Abstract
Cationic bactericidal peptides are components of natural host defenses against infections. While the mode of antibacterial action of cationic peptides remains controversial, several targets, including the cytoplasmic membrane and macromolecular synthesis, have been identified for peptides acting at high concentrations. The present study identified peptide effects at lower, near-lethal inhibitory concentrations. An amidated hybrid of the flounder pleurocidin and the frog dermaseptin (P-Der), two other pleurocidin derivatives, and pleurocidin itself were studied. At 2 μg/ml, the MIC, P-Der inhibited Escherichia coli growth in a broth dilution assay but did not cause bacterial death within 30 min, as estimated by viable count analysis. Consistent with this, P-Der demonstrated a weak ability to permeabilize membranes but was able to translocate across the lipid bilayer of unilamellar liposomes. Doses of 20 μg/ml or more reduced bacterial viable counts by about 2 log orders of magnitude within 5 min after peptide treatment. Abrupt loss of cell membrane potential, observed with a fluorescent dye, dipropylthiacarbocyanine, paralleled bacterial death but did not occur at the sublethal, inhibitory concentrations. Both lethal and sublethal concentrations of P-Der affected macromolecular synthesis within 5 min, as demonstrated by incorporation of [3H]thymidine, [3H]uridine, and [3H]histidine, but the effects were qualitatively distinct at the two concentrations. Variations of the inhibition pattern described above were observed for pleurocidin and two other derivatives. Our results indicate that peptides at their lowest inhibitory concentrations may be less capable of damaging cell membranes, while they maintain their ability to inhibit macromolecular synthesis. Better understanding of the effects of peptides acting at their MICs will contribute to the design of new peptides effective at lower, less toxic concentrations.
Short, positively charged, amphipathic peptides are being considered as a novel class of antimicrobials. Most of these are based on natural templates present in virtually all species of life. Indeed, hundreds of polycationic peptides with broad spectra of antimicrobial activity have been isolated from a multitude of organisms, and their roles in preventing the onset of infections have been recognized (2, 13).
Pleurocidin, an α-helical cationic peptide, is derived from winter flounder (7), and its processing and expression pattern in flounder tissues have recently been described (6, 9). Several variants of the 25-amino-acid pleurocidin and its closest homologues, frog-derived dermaseptin (23) and insect-derived ceratotoxin (19), have been constructed and tested for their antimicrobial activities (15). Of those, pleurocidin amidated at its C terminus (P-CN) and a C-terminally amidated hybrid of pleurocidin and dermaseptin (P-Der) exhibited improved activity against Vibrio anguillarum in vitro (15). In addition, P-CN was shown to protect coho salmon from V. anguillarum infections (15). Also, the activity of pleurocidin against V. anguillarum and Aeromonas salmonicida was shown to be potentiated by salmon histone H1 peptides (26).
The major purpose of modifying natural cationic antimicrobial peptides is to increase their antimicrobial effects and decrease their toxicities. To do that in a rational rather than an empirical manner, some understanding of the peptide mode of action and structure-function relationships is required. There are numerous hypotheses to explain the mode of action of these peptides. Cationic peptides are well suited to interaction with bacterial membranes, and many, including peptides like pleurocidin, only fold into their characteristic secondary structures upon insertion into these membranes. However, how this interaction with membranes leads to bacterial cell death is somewhat controversial and indeed may vary from peptide to peptide. Different investigators have proposed destruction of the cytoplasmic membrane permeability barrier, inhibition of cytoplasmic targets, and lysis as possible mechanisms of killing, although lysis is, in fact, rarely observed.
The initial interaction of cationic antimicrobial peptides with the cytoplasmic membrane involves the insertion of the peptides parallel to the membrane surface into the interface between the phospholipid head groups and fatty acid chains of the outer monolayer of this membrane. Thereafter, diverse models propose how the peptides reorient perpendicular to the plane of the membrane and the consequences thereof.
In the barrel-stave model, amphipathic peptides are proposed to align perpendicular to the membrane to form the staves of a transient barrel of various sizes (depending on the number of peptide subunits or staves) that contains in its center a hydrophilic pore traversing the cytoplasmic membrane (28). This would then lead to leakage of cytoplasmic contents, which might then lead to cell death. It has been shown, however, that pore formation does not always accompany the antimicrobial activity of peptides (32). Conversely, in the carpet model (29), it was proposed that loss of cell membrane integrity occurs when the membrane becomes covered by a “carpet” of peptides oriented parallel to the membrane, which then collapses inward to destroy the permeability barrier of the membrane. Both of the models described above would lead to the prediction that antimicrobial activity would be accompanied by a loss of the membrane potential. A third model, referred to as the “aggregate model,” suggests that cationic peptides aligned at the membrane interface reversibly and randomly cluster to form transient water-containing micellar aggregates, which span the membrane and which can collapse to either side of the membrane, permitting peptide translocation to the cytoplasm (12). Other models envision transient toroidal channels composed of peptide and lipid (14, 21).
There are several reports that peptides affect intracellular activities (3, 31). Only recently have research interests been directed toward attempting to link and explain the membrane and nonmembrane effects of cationic peptides (4, 8, 11, 17, 24, 25, 27). Most research to date has been conducted at high multiples of the MICs or, in the case of model studies, high peptide-to-lipid ratios and has identified cell membranes as the target for cationic peptide action. The present study examined the effects of four previously described pleurocidin-based peptides, both at their MICs and at higher multiples of their MICs, on the Escherichia coli cytoplasmic membrane and on macromolecular synthesis.
MATERIALS AND METHODS
Materials and bacterial strains.
All peptides were synthesized by N-(9-fluorenyl)methoxy carbonyl chemistry at the Nucleic Acid Protein Service unit at the University of British Columbia. Peptide sequences are shown in Fig. 1. Peptide purity was confirmed by high-pressure liquid chromatography and mass spectrometry analysis. Bovine serum albumin fraction V lyophilysate was purchased from Boehringer Mannheim (Mannheim, Germany). E. coli strain CGSC 4908 (his-67 thyA43 pyr-37), auxotrophic for thymidine, uridine, and l-histidine (5), was kindly supplied by the E. coli Genetic Stock Centre (Yale University, New Haven, Conn.). Defined M9 minimal medium, supplemented with 5 mg of thymidine per liter, 10 mg of uridine per liter, and 20 mg of l-histidine per liter (Sigma Chemical Co., St. Louis, Mo.), was used to grow E. coli CGSC 4908 unless otherwise specified. E. coli UB1005 was a wild-type strain from our laboratory stock collection and was grown in standard Luria-Bertani medium at 37°C. Dipropylthiacarbocyanine (diSC35) was purchased from Molecular Probes (Eugene, Oreg.). The tritiated precursors [methyl-3H]thymidine (25.0 Ci/mmol), [5-3H]uridine (26.0 Ci/mmol), and l-[2,5-3H]histidine (46.0 Ci/mmol) were purchased from Amersham Pharmacia Biotech UK Limited. The lipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (PG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE), phosphatidylglycerol from egg yolk (ePG), cardiolipin, and 1-palmitoyl-2-{6-[(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-caproyl}-l-α-phosphatidylcholine (C6-NBD-PC) were purchased from Avanti Polar Lipids Inc. (Alabaster, Ala.). l-α-Phosphatidy-dl-choline from egg-yolk (ePC) and calcein were purchased from Sigma.
FIG. 1.
Peptide sequences. One-letter symbols are used for amino acids. Amidation of the C terminus is shown as NH2.
MICs.
The MICs of the peptides were determined by a broth microdilution assay modified from the method of Amsterdam (1). Briefly, serial dilutions of the peptide were made in 0.2% BSA bovine serum albumin-0.01% acetic acid solution in 96-well polypropylene (Costar; Corning Incorporated, Corning, N.Y.) microtiter plates. Each well was inoculated with 100 μl of E. coli CGSC 4908 in supplemented M9 minimal medium to a final concentration of approximately 105 CFU/ml. The MIC was taken as the lowest peptide concentration at which growth was inhibited after 24 h of incubation at 37°C.
Cytoplasmic membrane depolarization and bacterial killing assay.
The depolarization of the cytoplasmic membrane of E. coli CGSC 4908 by the peptides was determined with membrane potential-sensitive cyanine dye diSC35 (30) by the method of Wu et al. (32), as described by Friedrich et al (10). Briefly, an equilibrated 1.5-ml suspension of bacterial cells in HEPES (5 mM)-diSC35 (0.4 μM)-KCl (100 mM) was placed in a 1-cm cuvette, and the desired concentration of the peptide to be tested was added. Changes in fluorescence due to the disruption of the cytoplasmic membrane potential were continuously recorded with a spectrofluorimeter at an excitation wavelength of 622 nm and an emission wavelength of 670 nm. Cell viability was assessed by plating aliquots withdrawn from a cell suspension identical to that used in the permeabilization experiment and prepared with the same bacterial culture.
Macromolecular synthesis and bacterial killing assays.
Overnight cultures of E. coli CGSC 4908 were diluted 10−3 in synthetic media and allowed to grow to the exponential phase (optical density at 600 nm, 0.3). The cultures were spun down and resuspended in warm synthetic M9 medium, and 500-μl aliquots were incubated with 15 μl of either [3H]thymidine, [3H]uridine, or l-[3H]histidine and an excess of the remaining two supplements. After 5 min of incubation at 37°C, the peptides were added at the specified concentrations. Samples of 50 μl were removed immediately before peptide addition (0 min) and at 5, 10, 20, and 40 min after peptide addition and added to 5 ml of ice-cold 10% trichloroacetic acid (Fisher Scientific, Fair Lawn, N.J.) with excess unlabeled precursors in order to precipitate the macromolecules. After 40 min on ice and 15 min at 37°C the samples were collected over vacuum on Whatman 47 mm GF/C glass microfiber filters (VWR Canlab, Mississauga, Ontario, Canada) and washed twice with 10 ml of ice-cold 10% trichloroacetic acid. The filters were dried and placed in 7-ml scintillation vials with ReadySafe liquid scintillation cocktail (Beckman, Fullerton, Calif.), and counts were obtained in a Beckman LS600IC scintillation counter for 5 min for each filter. At the same time points, 5-μl aliquots were removed from nonradioactive parallel cultures otherwise identical to those containing a radioactive precursor, diluted in 1 ml of buffer, and plated onto plates with Luria-Bertani medium and supplements in order to obtain a viable count. The aliquots were diluted immediately upon withdrawal from the experimental samples, thus preventing further peptide action by radically reducing its concentration. This ensured that the viable counts corresponded to the respective time points.
Dansyl polymyxin B displacement assay.
The relative binding affinity of each peptide for lipopolysaccharide (LPS) was determined by the dansyl polymyxin B displacement assay of Moore et al. (22) with LPS isolated from E. coli UB1005. Maximal displacement of LPS was expressed as a percentage, in which 100% displacement of dansyl polymyxin B was taken as that observed with excess polymyxin B.
Outer membrane permeabilization assay.
The outer membrane permeabilization activities of the peptide variants were determined by the 1-N-phenylnaphthylamine uptake assay of Loh et al. (18) with intact cells of E. coli UB1005.
Liposome preparation.
Symmetrically labeled unilamellar liposomes were made from a 1:1 mixture of PC and PG. The mixture contained 0.5 mol% C6-NBD-PC. The lipids were dissolved in chloroform and dried under a stream of nitrogen, followed by 2 h of vacuum drying. The lipid film was rehydrated with TSE buffer (10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA [pH 7.5]) and extruded 10 times through two stacked filters with a pore size of 100 nm. For asymmetrically labeled liposomes (inner leaflet only), the symmetrically labeled unilamellar liposomes were mixed with 1 M sodium dithionite in 1 M Tris-HCl (pH 7.5), and the mixture was incubated for 15 min at 23°C, which resulted in chemical quenching of the nitrobenzoxadiazolyl (NBD) groups in the outer leaflet of the bilayers. The liposomes were immediately separated from dithionite by gel filtration with Bio-Gel A (1.5 m by 10 cm; Bio-Rad, Hercules, Calif.) at 23°C.
For calcein-encapsulated unilamellar liposomes, a PC-PG (1:1) lipid film was rehydrated with 5 mM sodium HEPES (pH 7.5) containing 100 mM calcein. The liposome suspension was extruded 10 times through two stacked polycarbonate filters (pore size, 100 nm), and free calcein was removed by passage of the liposome suspension through a Sephadex G-50 column (1.5 by 10 cm; Pharmacia Biotech AB, Uppsala Sweden) at 23°C and elution with a buffer containing 20 mM sodium HEPES, 150 mM NaCl and 1 mM sodium EDTA (pH 7.5). Calcein-free PC-PG (1:1) unilamellar liposomes made in the same elution buffer were added to adjust the final liposome concentrations.
P-Der translocation experiments.
Unilamellar liposomes (ePC, ePG, and dansyl-PE [50/45/5]) containing 200 mM chymotrypsin solution in buffer containing 150 mM NaCl and 20 mM HEPES (pH 7.4) were made as described by Kobayashi et al. (16). Trypsin-chymotrypsin inhibitor (final concentration, 200 mM; Sigma) was added to the liposomes to inactivate any enzyme present outside the unilamellar liposomes. Excitation of tryptophan residues in P-Der at 280 nm leads to fluorescence transfer to the dansyl group in DNS-PE, leading to an emission recorded at 510 nm. A decrease in fluorescence after peptide addition indicated digestion of the internalized peptide by the enzyme within the liposomes.
RESULTS
Effects of peptides on the E. coli cytoplasmic membrane.
To evaluate the effects of the selected peptides on E. coli cytoplasmic membranes, fluorescent probe diSC35 was used. diSC35 distributes between the cells and the medium, depending on the cytoplasmic membrane potential, and self-quenches when it is concentrated inside the bacterial cells. If the membrane is depolarized, the probe will be released into the medium, causing a measurable increase in fluorescence.
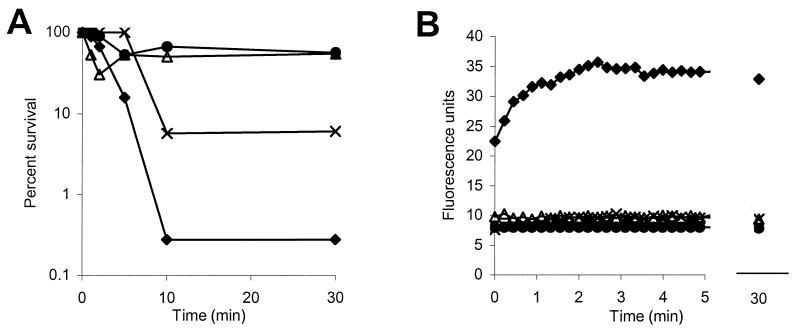
P-Der, applied at five-fold its MIC, did not cause E. coli cytoplasmic membrane permeabilization, estimated by the diSC35 assay, within 30 min of the addition of the peptide (Fig. 2B), even though it caused a 12-fold decrease in the viable count (Fig. 2A). Conversely, 10-fold the MIC of P-Der caused immediate maximal membrane depolarization (Fig. 2B), accompanied by a 500-fold decrease in viable cell counts within 10 min (Fig. 2A).
FIG. 2.
Ability of P-Der to permeabilize the cytoplasmic membrane of E. coli CGSC 4908. Bacterial survival (A) and permeabilization of the cytoplasmic membrane (B) are shown. The fluorescence of a membrane potential-sensitive dye, diSC35, was measured. The peptide was added at its MIC (▵), 5 times its MIC (×), and 10 times its MIC (⧫). The results for a control sample to which no peptide was added are also shown (•). Data representative of three separate experiments are shown.
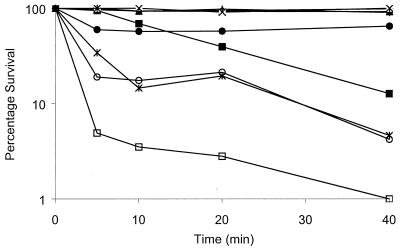
Indeed, 10-fold the MICs of all peptides tested permeabilized the E. coli cell membrane within the initial 3 min after peptide addition, as summarized in Table 1. Rapid cell membrane permeabilization was accompanied by the initiation of cell death either immediately or within 5 min (Fig. 3). Delayed cell death, as observed in the case of P-M (Fig. 3), indicated that factors other than membrane depolarization per se may have triggered or caused the lethal event.
TABLE 1.
Correlation between inhibitory concentration, cytoplasmic membrane depolarization, and inhibition of macromolecular synthesisa
| Peptide | MIC (μg/ml) | Concn applied | Maximal cytoplasmic membrane depolarization (%)b | Time (min) of initiation of inhibition of synthesis
|
||
|---|---|---|---|---|---|---|
| DNA | RNA | Protein | ||||
| Pleurocidin | 2 | MIC | 40 | 20 | —d | 10 |
| 10 times the MIC | 100 | 5 | 10 | 0 | ||
| P-CN | 1 | MIC | 75 | 10 | 5 | 5 |
| 10 times the MIC | 100 | 0 | 0 | 0 | ||
| P-M | 4 | MIC | 30 | 20 | 10 | — |
| 10 times the MIC | 100 | 5 | 0 | 5 | ||
| P-Der | 2 | MIC | 0 | 20 | 5 | — |
| 10 times the MIC | 100 | 10 | 0 | 5 | ||
| CCCP | 50 μM | NDc | — | — | 20 | |
| Rifampin | 32 | 2 times the MIC | ND | — | 5 | 20 |
Maximal cytoplasmic membrane depolarizations caused by each peptide at its MIC and 10 times the MIC within 3 min from the addition of peptide are shown. The times between the addition of peptides until the initiation of macromolecular synthesis inhibition are shown.
Recorded 3 min after peptide addition to cells equilibrated with diSC35.
ND, not determined (CCCP interferes with diSC35 fluorescence).
—, no inhibition observed during the course of the experiment.
FIG. 3.
Effects of pleurocidin, P-CN, and P-M on survival of E. coli CGSC 4908. Peptides were added to specified concentrations, as follows: P-M at its MIC (▴), P-M at 10 times its MIC (▪), pleurocidin at its MIC (•), pleurocidin at 10 times its MIC (∗), P-CN at its MIC (○), and P-CN at 10 times its MIC (□). In addition, the results for the no peptide controls for P-M (⧫), pleurocidin (×), and P-CN (+) are shown. Since the data for each peptide were obtained in a separate set of experiments, the results for all three no-peptide controls are shown for comparison. Samples of 10 μl were removed at the indicated intervals and plated. Percent survival relative to the number of bacteria prior to peptide addition was determined. Data representative of three separate experiments are shown.
Addition of pleurocidin, P-CN, and P-M at the MICs resulted in only 40, 75, and 30% of the maximal membrane permeabilization, respectively (Table 1). Our data thus show that although pleurocidin and its derivatives exert effects on the E. coli cytoplasmic membrane, especially when they are added at high multiples of their MICs, not all antimicrobial activity can be attributed to the failure of the cytoplasmic membrane barrier.
Effects of peptides on DNA, RNA, and protein synthesis in E. coli CGSC 4908.
To assess the effects of pleurocidin peptides on the synthesis of macromolecules in bacteria, the incorporation of the radioactive precursors [methyl-3H]thymidine, [5-3H]uridine, and l-[2,5-3H]histidine into DNA, RNA, and proteins, respectively, was monitored in an E. coli strain auxotrophic for these three precursors.
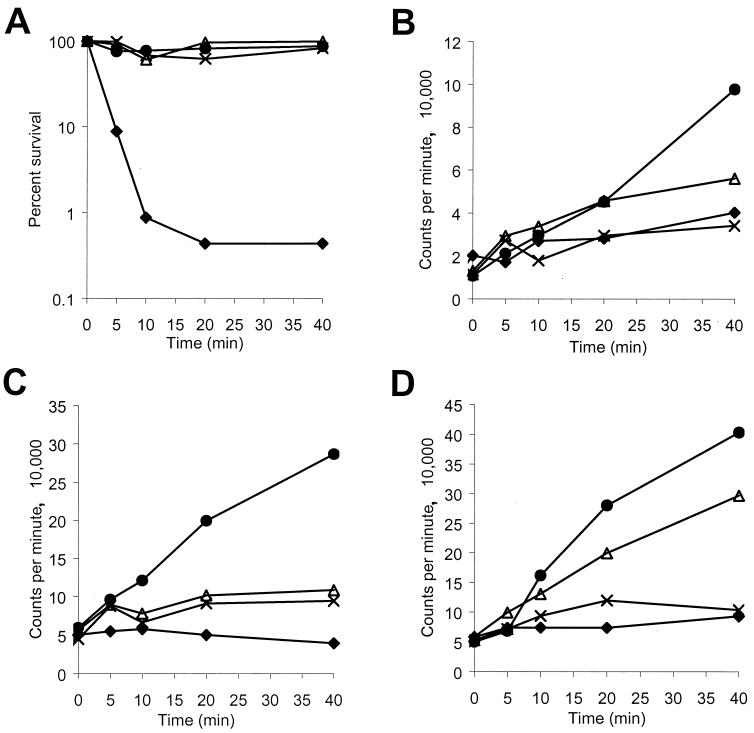
P-Der, added at 10-fold its MIC, caused immediate inhibition of RNA synthesis (Fig. 4C), followed by inhibition of protein synthesis (Fig. 4D) and DNA synthesis (Fig. 4B). When added at its MIC, P-Der inhibited RNA synthesis within 5 min of peptide addition (Fig. 4C), followed by the inhibition of DNA synthesis (Fig. 4B). Protein synthesis did not appear to be inhibited (Fig. 4D), and cell viability was not affected (Fig. 4A) at that concentration of P-Der within the 40-min time course of the experiment.
FIG. 4.
Effects of P-Der on macromolecular synthesis in E. coli CGSC 4908. Bacterial survival (A), [3H]thymidine incorporation into DNA (B), [3H]uridine incorporation into RNA (C), and l-[3H]histidine incorporation into protein (D) were measured. The peptide was added at its MIC (▵), 5 times its MIC (×), and 10 times its MIC (⧫). The results for control sample with no peptide are also shown (•). Data representative of three separate experiments are shown.
A summary of macromolecular synthesis inhibition data for all peptides correlated with the cytoplasmic membrane depolarization data described above is shown in Table 1. All peptides at concentrations 10-fold their MICs inhibited the incorporation of radioactive precursors into RNA, DNA, and protein in E. coli CGSC 4908. In the case of P-CN, the synthesis of all three macromolecules was inhibited within 5 min after peptide addition, while the time patterns of inhibition varied for the other three peptides. Protein synthesis appeared to be inhibited first in the case of pleurocidin, while RNA synthesis appeared to be inhibited first upon addition of high concentrations of P-M and P-Der.
Addition of all peptides at their MICs still resulted in specific inhibition of macromolecular synthesis, even though only partial or no membrane depolarization (Table 1) and little or no killing (Fig. 3) were observed. The pattern of inhibition was similar, but not identical, to that observed at high multiples of the MIC. RNA synthesis was inhibited first upon addition of P-Der and P-M, protein synthesis was inhibited first upon addition of pleurocidin, and the synthesis of all three macromolecules was inhibited within 10 min upon addition of P-CN. Protein synthesis and RNA synthesis did not appear to be inhibited by P-Der and pleurocidin at their MICs, respectively, during the 40-min course of the experiment. This finding is particularly important in the case of P-Der, which appeared to selectively inhibit RNA synthesis (Fig. 4C) at concentrations which had no effect on cytoplasmic membrane permeabilization (Fig. 2B). The effect of P-Der was thus similar to that observed for the RNA polymerase-specific, bacteriostatic inhibitor rifampin, but not for carbonyl cyanide m-chloraphenylhydrazone (CCCP), which is bacteriostatic due to its ability to equilibrate protons across, and thus depolarize, the cytoplasmic membrane (Table 1). Norfloxacin and chloramphenicol were used as other controls. The former inhibited DNA synthesis, while the latter inhibited protein synthesis (data not shown).
Interaction of peptides with the outer membrane.
All four peptides were able to displace dansyl polymyxin B which had been prebound to LPS (Table 2). The affinity and avidity of the interaction varied. For example, P-M demonstrated a maximal displacement of dansyl polymyxin B compared to the displacement caused by nondansylated polymyxin B of 95% and the concentration of P-M required for maximal LPS binding was 4 μg/ml, making it the best LPS binder. Interestingly, P-M was the least active of all four peptides in antimicrobial assays. However, the results obtained for the remaining three peptides argued against any relationship between LPS binding and antimicrobial activity.
TABLE 2.
Ability of pleurocidin, P-CN, P-M, and P-Der to bind to E. coli LPS and permeabilize E. coli outer membrane
| Peptide | LPS bindinga
|
Time (min) required until maximal (100%) outer membrane permeabilization was achieved | |
|---|---|---|---|
| Imax (%) | IC50 (μg/ml) | ||
| Pleurocidin | 78 | 32 | 5 |
| P-CN | 85 | 24 | 4 |
| P-M | 95 | 7 | 6 |
| P-Der | 67 | 16 | 5 |
| Polymyxin B | 100 | 5 | 2 |
| Mg2+ | 60 | 140 | NDb |
Imax, maximal displacement of dansyl polymyxin B by the various peptides compared to the displacement caused by nondansylated polymyxin B, which is taken as 100%; IC50, one-half of the peptide concentration required for maximal LPS binding.
ND, not determined.
All four peptides, when applied at 2.6 μg/ml, fully permeabilized the E. coli outer membrane within 4 to 6 min (Table 2). This indicated that the peptides studied here were able to reach the cytoplasmic membrane.
Ability of P-Der to stimulate lipid flip-flop and lyse liposomes.
The observation that cationic antimicrobial peptides, especially P-Der, were able to inhibit macromolecular synthesis without fully permeabilizing the cytoplasmic membrane (and are able to do so in a manner distinctly different from that of the depolarizing uncoupler CCCP; Table 1) indicated that they could translocate across the cytoplasmic membrane. Thus, we investigated the mode of interaction of P-Der with membranes using a series of liposome-based assays (20, 34). The induction by P-Der of lipid translocation between the inner and the outer leaflets of a membrane bilayer (i.e., lipid flip-flop) was assessed with large unilamellar liposomes with their inner leaflet labeled with C6-NBD-PC. The rate of NBD-labeled lipid transfer from the inner leaflet to the outer leaflet was measured after addition of peptide in the presence of extraliposomal sodium dithionite as a quencher, since transfer of the NBD-labeled lipid into the outer leaflet resulted in its being quenched by sodium dithionite and a decrease in fluorescence. The spontaneous rate of lipid flip-flop was monitored in the course of the experiment (Fig. 5) and was found to be negligible.
FIG. 5.
Ability of P-Der to induce lipid flip-flop across a liposome bilayer. Unilamellar liposomes composed of PC-PG (1:1) were asymmetrically labeled with the inner leaflet containing fluorescent PC (C6-NBD-PC) and a quencher present outside. P-Der was added at the time indicated by the arrow. The ability of peptides to induce the translocation of the fluorescently labeled lipids from the inner leaflet to the outer leaflet, leading to quenching of the fluorescence, was measured. The decrease in fluorescence is thus representative of lipid flip-flop. The results for a control sample to which no peptide was added until 500 s and to which Triton X-100 was added at that point are also shown. A representative of three independent experiments is shown.
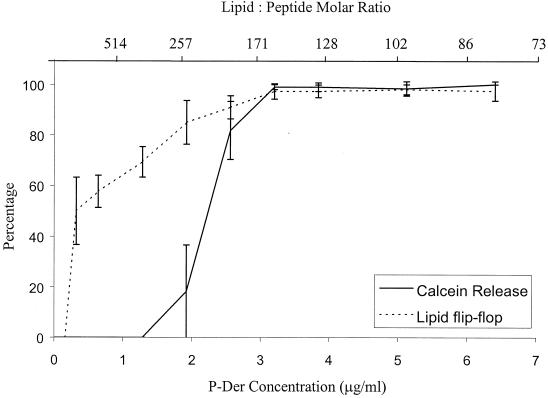
P-Der caused an increasing rate and extent of lipid flip-flop as its concentration increased (Fig. 5). At approximately two times the MIC, P-Der caused 100% lipid flip-flop in 3 min. The percent flip-flop caused by each peptide concentration was calculated as follows: 100 × (F0 − FP)/(F0 − FT), where F0, FP, and FT represent the fluorescence intensities in the labeled liposomes without peptide, with peptide, and with Triton X-100, respectively. A plot, averaged over three independent experiments, of percent flip-flop as a function of peptide concentration is shown in Fig. 6.
FIG. 6.
Correlation between lipid flip-flop and calcein release caused by P-Der. Percentages of maximal lipid flip-flop and calcein release are shown as a function of peptide concentration. Averages of three experiments and standard deviations are shown.
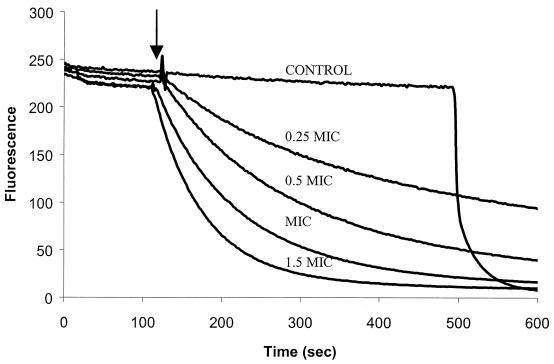
To ensure that the observed flip-flop was not caused by a disruption of the bilayer, the leakage of calcein from unilamellar liposomes was measured. As calcein is self-quenching at the high concentrations captured within the liposomes, an increase in detectable fluorescence is indicative of calcein leakage. As shown in Fig. 7, which shows representative results of an experiment with P-Der, the peptide at concentrations equal to or greater than 1.3 times its MIC caused calcein leakage to various extents. The percent leakage at each peptide concentration was calculated as follows: 100 × (FP − F0)/(FT − F0), where F0 and FP denote the fluorescence intensity before and after peptide addition, respectively, and FT represents the fluorescence intensity after addition of Triton X-100. A plot of percent leakage, averaged over three independent experiments, as a function of peptide concentration is shown in Fig. 6. The results show that while P-Der was able to induce calcein release at 3.8 μg/ml and relatively slow calcein release at 2.6 μg/ml, it did not cause calcein release at concentrations lower than 1 μg/ml. It is evident from the plots of percent flip-flop and percent calcein release as a function of peptide concentration (Fig. 6) that there was a range of concentrations around the MIC at which measurable lipid flip-flop occurred without measurable calcein release. This indicated that the peptide can stimulate lipid translocation between the leaflets of the bilayer without causing liposome lysis or gross permeabilization.
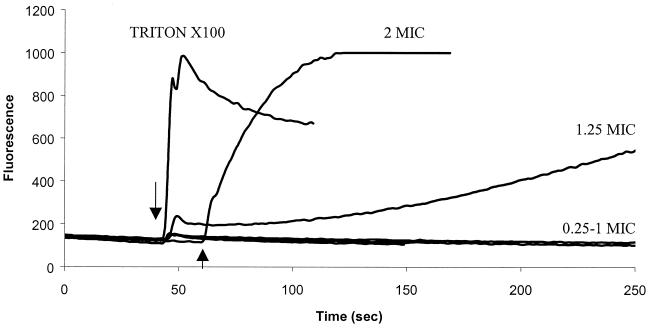
FIG. 7.
Ability of P-Der to induce calcein release from liposomes. Calcein-filled liposomes composed of PC-PG (1:1) were used. P-Der was added at the time indicated by the arrow. An increase in fluorescence corresponds to the release of calcein and is indicative of liposome leakage or lysis. Triton X-100 was used as a control to induce lysis. A result representative of three independent experiments is shown.
Ability of P-Der to translocate into liposomes.
While the data presented above show that P-Der can stimulate the translocation of lipids between bilayers, they do not indicate whether the peptide itself can translocate. The ability of P-Der to access the interiors of large unilamellar liposomes, which were made of an equimolar mixture ePC and ePG and which contained the fluorescently tagged lipid DNS-PE, was measured. The liposomes also contained active α-chymotrypsin. Resonance energy transfer from the P-Der tryptophan residue to DNS-PE resulted in an initial increase in fluorescence upon binding of the peptide to the membrane. Upon translocation, the peptide was digested by the α-chymotrypsin encapsulated in the liposomes, leading to a decrease in fluorescence intensity.
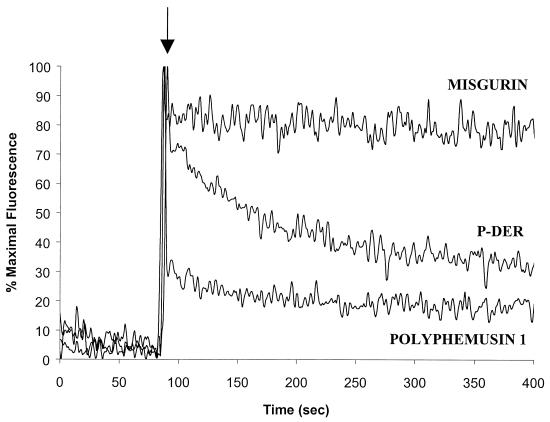
Evidence for peptide translocation is shown in Fig. 8. P-Der at 1.3 μg/ml caused a decrease in fluorescence, which is characteristic of translocation. Translocation was not as rapid as that observed for the positive control peptide, polyphemusin, which was the most effectively translocated peptide in a prior study (34). Misgurin, which in our hands has no antimicrobial activity (15), failed to translocate.
FIG. 8.
Ability of P-Der to translocate into liposomes. A decrease in fluorescence after addition of peptide at 1.3 μg/ml is indicative of digestion of the internalized peptide by liposome-entrapped chymotrypsin. Peptide internalization is thus shown. Polyphemusin and misgurin were used as a positive control and a negative control, respectively. A result representative of three separate experiments is shown.
DISCUSSION
Much of today's research on the mode of action of cationic antimicrobial peptides has been conducted at high multiples of the MICs or high peptide-to-lipid ratios. As a consequence, most research identifies cell membranes as the major target for cationic peptide action. The present study examined the effects of near-MICs of pleurocidin and three of its derivatives on the E. coli cytoplasmic membrane and on macromolecular synthesis. P-Der, a hybrid of pleurocidin and dermaseptin, demonstrated the ability to inhibit intracellular functions without damaging the E. coli cytoplasmic membrane at concentrations up to fivefold its MIC. When applied at 1.28μg/ml, P-Der was also able to enter large unilamellar liposomes, but it did not cause calcein leakage from the liposomes. The data are consistent with translocation of the peptide into bacterial cells without causing damage to the cytoplasmic membrane.
Yoshida et al. (33) recently reported that pleurocidin can translocate into PC-PG (3:1) liposomes but causes calcein leakage in that system. One hundred percent calcein release and peptide translocation were indeed reported when 10 μM pleurocidin was applied. However, the data of Yoshida et al. (33) also indicate that at pleurocidin concentrations lower than 2.5 μM, the peptide translocated across membranes with reasonable efficiency (up to 60%), while it caused less than 30% calcein release.
Kobayashi et al. (16) used a similar experimental design to describe the activities of the α-helical peptides magainin II and buforin II. Magainin II was an efficient promoter of lipid flip-flop and leakage, but it did not translocate well across the bilayer; and buforin II was not very effective at promoting leakage and flip-flop, but it translocated relatively well across lipid bilayers.
A significantly greater complexity of membrane interaction patterns was described by Zhang et al. (34), who characterized a number of α-helical, β-sheet, extended, and cyclic peptides. In their study the peptides fell into three broad groups with respect to their interactions with model membranes: (i) weak inducers of all membrane activities tested; (ii) gramicidin S, with its limited ability to induce flip-flop and calcein release but its good ability to insert into lipid monolayers, depolarize cytoplasmic membranes, and form channels in planar lipid bilayers; and (iii) strong inducers of most, but not all (depending on the peptide), membrane activities. Indeed, Zhang et al. (34) reported significant variations within the last group, and in most cases the relationship between model membrane activities and bactericidal action was complicated.
The results of our study add to the complexity reported above. Three separate activities of P-Der were measured in our liposome assays. First, the movement of lipids from one monolayer of the liposome bilayer to the other, also referred as lipid flip-flop, was assessed. Second, the ability of the peptide to cause the release of a fluorescent dye, calcein, trapped in the intravesicular space of the liposome was assessed. Lastly, the ability of the peptide to translocate across the liposome membrane was assessed. The results of these experiments revealed that lipid flip-flop occurred at peptide concentrations that were two- to three-fold lower than those that cause calcein release. Also, P-Der at 1.3 μg/ml was able to enter large unilamellar liposomes, but it did not cause calcein leakage from the liposomes. These data were consistent with the ability of the peptides to permeabilize the cytoplasmic membrane of E. coli and inhibit its intracellular functions.
At 10-fold their MICs, all four peptides tested in the present study caused rapid depolarization of the cytoplasmic membrane, cessation of macromolecular synthesis, and cell death. These results are thus consistent with data published for many other peptides, and they could be interpreted in terms of the cell membrane being the lethal target for cationic peptides applied at high concentrations. Although the destruction of the cytoplasmic membrane potential gradient is not per se a lethal event (e.g., CCCP is bacteriostatic rather than bactericidal), it might be argued that the damage caused to the cell membrane could cause leakage of essential cell molecules and thus be responsible for the rapid cell death and cessation of macromolecular synthesis.
However, a significantly different pattern was observed for pleurocidin, P-M, and especially P-Der. When used at its MIC, P-Der did not cause notable depolarization of the cell membrane or rapid killing. Even without rapid killing and cell membrane permeabilization, however, macromolecular synthesis and, in particular, RNA synthesis were inhibited. The P-Der hybrid, when used at its MIC, inhibited RNA synthesis about 10 min before other effects were observed. In fact, the pattern of macromolecular synthesis in the presence of P-Der at its MIC appeared to be similar to that observed in the presence of the RNA polymerase inhibitor rifampin. While it is too early to speculate about whether RNA inhibition is the primary target at sublethal concentrations of P-Der, our results suggest that this peptide can translocate into cells without substantial permeabilization. Therefore, internal targets must be considered for P-Der.
This is consistent with recently published data for lysozyme-derived fragments (27). Pellegrini et al. (27) showed that short, positively charged peptides inhibited DNA and RNA synthesis before they caused inner membrane permeabilization in E. coli. While in the experiments reported here P-Der was the only peptide which did not cause any inner membrane permeabilization while exerting intracellular effects, the results for all four peptides support the contention that intracellular targets may be a part of the antimicrobial mechanism of action when the peptides are used at their MICs. In this model, the incomplete or minimal cytoplasmic membrane depolarizations which were observed would be a consequence of the membrane disturbance and of ions being carried by the interstitial water while temporary peptide aggregates form as an intermediate during translocation across the cytoplasmic membrane. Recent reports (4, 8, 17, 24, 25) establish a strong precedent for a process in which cationic peptides enter bacterial cells (25) and target specific molecules, such as heat shock proteins (24).
It is exceedingly difficult to correlate primary sequence variations with the distinct activities of individual cationic antimicrobial peptides. Indeed, we contend that differences in the ways in which individual peptides interact with bacterial cells result from their diverse secondary and tertiary structures, which are in turn determined by variations in the primary sequences as well as other factors, such as hydrophobicity and charge characteristics of the environment. The variations in membrane and intracellular activities observed among the four closely related pleurocidin-based peptides and the parallels in the action of P-Der and buforin II, which are less similar, indicate that until nuclear magetic resonance imaging-quality structural comparisons are available, one should discuss peptide mode of action on a case-by-case basis.
Collectively, our data indicate that, at their MICs, cationic peptides can translocate into bacterial cells to exert their antimicrobial effects intracellularly. Such translocation is not unprecedented and has, in fact, been shown by Park et al. (25) for buforin II, although the specific mechanism of translocation in that particular case still remains unknown. Better understanding of the various aspects of peptide action on bacteria, especially those involving peptides at low multiples of their MICs, will provide the tools required to design more potent and less toxic antimicrobials.
Acknowledgments
This work was funded by grants from the Natural Sciences and Engineering Research Council and Canadian Bacterial Diseases Network (to R.E.W.H.). Also, R.E.W.H. is the holder of a Canada Research Chair.
REFERENCES
- 1.Amsterdam, D. 1996. Susceptibility testing of antimicrobials in liquid media. In V. Lorian (ed.), Antibiotics in laboratory medicine. The Williams & Wilkins Co., Baltimore, Md.
- 2.Boman, H. G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13:61-92. [DOI] [PubMed] [Google Scholar]
- 3.Boman, H. G., B. Agerberth, and A. Boman. 1993. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 61:2978-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castle, M., A. Nazarian, S. S. Yi, and P. Tempst. 1999. Lethal effects of apidaecin on Escherichia coli involve sequential molecular interactions with diverse targets. J. Biol. Chem. 274:32555-32564. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, S., M. Skiguchi, J. Stern, and H. Barner. 1963. The synthesis of messenger RNA without protein synthesis in normal and phage-infected thymineless strains of Escherichia coli. Proc. Natl. Acad. Sci. USA. 49:699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, A. M., R. O. Darouiche, D. Legarda, N. Connell, and G. Diamond. 2000. Characterization of a fish antimicrobial peptide: gene expression, subcellular localization, and spectrum of activity. Antimicrob. Agents Chemother. 44:2039-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, A. M., P. Weis, and G. Diamond. 1997. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 272:12008-12013. [DOI] [PubMed] [Google Scholar]
- 8.Daugelavicius, R., E. Bakiene, and D. H. Bamford. 2000. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob. Agents Chemother. 44:2969-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas, S. E., J. W. Gallant, Z. Gong, and C. Hew. 2001. Cloning and developmental expression of a family of pleurocidin-like antimicrobial peptides from winter flounder, Pleuronectes americanus (Walbaum). Dev. Comp. Immunol. 25:137-147. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich, C. L., D. Moyles, T. J. Beveridge, and R. E. W. Hancock. 2000. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob. Agents Chemother. 44:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich, C. L., A. Rozek, A. Patrzykat, and R. E. W. Hancock. 2001. Structure and mechanism of action of an indolicidin peptide derivative with improved activity against gram-positive bacteria. J. Biol. Chem. 276:24015-24022. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E. W., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, R. E. W., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 14.Huang, H. W. 2000. Action of antimicrobial peptides: two-state model. Biochem. 39:8347-8352. [DOI] [PubMed] [Google Scholar]
- 15.Jia, X., A. Patrzykat, R. H. Devlin, P. A. Ackerman, G. K. Iwama, and R. E. W. Hancock. 2000. Antimicrobial peptides protect coho salmon from Vibrio anguillarum infections. Appl. Environ. Microbiol. 66:1928-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi, S., K. Takeshima, C. B. Park, S. C. Kim, and K. Matsuzaki. 2000. Interactions of the novel antimicrobial peptide buforin 2 with lipid bilayers: proline as a translocation promoting factor. Biochemistry 39:8648-8654. [DOI] [PubMed] [Google Scholar]
- 17.Kragol, G., S. Lovas, G. Varadi, B. A. Condie, R. Hoffmann, and L. Otvos, Jr. 2001. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40:3016-3026. [DOI] [PubMed] [Google Scholar]
- 18.Loh, B., C. Grant, and R. E. W. Hancock. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchini, D., P. C. Giordano, R. Amons, L. F. Bernini, and R. Dallai. 1993. Purification and primary structure of ceratotoxin A and B, two antibacterial peptides from the female reproductive accessory glands of the medfly Ceratitis capitata (Insecta:Diptera). Insect Biochem. Mol. Biol. 23:591-598. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1996. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 35:11361-11368. [DOI] [PubMed] [Google Scholar]
- 21.Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1995. Translocation of a channel-forming antimicrobial peptide, magainin 2, across lipid bilayers by forming a pore. Biochemistry 34:6521-6526. [DOI] [PubMed] [Google Scholar]
- 22.Moore, R. A., N. C. Bates, and R. E. W. Hancock. 1986. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob. Agents Chemother. 29:496-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mor, A., V. H. Nguyen, A. Delfour, D. Migliore-Samour, and P. Nicolas. 1991. Isolation, amino acid sequence, and synthesis of dermaseptin, a novel antimicrobial peptide of amphibian skin. Biochemistry 30:8824-8830. [DOI] [PubMed] [Google Scholar]
- 24.Otvos, L., I. O, M. Rogers, P. Consolvo, B. Condie, S. Lovas, P. Bulet, and M. Blaszczyk-Thurin. 2000. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 39:14150-14159. [DOI] [PubMed] [Google Scholar]
- 25.Park, C. B., H. S. Kim, and S. C. Kim. 1998. Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 244:253-257. [DOI] [PubMed] [Google Scholar]
- 26.Patrzykat, A., L. Zhang, V. Mendoza, G. K. Iwama, and R. E. W. Hancock. 2001. Synergy of histone-derived peptides of coho salmon with lysozyme and flounder pleurocidin. Antimicrob. Agents Chemother. 45:1337-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellegrini, A., U. Thomas, P. Wild, E. Schraner, and R. von Fellenberg. 2000. Effect of lysozyme or modified lysozyme fragments on DNA and RNA synthesis and membrane permeability of Escherichia coli. Microbiol. Res. 155:69-77. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Paya, E., R. A. Houghten, and S. E. Blondelle. 1995. The role of amphipathicity in the folding, self-association and biological activity of multiple subunit small proteins. J. Biol. Chem. 270:1048-1056. [DOI] [PubMed] [Google Scholar]
- 29.Shai, Y. 1995. Molecular recognition between membrane-spanning polypeptides. Trends Biochem. Sci. 20:460-464. [DOI] [PubMed] [Google Scholar]
- 30.Sims, P. J., A. S. Waggoner, C. H. Wang, and J. F. Hoffman. 1974. Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. Biochemistry 13:3315-3330. [DOI] [PubMed] [Google Scholar]
- 31.Subbalakshmi, C., and N. Sitaram. 1998. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 160:91-96. [DOI] [PubMed] [Google Scholar]
- 32.Wu, M., E. Maier, R. Benz, and R. E. W. Hancock. 1999. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38:7235-7242. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida, K., Y. Mukai, T. Niidome, C. Takashi, Y. Tokunaga, T. Hatakeyama, and H. Aoyagi. 2001. Interaction of pleurocidin and its analogs with phospholipid membrane and their antibacterial activity. J. Pept. Res. 57:119-126. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, L., A. Rozek, and R. E. W. Hancock. 2001. Interaction of cationic antimicrobial peptides with model membranes. J. Biol. Chem. 276:35714-35722. [DOI] [PubMed] [Google Scholar]