1×), double copy (2×), or triple copies (3×) of A. fumigatus PMA1. (B) Approximately 1,000 spores from either wild-type or polyploid PMA1 cells, as described above for panel A, were used to inoculate microdilution medium (YG) containing 50 mM acetate adjusted from pH 3.5 to 7.0. Each assay was performed in duplicate, and growth was determined after 48 h.
The polyploid A. fumigatus PMA1 strains were evaluated to determine whether increased gene copy number altered the physiological properties of the cells. An important role of PMA1 in most fungi is the regulation of cytoplasmic pH in response to acid loading of the cytoplasm. In theory, cells that express larger amounts of PMA1 relative to the amount expressed in the wild type should have a greater acid efflux capacity, resulting in reduced susceptibility to acidic medium conditions. To examine this possibility, the weak acid acetate, which acidifies the cytoplasm under acidic conditions, was added to standard growth medium and the pH was adjusted from pH 3.5 to 7.0. Wild-type and PMA1 polyploid cells were added to the media in a microdilution format. Figure 4B illustrates that a larger gene copy number results in enhanced growth at pH <5 in contrast to the growth of the wild type, supporting the role of the H+-ATPase as a high-capacity proton pump.
Biochemical properties of A. fumigatus H+-ATPase.
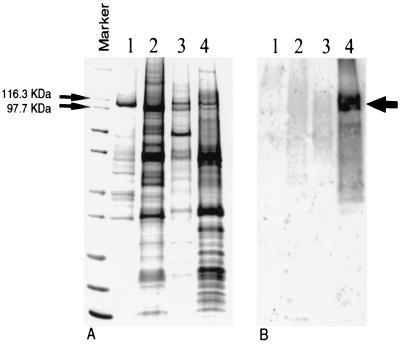
Plasma membranes were purified from A. fumigatus strain H11-20 to determine the biochemical properties of the H+-ATPase. SDS-polyacrylamide gel electrophoresis indicated that the A. fumigatus H+-ATPase is slightly larger than H+-ATPases from S. cerevisiae and Candida albicans but is equivalent in size to the enzyme from Cryptococcus neoformans (Fig. 5) Western blot analysis was used to validate the presence of the H+-ATPase by using a peptide-derived polyclonal antibody specific for an Aspergillus-specific amino acid stretch (K226PEMFETYKEYLATAN) between TM2 and TM3 of the A. fumigatus PMA1-predicted protein. As shown in Fig. 5B, the antibody was specific for the A. fumigatus H+-ATPase but did not cross-react with H+-ATPases from S. cerevisiae, C. albicans, and C. neoformans (Fig. 5B). The rather diffuse nature of the band may suggest some minor posttranslational modification, perhaps phosphorylation or glycosylation.
FIG. 5.
Electrophoretic and Western blot analyses of the A. fumigatus plasma membrane H+-ATPase. (A) Plasma membranes from A. fumigatus and other fungi were purified as described in Materials and Methods and were resolved by SDS-gel electrophoresis. Molecular mass markers designated by the lines show the region containing PMA1p from S. cerevisiae (lane 1; 100 kDa), C. albicans (lane 2; 97 kDa), C. neoformans (lane 3; 108 kDa), and A. fumigatus (lane 4; 108 kDa). The arrow in lane 4 shows the exact position of PMA1p from A. fumigatus (first main band). (B) Western blotting was performed with a peptide-derived polyclonal antibody specific for a unique amino acid stretch of the predicted A. fumigatus PMA1 protein. The position of the PMA1 band is indicated by the arrow.
The A. fumigatus enzyme was found to have an optimal pH for ATP hydrolysis of 6.5 and displayed Km and Vmax values of 0.5 ± 0.15 mM (n = 3) and 0.2 ± 0.06 μmol of inorganic phosphate released per mg of protein per min (n = 3), respectively. An apparent 50% inhibitory concentration (IC50) of 1.8 ± 0.1 μM (n = 6) was found for the transition state inhibitor orthovanadate, which classically inhibits P-type enzymes.
DISCUSSION
Few antifungal agents are available for the treatment of life-threatening infections caused by A. fumigatus, and the identification of new antifungal targets remains an important goal. As a family, the P-type ATPases are well-established therapeutic targets. The cardiac Na+, K+-ATPase and gastric H+, K+-ATPase serve as targets for two of the most clinically successful therapeutics, digoxin (16) and omeprazole (35), respectively. These drugs, which are used to treat heart disease and gastrointestinal ulcers, exhibit a high degree of enzyme selectivity that is essential to their clinical success. The fungal plasma membrane H+-ATPase has potential as a new antifungal target; and the large number of P-type enzymes in plants, parasites, and bacteria that remain unexploited offers promise for expanded development as therapeutic agents (25, 31).
In this report, the genetic and biochemical properties of the plasma membrane H+-ATPase from A. fumigatus have been described. The A. fumigatus PMA1 protein product consists of 989 amino acids with a putative molecular mass of ∼108,862 Da. SDS-polyacrylamide gel electrophoresis and Western blot analysis with a sequence-specific peptide antibody confirmed the size of the enzyme (Fig. 5) and indicated that it represents approximately 2% ± 1% (n = 6) of the total membrane protein. Previous studies have shown that fungal plasma membrane H+-ATPases are predominant membrane proteins, representing up to 25% of the total plasma membrane protein (23, 37). The protein amino acid sequence predicts a typical membrane topology with 10 transmembrane segments (Fig. 3). It contains the highly conserved signature sequences CSDKTGT, TGES, MLTGD, and GDGVN that are involved in ATP binding, phosphorylation, and ATP hydrolysis and that are characteristic of the non-heavy metal-transporting P-type ATPase family (20). The A. fumigatus enzyme in plasma membranes was found to have an optimal pH of 6.5 and displayed Km and Vmax values of 0.5 mM and 0.2 μmol of Pi released per mg of protein per min, respectively, which are typical values for fungal H+-ATPases (21, 26). An IC50 of 1.8 μM was found for the mechanism-specific inhibitor vanadate (5), which compares favorably with VO4 IC50s of 0.8, 1.6, and 9 μM for H+-ATPases from S. cerevisiae (15), C. neoformans (40), and C. albicans (23), respectively.
A. fumigatus PMA1 has two introns of 66 and 76 nucleotides near the N terminus, with typical splice sequences defining these sites (Table 1). The introns do not appear to define any significant functional boundaries in the enzyme. The A. fumigatus PMA1 cDNA and genomic DNA sequences of both strains showed 100% identity to one another. A high level of identity (99%) to A. nidulans PMA1 was seen as well (33). The amino acid sequence of A. fumigatus PMA1 shows a similarity of 40 to 63% with other (non-Aspergillus) fungal H+-ATPases and an overall 30% similarity with its animal cell counterparts (Table 2). A. fumigatus PMA1 showed 47% (38%) and 63% (52%) similarities (identities) with PMA1 genes from the prominent fungal pathogens C. neoformans and C. albicans, respectively (Table 2). In all cases, the catalytic ATP hydrolysis domain and closely linked transmembrane segments display the highest level of conservation. Regions at the N and C termini of the P-type ATPases are most divergent, while the extracellular loop domains linking transmembrane segments are more highly conserved in the fungal pumps but are more diverse with animal enzymes. This loop divergence helps account for the differential responses of the animal cell Na+, K+-ATPase to cardiac glycosides and for the specificity of antiulcer drugs like omeprazole to the gastric H+, K+-ATPase (25, 28, 29). It is this well-documented diversity that has facilitated the development of drug specificity between P-type enzymes. In principle, a drug targeted to the A. fumigatus H+-ATPase has the potential either to be specific for this fungal pathogen and related organisms because of the >90% similarity observed for Aspergillus spp. or to have a broad spectrum of antifungal activity because of a high degree (∼50%) of similarity with enzymes from other fungal pathogens. Nonetheless, the decreased similarity with related animal enzymes, especially in known therapeutic interaction regions, should promote the necessary specificity.
The A. fumigatus H+-ATPase contains three unique amino acid insertion sequences in the conserved central region of the protein consisting of 39, 7, and 16 amino acids at positions 224, 364, and 886, respectively (Fig. 2). These insertions are absent from all other fungal membrane H+-ATPases but were present in A. nidulans pmaA. The role of these insertions is unclear. However, they do not appear to play an important functional role in catalysis because binding of the peptide-derived antibody to this region had no effect on ATP hydrolysis (data not shown).
The H+-ATPase plays a critical role in fungal cell physiology, and it is one of the few antifungal targets that have been demonstrated to be essential by gene disruption (38). In this work, we attempted to disrupt A. fumigatus PMA1 by replacement with a disrupted copy of the gene containing the pyrG cassette. All of the transformants contained both the intact PMA1 and the disrupted copy. Alternatively, an unsuccessful attempt was made to introduce well-characterized mutations in conserved residues which are know to attenuate the catalytic properties of other H+-ATPases (30). We cannot rule out the possibility that homologous recombination at A. fumigatus PMA1 is a low-frequency event, although PMA1 genes typically show high levels of recombination in other fungi (14). Homologous recombination levels approaching 45% have been observed in A. fumigatus following electroporation or spheroplast transformation with linearized DNA (7, 43). It was possible to show that when a second A. fumigatus PMA1 gene was introduced into the chromosome, it was a target for homologous recombination. Collectively, these data are consistent with A. fumigatus PMA1 being an essential gene. A final confirmation of essentiality will have to await the development of a conditional expression system for lethal genes. Yet, it was possible to show functionally the importance of the H+-ATPase in intracellular pH regulation by demonstrating that an increase in the gene copy number (and, presumably, increases in the levels of gene expression and enzyme assembly) confers growth resistance in low-pH medium with the weak acid acetate (Fig. 4). As the medium is made acidic, more protons are released into the neutral cytoplasm due to changes in acetate equilibrium across the cell membrane. Wild-type cells cannot handle the acid loading below pH 5, but cells expressing larger amounts of the high-capacity A. fumigatus PMA1 proton efflux pump are more resistant to the acid conditions as protons are efficiently exported.
Overall, the fungal H+-ATPase has well-defined properties that should facilitate drug discovery. The enzyme is fully amenable to detailed genetic and biochemical analyses, which should enable an evaluation of drug-target interactions.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andriole, V. T. 1996. Aspergillus infections: problems in diagnosis and treatment. Infect. Agents Dis. 5:47-54. [PubMed] [Google Scholar]
- 3.Andriole, V. T. 1993. Infections with Aspergillus species. Clin. Infect. Dis. 17(Suppl. 2):S481-S486. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, D. 1993. Treatment of opportunistic fungal infections. Clin. Infect. Dis. 16:1-7. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, B. J., and C. W. Slayman. 1979. The effects of vanadate on the plasma membrane ATPase of Neurospora crassa. J. Biol. Chem. 254:2928-2934. [PubMed] [Google Scholar]
- 6.d'Enfert, C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5"-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76-82. [DOI] [PubMed] [Google Scholar]
- 7.d'Enfert, C., G. Weidner, P. C. Mol, and A. A. Brakhage. 1999. Transformation systems of Aspergillus fumigatus. New tools to investigate fungal virulence. Contrib. Microbiol. 2:149-166. [DOI] [PubMed] [Google Scholar]
- 8.Denning, D. W. 1996. Diagnosis and management of invasive aspergillosis. Curr. Clin. Top. Infect. Dis. 16:277-299. [PubMed] [Google Scholar]
- 9.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 10.Denning, D. W. 1994. Treatment of invasive aspergillosis. J. Infect. 28(Suppl. 1):25-33. [DOI] [PubMed] [Google Scholar]
- 11.Denning, D. W., R. M. Tucker, L. H. Hanson, and D. A. Stevens. 1990. Itraconazole in opportunistic mycoses: cryptococcosis and aspergillosis. J. Am. Acad. Dermatol. 23(3 Pt 2):602-607. [DOI] [PubMed] [Google Scholar]
- 12.Feng, D. F., and R. F. Doolittle. 1987. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol. 25:351-360. [DOI] [PubMed] [Google Scholar]
- 13.Goffeau, A. 1998. The inventory of all ion and drug ATPases encoded by the yeast genome. Acta Physiol. Scand. Suppl. 643:297-300. [PubMed] [Google Scholar]
- 14.Harris, S., K. S. Rudnicki, and J. E. Haber. 1993. Gene conversions and crossing over during homologous and homologous ectopic recombination in Saccharomyces cerevisiae. Genetics 135:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, S. L., D. S. Perlin, D. Seto-Young, and J. E. Haber. 1991. Evidence for coupling between membrane and cytoplasmic domains of the yeast plasma membrane H+-ATPase. An analysis of intragenic revertants of PMA1-105. J. Biol. Chem. 266:24439-24445. [PubMed] [Google Scholar]
- 16.Hauptman, P. J., and R. A. Kelly. 1999. Digitalis. Circulation 99:1265-1270. [DOI] [PubMed] [Google Scholar]
- 17.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 18.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latge, J. P. 1995. Tools and trends in the detection of Aspergillus fumigatus. Curr. Top. Med. Mycol. 6:245-281. [PubMed] [Google Scholar]
- 20.Lutsenko, S., and J. H. Kaplan. 1995. Organization of P-type ATPases: significance of structural diversity. Biochemistry 34:15607-15613. [DOI] [PubMed] [Google Scholar]
- 21.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith.1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 22.Moller, J. V., B. Juul, and M. le Maire. 1996. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta 1286:1-51. [DOI] [PubMed] [Google Scholar]
- 23.Monk, B. C., M. B. Kurtz, J. A. Marrinan, and D. S. Perlin. 1991. Cloning and characterization of the plasma membrane H+-ATPase from Candida albicans. J. Bacteriol. 173:6826-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monk, B. C., C. Montesinos, C. Ferguson, K. Leonard, and R. Serrano. 1991. Immunological approaches to the transmembrane topology and conformational changes of the carboxyl-terminal regulatory domain of yeast plasma membrane H(+)-ATPase. J. Biol. Chem. 266:18097-18103. [PubMed] [Google Scholar]
- 25.Monk, B. C., and D. S. Perlin. 1994. Fungal plasma membrane proton pumps as promising new antifungal targets. Crit. Rev. Microbiol. 20:209-223. [DOI] [PubMed] [Google Scholar]
- 26.Na, S., D. S. Perlin, D. Seto-Young, G. Wang, and J. E. Haber. 1993. Characterization of yeast plasma membrane H+-ATPase mutant PMA1-A135V and its revertants. J. Biol. Chem. 268:11792-11797. [PubMed] [Google Scholar]
- 27.Ng, T. T., and D. W. Denning. 1995. Liposomal amphotericin B (AmBisome) therapy in invasive fungal infections. Evaluation of United Kingdom compassionate use data. Arch. Intern. Med. 155:1093-1098. [PubMed] [Google Scholar]
- 28.Palmgren, M. G., and K. B. Axelsen. 1998. Evolution of P-type ATPases. Biochim. Biophys. Acta 1365:37-45. [DOI] [PubMed] [Google Scholar]
- 29.Perlin, D. S. 1998. Ion pumps as targets for therapeutic intervention: old and new paradigms. Elec. J. Biotech. 1:1-10.
- 30.Perlin, D. S., and J. E. Haber. 1998. Genetic approaches to structure-function analysis in the yeast plasma membrane H+-ATPase. Adv. Mol. Cell. Biol. 23A:143-166.
- 31.Perlin, D. S., D. Seto-Young, and B. C. Monk. 1997. The plasma membrane H+-ATPase of fungi. A candidate drug target? Ann. N. Y. Acad. Sci. 834:609-617. [DOI] [PubMed] [Google Scholar]
- 32.Rambosek, J., and J. Leach. 1987. Recombinant DNA in filamentous fungi: progress and prospects. Crit. Rev. Biotechnol. 6:357-393. [DOI] [PubMed] [Google Scholar]
- 33.Reoyo, E., E. A. Espeso, M. A. Penalva, and T. Suarez. 1998. The essential Aspergillus nidulans gene pmaA encodes an homologue of fungal plasma membrane H+-ATPases. Fungal Genet. Biol. 23:288-299. [DOI] [PubMed] [Google Scholar]
- 34.Robinson, R. F., and M. C. Nahata. 1999. A comparative review of conventional and lipid formulations of amphotericin B. J. Clin. Pharmacol. Ther. 24:249-257. [DOI] [PubMed] [Google Scholar]
- 35.Sachs, G. 1997. Proton pump inhibitors and acid-related diseases. Pharmacotherapy 17:22-37. [PubMed] [Google Scholar]
- 36.Sawaya, B. P., J. P. Briggs, and J. Schnermann. 1995. Amphotericin B nephrotoxicity: the adverse consequences of altered membrane properties. J. Am. Soc. Nephrol. 6:154-164. [DOI] [PubMed] [Google Scholar]
- 37.Serrano, R. 1988. Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim. Biophys. Acta 947:1-28. [DOI] [PubMed] [Google Scholar]
- 38.Serrano, R., M. C. Kielland-Brandt, and G. R. Fink. 1986. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+K+), K+- and Ca2+-ATPase. Nature 319:689-693. [DOI] [PubMed] [Google Scholar]
- 39.Seto-Young, D., B. C. Monk, and D. S. Perlin. 1992. Assessing hydrophobic regions of the plasma membrane H+-ATPase from Saccharomyces cerevisiae. Biochim. Biophys. Acta 1102:213-219. [DOI] [PubMed] [Google Scholar]
- 40.Soteropoulos, P., T. Vaz, R. Santangelo, P. Paderu, D. Y. Huang, M. J. Tamas, and D. S. Perlin. 2000. Molecular characterization of the plasma membrane H+-ATPase: a new antifungal target in Cryptococcus neoformans. Antimicrob. Agents Chemother. 44:2349-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wach, A., A. Schlesser, and A. Goffeau. 1992. An alignment of 17 deduced protein sequences from plant, fungi, and ciliate H+-ATPase genes. J. Bioenerg. Biomembr. 24:309-317. [DOI] [PubMed] [Google Scholar]
- 42.Wang, G., M. J. Tamas, M. J. Hall, A. Pascual-Ahuir, and D. S. Perlin. 1996. Probing conserved regions of the cytoplasmic LOOP1 segment linking transmembrane segments 2 and 3 of the Saccharomyces cerevisiae plasma membrane H+-ATPase. J. Biol. Chem. 271:25438-25445. [DOI] [PubMed] [Google Scholar]
- 43.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5"-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]