Abstract
Slaughterhouse pig samples were analyzed by PCR for Enterocytozoon bieneusi infection. Thirty-two percent were found to be positive, with rates being higher over the summer months. Three isolates from pigs were identical in their ribosomal internal transcribed spacer sequence to human E. bieneusi type D, two were identical to type F (from a pig), and nine were previously unreported. The viability of these spores was demonstrated by their ability to infect gnotobiotic piglets. The presence of the infection in liver was shown by in situ hybridization.
Microsporidia, a phylum representing over 1,000 species in 100 genera, are intracellular, obligate, protozoan parasites that infect every major animal group (6). Six of these genera, including at least 12 species, cause disease in humans and in animals (6). Emerging with the AIDS epidemic, Enterocytozoon bieneusi is the most prevalent human-pathogenic species. E. bieneusi causes chronic diarrhea and wasting and is responsible for 30 to 50% of all cases of diarrhea in people with AIDS (9). Little is known about the mode of transmission, zoonotic potential, and possible reservoir of E. bieneusi. Some evidence suggests that transmission occurs via contaminated water (7, 12, 17). A number of possible nonhuman reservoirs of E. bieneusi exist. E. bieneusi infects pigs, monkeys, cattle, dogs, cats, llamas, and rabbits (1, 4, 5, 10, 13, 16). In our laboratory, E. bieneusi from infected humans has been transmitted experimentally to macaques (18) and to pigs (8). These observations reflect indirectly the zoonotic nature of E. bieneusi and indicate that cross-species transmission is a real possibility.
The molecular characterization of the 243-bp internal transcribed spacer (ITS) region of the E. bieneusi rRNA reveals differences among E. bieneusi strains in relation to the host type. Currently, there are 19 different E. bieneusi ITS types from humans, pigs, cattle, and cats (1, 5, 10, 13, 14, 15, 16). To date, the only strains found in more than one host type is type D, found in humans and macaques, and types K and IV, which are of cat and human origins. No other crossover between hosts has been observed. This study was undertaken to determine the prevalence of E. bieneusi in swine shipped to a slaughterhouse in Massachusetts and to determine whether they are a potential source of human infection and of experimental material.
Fecal, bile (11), gallbladder, and liver tissue samples were collected monthly at a local slaughterhouse over 18 months. The bulk of the animals sampled, 176, were from the area around New Holland, Pa. The rest were from Massachusetts (four pigs), New Hampshire (eight pigs), and Vermont (14 pigs). All pigs appeared clinically healthy.
DNA was extracted from pig feces or bile for PCR detection of E. bieneusi as described by Kondova et al. (8) with the following modifications. Bile was washed three times by serial centrifugation at 4,000 × g for 20 min and resuspension in phosphate-buffered saline (PBS). The final resuspension was in 1/10 of the original volume of PBS. Feces were used directly. Approximately 200 μl of washed bile or feces was transferred to a 2-ml screw-cap conical tube containing 200 μl of 0.5-mm-diameter glass beads (Biospec Products, Inc.), 400 μl of digestion buffer (100 mM NaCl, 25 mM EDTA, 10 mM Tris-Cl [pH 8.0], 1% sodium dodecyl sulfate), and 600 μl of Tris-buffered phenol-chloroform-isoamyl alcohol (pH 8.0, 25:24:1). The sample was then placed in a mini-bead beater (Biospec Products, Inc.) at 5,000 rpm for 2 min. Samples were spun in a microcentrifuge for 2 min at top speed, and the supernatant was transferred to a new tube. Next, 300 μl of supernatant was adjusted to a 0.7 M concentration with 5 M NaCl, and 40 μl of CTAB mix (10% cetyltrimethylammonium bromide, 0.7 M NaCl) was added. The sample was incubated for 10 min at 65°C. After incubation, the solution was extracted with an equal volume of chloroform and the DNA was recovered from the resulting supernatant using the Geneclean system (Bio 101) by following the manufacturer's protocol for liquid samples, except that the DNA was resuspended in 20 μl of TE (10 mM Tris, 1 mM EDTA [pH 8.0]).
PCR amplification was performed on 1 μl of the above-described preparation with two different sets of nested primers. One set used the EBIER1 and EBIEF1 primers as described by da Silva et al. (3) as the outer primer pair and EBIER6 (5′-GCGACACTCTTAGACGTATC) and EBIEF5 (5′-TGTCCTTCCGTCAATTTC) as the inner primer pair. Cycling parameters for the first PCR were 35 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 40 s. One microliter of the first reaction mixture was used as the template for the nested reaction with cycling parameters of 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 40 s. A 350-bp internal control amplified by EBIER1 and EBIEF1 was included in each reaction mixture to test for false-negative results due to inhibition of PCR. Positive reactions were confirmed by using a second set of nested primers that amplify the ITS region. These primers were used so that a PCR product could be obtained for sequence analysis of the ITS. The outer primers were EBITS3 (5′-GGTCATAGGGATGAAGAG) and EBITS4 (5′-TTCGAGTTCTTTCGCGCTC), and the cycling parameters were 35 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 40 s; the inner primers were EBITS1 (5′-GCTCTGAATATCTATGGCT) and EBITS2.4 (5′-ATCGCCGACGGATCCAAGTG), and the cycling parameters were 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 40 s. These reactions produced fragments of 435 and 390 bp, respectively. The identities of the PCR products were further confirmed by digestion with MspA1I restriction enzyme (8).
Two hundred two pigs were sampled during the 18-month period. Thirty-two percent were positive for E. bieneusi. The feces of 18% were positive, and the bile specimens of 17% were positive. For 3%, both the bile and feces specimens were positive. Table 1 shows the percentages of positive samples per collection date. Surprisingly, the percentage of infected animals was lower (16%) for pigs weighing less than 10 kg and greater (37%) for pigs weighing more than 10 kg. These results are in contrast to results of another report (1), where weaned piglets and feeder pigs had infection rates of 90 and 58%, respectively.
TABLE 1.
Prevalence of E. bieneusi spores in swine feces and/or bile specimensa
| Sample date (mo/yr) | No. of pigs | No. (%) of:
|
|||
|---|---|---|---|---|---|
| Positive pigs | Pigs with positive bile specimens | Pigs with positive fecal specimens | Pigs with positive bile and fecal specimens | ||
| 11/98 | 11 | 1 (9) | 0 | 1 (9) | 0 (0) |
| 1/99 | 11 | 1 (9) | 0 | 1 (9) | 0 (0) |
| 3/99 | 14 | 2 (14) | 0 | 2 (14) | 0 (0) |
| 4/99 | 29 | 5 (17) | 1 (3) | 4 (14) | 0 (0) |
| 5/99 | 11 | 9 (82) | 5 (45) | 5 (45) | 1 (9) |
| 6/99 | 16 | 8 (50) | 3 (19) | 5 (31) | 0 (0) |
| 8/99 | 13 | 7 (54) | 5 (38) | 2 (15) | 0 (0) |
| 9/99 | 11 | 5 (45) | 3 (27) | 3 (27) | 1 (9) |
| 11/99 | 15 | 5 (33) | 3 (20) | 3 (20) | 1 (7) |
| 12/99 | 17 | 4 (24) | 2 (12) | 2 (12) | 0 (0) |
| 02/00 | 26 | 6 (23) | 3 (11) | 3 (11) | 0 (0) |
| 3/00 | 12 | 4 (33) | 3 (25) | 1 (8) | 0 (0) |
| 6/00 | 16 | 7 (44) | 6 (37) | 4 (25) | 3 (19) |
| Total | 202 | 64 (32) | 34 (17) | 36 (18) | 6 (3) |
Specimens were collected over 18 months at a slaughterhouse in the northeastern United States. The number of positive samples and the corresponding percentage are shown for each sampling date and type of sample.
A recent study of 109 healthy pigs in Switzerland by Breitenmoser et al. (1) also using PCR techniques found a similar percentage of E. bieneusi-positive pigs (35%); however, only fecal samples were tested. The differences in prevalence may reflect real geographic or seasonal variation or differences in the sensitivities of the techniques used. Our data indicate that there is a trend toward higher prevalence in the warmer spring and summer months (Table 1); however, due to the short duration of the study, no statistical conclusions can be drawn. The time of year in the Swiss study was not indicated. The contribution, if any, of E. bieneusi to disease in pigs remains to be determined. The clinical outcome of the infection in the slaughtered pigs was not investigated in this study. Experimental infections have so far indicated that in the short term (2 to 3 weeks [8]), there is no evidence of diarrhea. However, Rinder et al. (16) found that four of six pigs with diarrhea excreted E. bieneusi.
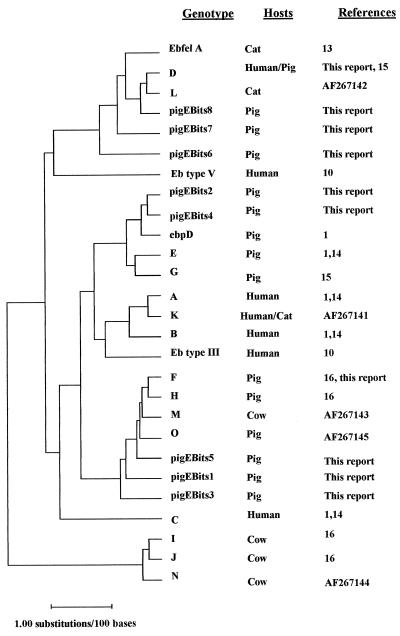
EBITS1 and -2.4 PCR products from 17 of the E. bieneusi-positive samples were randomly selected for sequencing to ensure the specificity of the PCR assay. Sequencing was performed on the products from two separate PCRs, which were sequenced in both directions. Sequences were edited and assembled by using the consensus sequence of these four sequences. Homology to published E. bieneusi sequences was 100% for the small- and large-subunit RNA portions of each PCR fragment flanking the ITS. ITS sequence homologies ranged from 97 to 100% of published E. bieneusi ITS sequences. Table 2 shows polymorphic sites revealed by comparison of previously reported ITS sequences with those resulting from this study. Our analysis revealed isolates from three different pigs that were identical to E. bieneusi ITS type D (15), originally isolated from humans. Two isolates were homologous to type F (16), originally isolated from pig, and nine previously unreported types with seven new polymorphic nucleotides were discovered. These new isolates have been designated PigEBITS1 to -9. Isolates from positive bile or feces samples from the same pig were identical. The relatedness of the ITS types from different animal origins are shown in the dendrogram in Fig. 1.
TABLE 2.
Nucleotides that vary in the rRNA ITSa
| Source | Genotype | Nucleotide at positiona:
|
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 31 | 34 | 35 | 76 | 77 | 80 | 81 | 88 | 93 | 95 | 97 | 103 | 113 | 117 | 118 | 124 | 129 | 130 | 131 | 134 | 136 | 137 | 141 | 143 | 145 | 147 | 148 | 149 | 157 | 158 | 178 | 190 | 193 | 196 | ||
| Consensus | G | A | G | T | C | G | G | C | G | T | G | C | T | T | G | G | G | G | G | G | C | G | C | T | A | A | G | T | G | G | T | G | T | G | A | |
| Human | A | - | G | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | - | - |
| Human | B | - | - | - | - | - | A | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | - | - |
| Human | C | T | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | A | - | - | - | - | - | - | - | - | - | - | - | T | - | - | A | - | - | - |
| Human and pig | D | - | G | - | - | - | - | - | - | - | C | - | - | - | C | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Pig | E | - | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - |
| Pig | F | - | - | - | - | - | - | - | T | - | - | T | - | - | - | - | - | - | - | - | - | - | A | - | - | G | - | - | - | - | - | - | - | - | - | G |
| Pig | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | G |
| Pig | H | - | G | - | - | - | - | - | T | - | - | T | - | - | - | - | - | - | - | - | - | - | A | - | - | G | - | - | - | - | - | - | - | - | - | G |
| Cow | I | - | G | A | - | - | - | - | T | - | - | - | - | - | - | - | - | - | A | - | C | - | - | T | - | - | - | A | - | - | - | A | - | - | - | - |
| Cow | J | - | G | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | A | - | C | - | - | T | - | - | - | A | - | - | - | A | - | - | - | - |
| Cat and human | K | - | G | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Cat | L | - | G | - | - | - | - | - | - | - | A | - | - | - | C | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Cow | M | - | - | - | - | - | - | - | T | - | - | T | - | - | - | - | - | - | - | - | - | - | A | T | - | G | - | - | - | - | - | - | - | - | - | G |
| Cow | N | - | G | - | - | - | - | - | T | - | - | - | T | - | - | - | - | - | A | - | C | - | - | T | - | - | - | A | - | - | - | A | - | - | - | - |
| Pig | O | - | - | - | - | - | - | - | T | - | - | T | - | - | - | - | A | - | - | - | - | - | A | C | - | G | - | - | - | - | - | - | - | - | - | G |
| Llama | P | - | G | A | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Human | Type III | - | G | - | - | - | - | - | - | - | - | G | - | - | - | - | - | - | - | - | - | T | - | - | T | - | - | - | - | - | - | - | - | - | - | - |
| Human | Type V | T | - | - | - | - | - | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | - | - |
| Cat | EbfelA | - | G | - | - | - | - | - | - | - | C | - | - | - | C | T | A | - | - | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Pig | EbpD | - | - | - | - | - | - | - | T | - | - | T | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - |
| Pig | EbpB | - | - | - | - | T | - | - | T | - | - | T | - | - | - | - | - | - | - | - | - | T | A | - | - | - | - | - | - | - | - | - | - | - | - | G |
| Pig | Pig EBITS9 | - | - | - | - | - | - | - | T | - | - | T | - | - | - | - | - | - | - | - | - | - | A | - | - | G | - | - | C | - | - | - | - | - | - | G |
| Pig | PigEBITS1 | - | G | - | - | - | - | - | T | A | - | T | - | - | - | - | - | - | - | - | - | - | A | - | - | G | - | - | - | - | - | - | - | - | - | G |
| Pig | PigEBITS2 | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | A | - | C | - | - | - | - | - | - | - | - | - | - | - |
| Pig | PigEBITS3 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | G | - | - | - | - | - | - | - | - | - | G |
| Pig | PigEBITS4 | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - |
| Pig | PigEBITS5 | - | - | - | - | - | - | - | T | - | - | T | - | - | - | - | - | - | - | - | - | - | - | - | - | G | - | - | - | - | - | - | - | - | - | - |
| Pig | PigEBITS8 | - | G | - | - | - | - | - | - | - | C | - | - | - | C | T | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - |
| Pig | PigEBITS6 | - | G | - | - | - | - | - | - | - | C | - | - | T | C | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | A | - |
| Pig | PigEBITS7 | - | - | - | C | - | - | - | - | - | C | - | - | - | C | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
Nucleotides found at 35 polymorphic sites in the E. bieneusi small-subunit rRNA ITSs of 30 representative sequences. A to P, type III, type V, EdpD, EbfelA, and EdpB are genotypes presently in GenBank. PigEBITS1 to -9 are new ITS sequence types discovered during this work in positive pig samples from a slaughterhouse. The type(s) of organism from which each sequence was derived is displayed. The accession numbers for the previously sequenced E. bieneusi types are as follows: AF10197 to AF101200 for types A to D, AF135832 to AF135837 for types E to J, AF267141 to AF267146 for types K to P, AF118144 for type EbfelA, AF076043 for type EbpD, and AF06041 for type EbpB.
FIG. 1.
Dendrogram of E. bieneusi genotypes. The plot was constructed by the Jukes-Cantor correction method, modified to consider gaps. Distances are estimated numbers of substitutions per 100 bases, as represented by the bar. Genotypes, hosts, and references are shown. All accession numbers are as given in footnote a of Table 2.
Table 2 shows that type D has many polymorphisms in common with isolates of pig origin. Figure 1 indicates that type D is more closely related to a number of pig isolates and even to the cat isolate than it is to the other human isolates. This may indicate that type D is more likely to be zoonotic.
As is evident from our work and that of others, there are already 30 different types of E. bieneusi ITS sequences with 35 polymorphic sites. It is conceivable that if enough isolates are sequenced, the number of types will rise dramatically and all 243 nucleotides in the ITS could be found to be polymorphic. Currently, genotypes that are present at low levels in samples can be missed by use of PCR-based detection, since the PCR may preferentially amplify the major genotypes. If this is true, the current method of letter designations for types will have to be revised. This trend in the data demonstrates the need to have other genetic markers for use in determining types. The reliance on one locus for a marker, particularly one as unstable as the ITS, is insufficient. However, sequencing more isolates from different sources may also reveal polymorphisms that can be closely correlated with particular locations or host types. Some trends are already apparent, as seen in Table 2. The A at ITS nucleotide 136 and the G at nucleotide 196 are found primarily in isolates of pig origin, though not in all pig isolates. Also, three of four isolates of bovine origin have four distinctive polymorphic sites (sites 129, 131, 147, and 158). These types of polymorphism, if found in restriction sites, could be used for restriction fragment length polymorphism assays to broadly screen isolates without the need to sequence them all, as was done by Liguory et al. (10) in screening human isolates.
Interestingly, some ITS types are geographically widespread. Type D and type F ITS patterns have now been detected in Germany, Switzerland, the United States, and Uganda (1, 14, 16; unpublished data). Type A has been shown in Germany, Switzerland, and Peru (1, 14; our unpublished data). This may mean either that they are highly conserved or that there is enough movement of organisms to maintain genetic homogeneity in other regions.
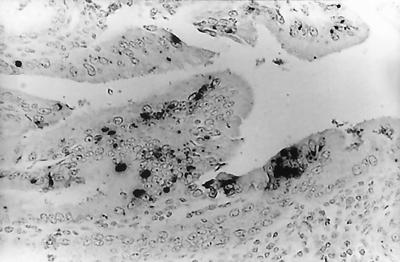
No antibody specific for E. bieneusi is available, so immunohistochemistry could not be used to detect parasites in tissue. In situ hybridization of formalin-fixed, paraffin-embedded pig liver and gallbladder sections, as previously described by members of our laboratory (2, 8) and others (19), was used to confirm the presence of E. bieneusi in tissue. The liver of only one pig was shown to be positive for E. bieneusi (Fig. 2). As previously shown with humans, macaques, and experimentally infected piglets, this result demonstrates that E. bieneusi infection can spread, although rarely, from the intestinal tract, even in supposedly immunologically normal animals.
FIG. 2.
In situ hybridization of liver tissue from a slaughterhouse pig bile sample positive for E. bieneusi. Black granular staining shows a positive hybridization.
The viability of detected spores was assessed by using the gnotobiotic piglet model developed by our laboratory (8). E. bieneusi spores concentrated from the bile specimens of several positive animals and washed by centrifugation at 3 × g and resuspension in PBS (final volume, 2 ml) were used to inoculate four piglets within 24 h after delivery. Due to the difficulty of obtaining and counting spores, the number used for inoculation is not known. Together with two control animals, the piglets were monitored daily for shedding of spores in feces. All piglets were immunosuppressed during the experiment by a daily oral cyclosporine dose of 15 mg/kg of body weight and by intramuscular administration of 25 mg of methylprednisolone sodium per kg. All piglets inoculated with the spores from the bile of a pig naturally infected with E. bieneusi became infected between days 6 and 12 after challenge, as determined by PCR and modified trichrome staining. They remained infected for the 3-week duration of the experiment, with no development of diarrhea. Control animals remained uninfected.
We report that a high percentage (30.5%) of apparently healthy pigs shipped to a slaughterhouse in the northeastern United States were infected with E. bieneusi. The occurrence of ITS types that are found in humans demonstrates the zoonotic potential for E. bieneusi from swine. The viability of the spores, the high prevalence of infection, and the large number of swine in the country indicate that swine may be a significant source of environmental contamination.
Nucleotide sequence accession numbers.
The GenBank accession numbers for PigEBITS1 to -9 are AF348469 to AF348477, respectively.
Acknowledgments
This work was supported by two NIH grants, R01-AI43196 and R01-AI41889.
We thank Julia Dilo for her technical assistance on this project (she performed a large portion of the DNA extractions and PCRs) and the owners and workers at the Adams Farm Slaughterhouse for their cooperation in obtaining samples for this project.
REFERENCES
- 1.Breitenmoser, A. C., A. Mathis, E. Burgi, R. Weber, and P. Deplazes. 1999. High prevalence of Enterocytozoon bieneusi in swine with four genotypes that differ from those identified in humans. Parasitology 118:447-453. [DOI] [PubMed] [Google Scholar]
- 2.Carville, A., K. Mansfield, G. Widmer, A. Lackner, D. Kotler, P. Wiest, T. Gumbo, S. Sarbah, and S. Tzipori. 1997. Development and application of genetic probes for detection of Enterocytozoon bieneusi in formalin-fixed stools and in intestinal biopsy specimens from infected patients. Clin. Diagn. Lab. Immunol. 4:405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.da Silva, A. J., D. A. Schwartz, G. S. Visvesvara, H. de Moura, S. B. Slemenda, and N. J. Pieniazek. 1996. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small-subunit rRNA. J. Clin. Microbiol. 34:986-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.del Aguila, C., F. Izquierdo, R. Navajas, N. J. Pieniazek, G. Miro, A. I. Alonso, A. J. Silva, and S. Fenoy. 1999. Enterocytozoon bieneusi in animals: rabbits and dogs as new hosts. J. Eukaryot. Microbiol. 46:8S-9S. [PubMed] [Google Scholar]
- 5.Deplazes, P., A. Mathis, C. Muller, and R. Weber. 1996. Molecular epidemiology of Encephalitozoon cuniculi and first detection of Enterocytozoon bieneusi in faecal samples of pigs. J. Eukaryot. Microbiol. 43:93S. [DOI] [PubMed] [Google Scholar]
- 6.Didier, E. S., K. F. Snowden, and J. A. Shadduck. 1998. Biology of microsporidian species infecting mammals. Adv. Parasitol. 40:283-320. [DOI] [PubMed] [Google Scholar]
- 7.Dowd, S. E., C. P. Gerba, and I. L. Pepper. 1998. Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl. Environ. Microbiol. 64:3332-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondova, I., K. Mansfield, M. A. Buckholt, B. Stein, G. Widmer, A. Carville, A. Lackner, and S. Tzipori. 1998. Transmission and serial propagation of Enterocytozoon bieneusi from humans and rhesus macaques in gnotobiotic piglets. Infect. Immun. 66:5515-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotler, D. P., and J. M. Orenstein. 1998. Clinical syndromes associated with microsporidiosis. Adv. Parasitol. 40:321-349. [DOI] [PubMed] [Google Scholar]
- 10.Liguory, O., F. David, C. Sarfati, F. Derouin, and J.-M. Molina. 1998. Determination of types of Enterocytozoon bieneusi strains isolated from patients with intestinal microsporidiosis. J. Clin. Microbiol. 36:1882-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansfield, K. G., A. Carville, D. Hebert, L. Chalifoux, D. Shvetz, K. C. Lin, S. Tzipori, and A. Lackner. 1998. Localization of persistent Enterocytozoon bieneusi infection in normal rhesus macaques (Macaca Mulatta) to the hepatobiliary tree. J. Clin. Microbiol. 36:2336-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall, M. M., D. Naumovitz, Y. Ortega, and C. R. Sterling. 1997. Waterborne protozoan pathogens. Clin. Microbiol. Rev. 10:67-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathis, A., A. C. Breitenmoser, and P. Deplazes. 1999. Detection of new Enterocytozoon genotypes in faecal samples of farm dogs and a cat. Parasite 6:189-193. [DOI] [PubMed] [Google Scholar]
- 14.Rinder, H., S. Katzwinkel-Wladarsch, and T. Loscher. 1997. Evidence for the existence of genetically distinct strains of Enterocytozoon bieneusi. Parasitol. Res. 83:670-672. [DOI] [PubMed] [Google Scholar]
- 15.Rinder, H., S. Katzwinkel-Waldarsch, A. Thomschke, and T. Loscher. 1999. Strain differentiation in microsporidia. Tokai J. Exp. Clin. Med. 23:433-437. [PubMed] [Google Scholar]
- 16.Rinder, H., A. Thomschke, B. Dengjel, R. Gothe, T. Loscher, and M. Zahler. 2000. Close genotype relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J. Parasitol. 86:185-188. [DOI] [PubMed] [Google Scholar]
- 17.Sparfel, J. M., et al. 1997. Detection of microsporidia and identification of Enterocytozoon bieneusi in surface water by filtration followed by specific PCR. J. Eukaryot. Microbiol. 44:78S. [DOI] [PubMed] [Google Scholar]
- 18.Tzipori, S., A. Carville, G. Widmer, D. Kotler, K. G. Mansfield, and A. A. Lackner. 1997. Transmission and establishment of a persistent infection of Enterocytozoon bieneusi, derived from a human with AIDS, in simian immunodeficiency virus-infected rhesus monkeys. J. Infect. Dis. 175:1016-1020. [DOI] [PubMed] [Google Scholar]
- 19.Velasquez, J. N., S. Carnevale, J. H. Labbe, A. Chertcoff, M. G. Cabrera, and W. Oelemann. 1999. In situ hybridization: a molecular approach for the diagnosis of the microsporidian parasite Enterocytozoon bieneusi. Hum. Pathol. 30:54-58. [DOI] [PubMed] [Google Scholar]