Abstract
The term endophyte refers to interior colonization of plants by microorganisms that do not have pathogenic effects on their hosts, and various endophytes have been found to play important roles in plant vitality. In this study, cultivation-independent terminal restriction fragment length polymorphism analysis of 16S ribosomal DNA directly amplified from plant tissue DNA was used in combination with molecular characterization of isolates to examine the influence of plant stress, achieved by infection with the blackleg pathogen Erwinia carotovora subsp. atroseptica, on the endophytic population in two different potato varieties. Community analysis clearly demonstrated increased bacterial diversity in infected plants compared to that in control plants. The results also indicated that the pathogen stress had a greater impact on the bacteria population than the plant genotype had. Partial sequencing of the 16S rRNA genes of isolated endophytes revealed a broad phylogenetic spectrum of bacteria, including members of the α, β, and γ subgroups of the Proteobacteria, high- and low-G+C-content gram-positive organisms, and microbes belonging to the Flexibacter-Cytophaga-Bacteroides group. Screening of the isolates for antagonistic activity against E. carotovora subsp. atroseptica revealed that 38% of the endophytes protected tissue culture plants from blackleg disease.
Blackleg of potatoes caused by Erwinia carotovora subsp. atroseptica is a severe field disease leading to the development of an inky black and slimy soft rot of stems. Severely affected plants die, and tubers from diseased plants may show a black soft rot during storage (29). The pathogen is carried within diseased potato tubers or other plant debris, but it is usually dormant and does not cause disease symptoms unless environmental conditions are favorable (29).
The term endophyte refers to interior colonization of plants by bacterial or fungal microorganisms. Endophytic bacteria colonize an ecological niche similar to that colonized by plant pathogens but do not cause damage to their hosts. Several bacterial endophytes have been reported to support growth and improve the health of plants (13, 14, 26, 33, 35) and therefore may be important sources of biocontrol agents. Erwinia carotovora, for example, is inhibited by numerous endophytic bacteria, including several strains of Pseudomonas sp. (16), Curtobacterium luteum, and Pantoea agglomerans (37). Furthermore, Wilhelm et al. (43) demonstrated that Bacillus subtilis strains isolated from the xylem sap of healthy chestnut trees exhibit antifungal effects against Cryphonectria parasitica causing chestnut blight.
The proposed mechanisms by which bacteria control phytopathogens are as varied as the bioactive microorganisms. Siderophores, which chelate iron or other metals, contribute to disease suppression by conferring a competitive advantage to biocontrol strains (17). Furthermore, the production of antimicrobial substances, such as antibiotics or HCN, is an important mechanism to fight phytopathogens (4). Indirect disease control is achieved by mechanisms modulating the plant immune response, including the induction of systemic acquired resistance (42).
Identification of endophytes has relied mainly upon cultivation-based methods (3, 36). However, due to the unknown growth requirements of many microbes and the presence of cells that are in a viable but not culturable state (39), cultivation-independent population analysis leads to a less biased determination of microbial diversity. Molecular techniques based on the 16S rRNA gene as a phylogenetic marker (2) provide a powerful approach to circumvent drawbacks related to cultivation. Techniques such as terminal restriction fragment length polymorphism (T-RFLP) analysis (19, 30) or denaturing gradient gel electrophoresis (24, 34) in combination with sequence analysis of 16S rRNA genes allow rapid characterization of microbial communities.
Recently, Garbeva et al. (12) monitored endophytic populations of potato by PCR-denaturing gradient gel electrophoresis, which revealed the occurrence of a range of organisms falling into several distinct phylogenetic groups. Their results also suggested the presence of nonculturable endophytes in potato. In previous experiments we found by T-RFLP analysis, as well as by sequencing of partial 16S rRNA genes, a broad phylogenetic spectrum of bacteria that are able to colonize potato plants endophytically, including members of the α subgroup of the Proteobacteria (α-Proteobacteria), the β-Proteobacteria, the γ-Proteobacteria, the high-G+C-content gram-positive bacteria, the Flexibacter-Cytophaga-Bacteroides group, and the Planctomycetales. Most of the bacterial endophytes originated from the corresponding rhizospheres, although some bacteria were exclusively detected as endophytes (31).
The objective of the present study was to analyze the response of endophytic populations in potato to the presence of a pathogen, E. carotovora subsp. atroseptica. For community profiling of endophytes of pathogen-infected and control plants a cultivation-independent approach was chosen, in which 16S rRNA genes were PCR amplified and subjected to T-RFLP analysis. In addition, endophytes were isolated, identified by 16S rRNA gene analysis, and tested for the ability to protect tissue culture plants from blackleg disease.
MATERIALS AND METHODS
Greenhouse experiment.
Two potato varieties, Agria and Bionta, were used for the analysis of bacterial endophytes in healthy and diseased plants. Potato tubers were planted in pots filled with standard growth substrate (TKS2 soil substrate; Knauf Perlite GmbH) containing 200 to 500 mg of N per liter, 200 to 500 mg of potassium oxide per liter, and 300 to 600 mg of phosphorpentoxide per liter. After 2 weeks plants were wounded with a sterile scalpel at the stem base and infected with 2 × 106 cells of a mid-exponential-phase culture of E. carotovora subsp. atroseptica SCRI 1039 I (= DSM 30184). Control plants were not wounded and not infected with E. carotovora subsp. atroseptica. Each plant genotype and treatment was replicated three times. Six weeks after planting lower stem (approximately 2 to 5 cm above the ground) and upper stem (approximately 20 to 25 cm above the ground) sections were harvested for subsequent DNA isolation.
DNA isolation.
In order to avoid isolation of surface bacterial DNA, stems were soaked in 5% bleach for 10 min, rinsed four times with sterile, distilled water, rinsed with ethanol, and finally flamed. Subsequently, the stems were aseptically peeled. DNA isolation from stem tissue was performed as described by Sessitsch et al. (31).
T-RFLP analysis.
Partial 16S rRNA gene sequences were amplified with a thermocycler (PTC-100; MJ Research, Inc.) by using an initial denaturing step of 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 1 min of annealing at 54°C, and 2 min of extension at 72°C. PCR mixtures (50 μl) contained 0.5 μl of extracted DNA, 1× reaction buffer (Gibco BRL), 200 μM (each) dATP, dCTP, dGTP, and dTTP, 2 mM MgCl2, 2.5 U of Taq DNA polymerase (Gibco BRL), 0.2 μM primer 8f (5′-AGAGTTTGATCCTGGCTCAG-3′) (11) labeled with 6-carboxyfluorescein (MWG) at the 5′ end, and 0.2 μM primer 926r (5′-CCGTCAATTCCTTT[AG]AGTTT-3′) (19). Three independent PCR mixtures for each sample were combined and used for subsequent T-RFLP analysis. Approximately 200 ng of fluorescently labeled PCR amplification products was digested with the restriction enzymes AluI (Gibco BRL), RsaI (Gibco BRL), and TaqI (Amersham, Slough, England) individually, as well as with a combination of HhaI and HaeIII (Gibco BRL). Aliquots (0.75 μl) were mixed with 1 μl of loading dye buffer (diluted 5:1 in deionized formamide [Flucka]) and 0.3 μl of a DNA fragment length standard (Rox 500; PE Applied Biosystems Inc., Foster City, Calif.). The mixtures were denatured for 2 min at 92°C and immediately chilled on ice prior to electrophoretic separation on 5% polyacrylamide gels. Fluorescently labeled terminal restriction fragments (T-RFs) were detected with an ABI 373A automated DNA sequencer (PE Applied Biosystems Inc.) operated in the GeneScan mode. Lengths of labeled fragments were determined by comparison with the internal standard using the GeneScan 2.5 software package (PE Applied Biosystems Inc.).
Analysis of T-RF profiles.
T-RFs between 35 and 500 bp long with heights of ≥50 fluorescence units were included in the analysis. Because of uncertainties of size determination with our automated DNA sequencer, T-RFs that differed by less than 1.5 bp were clustered. For all treatments three individual plants and three replicate T-RFLP profiles for each plant were analyzed individually, and representative sample profiles were determined as suggested by Dunbar et al. (10). Essentially, the sum of peak heights in each replicated profile was calculated, which indicated the total DNA quantity, and the peak intensity was adjusted to the smallest DNA quantity within a treatment. Representative sample profiles were composed of mean values of individual peak heights. In addition, in representative sample profiles only peaks showing mean values of ≥50 fluorescence units were included. Furthermore, T-RFs that were present in only one individual plant were not included in the analysis.
As a parameter for the structural diversity of the microbial community, the Shannon-Weaver index (32) (H) was calculated for representative T-RFLP profiles by using the following function: H = −ΣPilnPi., where Pi is the relative intensity of a peak in a profile. The relative intensity was calculated as follows: Pi = ni/N, where ni is the height of a peak and N is the sum of all peak heights in a T-RFLP profile. Statistical analysis was performed by using the Statistica software.
In order to determine similarities between T-RFLP profiles, a binary matrix recording the absence or presence of T-RFs was established. Cluster analysis was performed based on similarities calculated as described by Nei and Li (25) by using the unweighted pair group with mathematical average method. Tree generation was performed by using the TREECON software package (41) with 100 bootstrap replications.
Isolation and PCR-RFLP analysis of endophytic bacteria.
For isolation of endophytic bacteria, stem sections were surface sterilized as described above and subsequently aseptically peeled. Stems were tested for sterility on tryptic soy agar plates incubated for 2 days at 30°C. No growth was observed.
After addition of 3 ml of tryptic soy broth (Merck, Darmstadt, Germany) to 0.2 to 0.5 g of plant material, the tissue was macerated, and 50-, 100-, and 200-μl aliquots were plated on 10% tryptic soy agar. The plates were incubated for 24 h at 28°C. Colonies on each plate that could be distinguished based on colony morphology were picked, resulting in a total of 67 isolates which were analyzed further.
For isolation of genomic DNA, bacteria were grown overnight in 5 ml of tryptic soy broth in a rotatory shaker at 28°C. Cells were harvested by centrifugation for 10 min at 5,000 × g at 4°C. After the supernatant was decanted, 300 mg of glass beads was added, and DNA was isolated as described above.
Restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA gene was used to group isolates at the species level, whereas characterization of the 16S-23S rRNA intergenic spacer (IGS) region was used to distinguish different strains of the same species. PCR was carried out as described above by using 1 μl of extracted DNA and primers 8f (5′-AGAGTTTGATCCTGGCTCAG-3′) (11) and pH (5′-AAGGAGGTGATCCAGCCGCA-3′) (11). Primers pHr (5′-TGCGGCTGGATCACCTCCTT-3′) (21) and P23SR01 (5′-GGCTGCTTCTAAGCCAAC-3′) (21) were used for amplification of the 16S-23S rRNA IGS. Aliquots of a PCR product containing 200 ng of amplified DNA were digested with 5 U of endonucleases HhaI (Gibco BRL) and AluI (Gibco BRL) individually for 3 h at 37°C. The resulting DNA fragments were analyzed by gel electrophoresis in 2.5% agarose gels.
Screening of isolated bacteria for biocontrol activity in vivo.
Isolated endophytes were tested for the ability to protect plants from infection with E. carotovora subsp. atroseptica by using the potato lines Achirana Inta, Merkur, and Agria or Bionta. Plantlets grown in tissue culture were dipped in bacterial endophyte cultures (108 cells per ml) and subsequently transplanted onto MS agar (23). After incubation for 1 week in a growth chamber at 22°C with a light period of 14 h per day, plantlets were dipped in a culture of E. carotovora subsp. atroseptica (108 cells per ml) and again transplanted. After 2 weeks of incubation in the growth chamber, plants were scored for disease symptoms. Control plants were treated with either a 0.9% sterile NaCl solution or the pathogen only.
Antibiotic production assay.
Production of antibiotics by the isolated endophytes against E. carotovora subsp. atroseptica was tested on a Brown & Dilworth minimal medium containing 1% sucrose (6), as well as on 10% tryptic soy agar. A basal layer of agar was covered with 5 ml of soft agar containing about 106 CFU of the phytopathogen. Then, 10 μl of a well-grown overnight culture of the endophyte to be tested was spotted on the solidified agar surface. Plates were incubated at 28°C and checked for halo formation after 24 and 48 h.
DNA sequencing and computer analysis.
For sequence identification of isolated endophytes, total 16S ribosomal DNA (rDNA) genes were PCR amplified by using primers 8f (11) and pH (11) and the conditions described above. PCR products were purified by using a NucleoTraPCR kit (Macheroy-Nagel) according to the manufacturer's instructions and were used as templates in sequencing reactions. Partial DNA sequencing was performed by using either 16S rRNA gene primer 8f (11), primer 518r (5′-ATTACCGCGGCTGCTGG-3′) (19), or occasionally uni 360s (5′-GGAATCTTCCACAATGGGCG-3′) (15), the dideoxy chain termination method (28), an ABI 373A automated DNA sequencer, and an ABI PRISM Big Dye terminator cycle sequencing kit (PE Applied Biosystems Inc.). Sequences were subjected to BLAST analysis (1) with the National Center for Biotechnology Information database.
Nucleotide sequence numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AF406653 to AF406691.
RESULTS
T-RFLP analysis.
In all T-RFLP profiles there were two T-RFs that were derived from chloroplast and mitochondrial small-subunit rRNA sequences. In addition, a high number of fragments of bacterial origin were identified. A combination of restriction endonucleases HaeIII and HhaI produced the highest number of T-RFs. Digestion with these enzymes resulted in a total of 37 peaks, which was reduced to 28 peaks by the normalization procedure. The number of T-RFs in representative sample profiles obtained with HaeIII and HhaI ranged from 9 (Bionta, upper stem, E. carotovora subsp. atroseptica negative) to 20 (Agria, upper stem, E. carotovora subsp. atroseptica positive) (Table 1). In general, plants infected with E. carotovora subsp. atroseptica showed a higher number of T-RFs than control plants (Table 1). Ten HaeIII/HhaI T-RFs were detected exclusively in plants infected with E. carotovora subsp. atroseptica. Seven of them (66, 125, 153, 200, 204, 206, and 318 bp) were present in both varieties. One T-RF (149 bp) was observed only in Agria stems, whereas two fragments (175 and 224 bp) were detected exclusively in Bionta plants. Two T-RFs (131 and 294 bp) were detected only in control plants. Both were specifically detected in cultivar Agria. In addition, the two stem sections examined showed slightly different bacterial population structures. Upper sections contained three T-RFs that were not found in lower sections. A 125-bp T-RF was present in both varieties, whereas two T-RFs (149 and 294 bp) were found only in Agria plants. Lower stem sections possessed five specific T-RFs. Two of them (191 and 298 bp) were found in both cultivars. Two T-RFs, at 175 and 200 bp, were specifically detected in Bionta plants, whereas a 131-bp fragment was found only in Agria plants.
TABLE 1.
T-RFLP fragments and Shannon-Weaver indices obtained after HaeIII/HhaI digestion of 16S rRNA PCR products of DNA isolated from stems of E. carotovora-infected and control plants
| T-RF (bp)a | Agria plantsb
|
Bionta plantsb
|
||||||
|---|---|---|---|---|---|---|---|---|
| Without E. carotovora
|
With E. carotovora
|
Without E. carotovora
|
With E. carotovora
|
|||||
| Upper stem sections | Lower stem sections | Upper stem sections | Lower stem sections | Upper stem sections | Lower stem sections | Upper stem sections | Lower stem sections | |
| 39c | + | + | + | + | + | + | + | + |
| 41c | + | + | + | + | + | |||
| 43 | + | + | + | + | + | + | ||
| 60c | + | + | + | + | + | + | + | |
| 66c | + | + | + | |||||
| 125 | + | + | ||||||
| 131 | + | |||||||
| 134 | + | + | + | + | + | + | + | + |
| 137 | + | + | + | + | + | + | + | + |
| 141c | + | + | + | + | + | + | + | + |
| 144c | + | + | + | + | + | + | + | |
| 147 | + | + | + | + | + | + | + | + |
| 149 | + | |||||||
| 153 | + | + | ||||||
| 175 | + | |||||||
| 191c | + | + | + | |||||
| 200c | + | |||||||
| 204c | + | + | + | |||||
| 206 | + | + | + | + | ||||
| 224c | + | |||||||
| 294 | + | |||||||
| 298 | + | + | + | + | ||||
| 310 | + | + | + | + | + | |||
| 313 | + | + | + | + | + | + | ||
| 315 | + | + | + | |||||
| 318 | + | + | ||||||
| 335 | + | + | + | + | ||||
| 337 | + | + | + | + | + | + | + | + |
T-RFs derived from mitochondrial 18S rDNA (197 bp) and chloroplast 16S rDNA (303 bp) were not included in the analysis.
The Shannon-Weaver diversity indices for the treatments were calculated by determining the mean values for three replicates. The Shannon-Weaver diversity indices for Agria plants were as follows: upper stem sections without E. carotovora, 1.90; lower stem sections without E. carotovora, 1.79; upper stem sections with E. carotovora, 2.49; and lower stem sections with E. carotovora, 2.26. The Shannon-Weaver diversity indices for Bionta plants were as follows: upper stem sections without E. carotovora, 1.58; lower stem sections without E. carotovora, 1.92; upper stem sections with E. carotovora, 2.46; and lower stem sections with E. carotovora, 2.30. Within groups, the following values were not significantly different (P < 0.01): 1.90, 1.79, and 1.92; 1.79 and 1.58; and 2.49, 2.26, 2.46, and 2.30.
T-RFs for which we found corresponding isolates.
To determine the structural diversity. we calculated Shannon-Weaver diversity indices from the T-RFLP community fingerprints (Table 1). Plants infected with E. carotovora subsp. atroseptica showed significantly higher diversity than healthy plants. Similar diversities were found in infected plants, with Shannon-Weaver indices ranging from 2.26 to 2.49, whereas in control plants the Shannon-Weaver indices were considerably lower, ranging from 1.58 to 1.79.
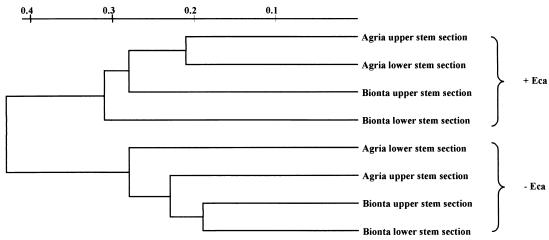
A cluster analysis based on T-RFLP profiles obtained with all four digests clearly demonstrated that plants infected with E. carotovora subsp. atroseptica had very different endophytic population structures than control plants (Fig. 1).
FIG. 1.
Unweighted pair group with mathematical average dendrogram generated from the 16S rDNA-based T-RFLP community profiles. The data obtained with four restriction digests (HaeIII plus HhaI, as well as AluI, RsaI, and TaqI individually) were combined. The scale indicates distance. +Eca, E. carotovora subsp. atroseptica positive; −Eca, E. carotovora subsp. atroseptica negative.
Isolation and sequence identification of bacterial endophytes.
The average number of bacteria cultured on 10% tryptic soy agar was 7 × 103 CFU per g (fresh weight) of potato stem. Based on colony morphology, we selected 34 colonies from Agria plants and 33 colonies from Bionta plants, which consisted of 17 and 20 16S rDNA PCR-RFLP types, respectively. Subsequent RFLP analysis of the 16S-23S rRNA IGS resulted in 20 and 25 IGS types for Agria and Bionta plants, respectively. From each of the 45 IGS types one isolate was chosen for further analysis. In control plants, only two (Agria) and nine (Bionta) morphologically and genetically different colonies were found. The remaining isolates were derived from pathogen-treated plants. No isolate that colonized plants subjected to both treatments was found.
Isolates were further characterized by partial sequencing of the 16S rRNA gene. Tables 2 and 3 summarize the results of the sequence analysis. One isolate, present in pathogen-infested plants of both lines (A8 and B11), was identified as E. carotovora subsp. atroseptica and was excluded from further analysis. Four additional isolates that showed the highest 16S rRNA gene sequence similarities to Agrobacterium tumefaciens, Pseudomonas fluorescens, Flavobacterium sp. strain B17, and Enterobacter cloacae were found to be common in both potato genotypes. The dominant eubacterial group among isolates obtained from Agria plants was the γ-Proteobacteria; seven IGS types fell into to this group. The remaining cultured endophytes were members of the α- and β-Proteobacteria (one and three IGS types, respectively), the Flexibacter-Cytophaga-Bacteroides group (two IGS types), the low-G+C-content gram-positive group (two IGS types), and the high-G+C-content gram-positive group (four IGS types). In contrast, isolates obtained from Bionta plants mainly belonged to the low-G+C-content gram-positive group (eight IGS types) and the γ-Proteobacteria (seven IGS types). Three endophytes belonged to the α-Proteobacteria, two belonged to the β-Proteobacteria, one belonged to the Flexibacter-Cytophaga-Bacteroides group, and three belonged to the high-G+C-content gram-positive group.
TABLE 2.
Sequence analysis of partial 16S rDNA (approximately 460 to 700 bp) of endophytic bacteria isolated from stems of potato variety Agria
| Endophyte | Origina | Theoretical T-RFs with HaeIII/HhaI (bp) | Biocontrol activity | Sequence analysis
|
||
|---|---|---|---|---|---|---|
| Closest NCBI database match (accession no.)b | % Similarity | Tentative phylogenetic group | ||||
| A1 | − | 233 | + | Bacillus megaterium (AF142677) | 98 | Firmicutes, Bacillus-Staphylococcus group |
| A2 | − | 229 | − | Arthrobacter oxidans (ASP243423) | 99 | Firmicutes, Actinomycetales, Micrococcaceae |
| A3 | + | 39 | − | Pseudomonas alcaligenes (AF094721) | 97 | gamma-Proteobacteria, Pseudomonadaceae |
| A4 | + | 39 | − | Pseudomonas alcaligenes (AF094721) | 98 | gamma-Proteobacteria, Pseudomonadaceae |
| A5c | + | 39 | − | Enterobacter cloacae (ECU65720) | 99 | gamma-Proteobacteria, Enterobacteriaceae |
| A7 | + | 39 | − | Pseudomonas putida (PSEGYRB3) | 99 | gamma-Proteobacteria, Pseudomonadaceae |
| A9c | + | 39 | − | Pseudomonas fluorescens (AF134705) | 99 | gamma-Proteobacteria, Pseudomonadaceae |
| A10c | + | 62 | − | Agrobacterium tumefaciens (ATU16SRDK) | 98 | alpha-Proteobacteria, Rhizobioceae |
| A11 | + | 320 | − | Uncultured potato root bacterium (PPL252736) | 92 | CFB group, Flavobacteriaceaec |
| A12 | + | 215 | + | Frateuria aurantia (FAU10481) | 94 | gamma-Proteobacteria, Xanthomonas group |
| A13 | + | 211 | − | Frateuria aurantia (FAU010481) | 94 | gamma-Proteobacteria, Xanthomonas group |
| A14c | + | 145 | + | Flavobacterium sp. strain B17 (AB027704) | 100 | CFB group, Flavobacteriaceae |
| A15 | + | 225 | + | Potato plant root bacterium (PPL252708) | 99 | alpha-Proteobacteria, Rhizobiaceae |
| A16 | + | 143 | + | Bacterium RSB-1 (AB032250) | 98 | Firmicutes, Actinomycetales |
| A17 | + | 225 | − | Potato plant root bacterium (PPL252708) | 99 | alpha-Proteobacteria, Rhizobiaceae |
| A18 | + | 222 | + | Clavibacter michiganensis (CLBSSRI) | 98 | Firmicutes, Actinomycetales, Microbacteriaceae |
| A19 | + | 65 | − | Exiguobacterium sp. (ESP16SRNA) | 99 | Firmicutes, Bacillus-Staphylococcus group |
| A20 | + | 141 | − | Aureobacterium kitamensis (AB013920) | 97 | Firmicutes, Actinomycetales |
| A21 | + | 199 | + | Rape rhizosphere bacterium (UBA295350) | 99 | beta-Proteobacteria, Comamonadaceae |
−, isolate derived from a control plant; +, isolate derived from a plant that was infected with the pathogen.
NCBI, National Center for Biotechnology Information.
Endophyte which was found in Agria plants as well as in Bionta plants.
CFB, Cytophaga-Flexibacter-Bacteroides.
TABLE 3.
Sequence analysis of partial 16S rDNA (approximately 460 to 700 bp) of endophytic bacteria isolated from stems of potato variety Bionta
| Endophyte | Origina | Theoretical T-RFs with HaeIII/HhaI (bp) | Biocontrol activity | Sequence analysis
|
||
|---|---|---|---|---|---|---|
| Closest NCBI database match (accession no.)b | % Identity | Tentative phylogenetic group | ||||
| B1 | − | 234 | + | Unidentified bacterium (UBA011425) | 98 | Firmicutes, Bacillus-Staphylococcus group |
| B2 | − | 219 | + | Achromobacter xylosoxidans (AF302097) | 98 | beta-Proteobacteria, Alcaligenaceae |
| B3 | − | 232 | + | Bacillus macroides (MB16RNAA) | 99 | Firmicutes, Bacillus-Staphylococcus group |
| B4 | − | 229 | + | Uncultured Paenibacillus sp. (AF245034) | 89 | Firmicutes, Bacillus-Staphylococcus group |
| B5 | − | 240 | − | Uncultured Paenibacillus sp. (AF245034) | 89 | Firmicutes, Bacillus-Staphylococcus group |
| B6 | − | 244 | − | Staphylococcus warneri (SW16SRRNA) | 96 | Firmicutes, Bacillus-Staphylococcus group |
| B7 | − | 181 | − | Clavibacter michiganensis (CLBSSRRI) | 99 | Firmicutes, Actinomycetales, Microbacteriaceae |
| B8 | − | 238 | − | Staphylococcus xylosus (SS1F0916S) | 99 | Firmicutes, Bacillus-Staphylococcus group |
| B9 | − | 227 | + | Uncultured Paenibacillus sp. (AF245034) | 89 | Firmicutes, Bacillus-Staphylococcus group |
| B10 | + | 216 | + | Stenotrophomonas maltophilia (SMA293464) | 97 | gamma-Proteobacteria, Xanthomonas group |
| B12c | + | 191 | − | Agrobacterium tumefaciens (ATU16SRDF) | 99 | alpha-Proteobacteria, Rhizobiaceae |
| B13 | + | 145 | − | Microbacterium sp. (AY017057S1) | 99 | Firmicutes, Actinomycetales, Microbacteriaceae |
| B14 | + | 61 | − | Rhizobium giardinii (AF345552) | 95 | alpha-Proteobacteria, Rhizobiaceae |
| B15 | + | 39 | − | Enterobacter amnigenus (EAQ23256S1) | 100 | gamma-Proteobacteria, Enterobacteriaceae |
| B16 | + | 40 | − | Stenotrophomonas maltophilia (AF100734) | 98 | gamma-Proteobacteria, Xanthomonas group |
| B17c | + | 39 | − | Pseudomonas fluorescens (AF134705) | 99 | gamma-Proteobacteria, Pseudomonadaceae |
| B18 | + | 204 | − | Unidentified eubacterium (EUB5871) | 95 | gamma-Proteobacteria, Enterobacteriaceae |
| B19c | + | 145 | − | Flavobacterium sp. strain B17 (AB027704) | 100 | CFB group, Flavobacteriaceaed |
| B20 | + | 145 | + | Microbacterium sp. (AB004713) | 98 | Firmicutes, Actinomycetales, Microbacteriaceae |
| B21 | + | 39 | + | Stenotrophomonas maltophilia (SMA293463) | 99 | gamma-Proteobacteria, Xanthomonas group |
| B22 | + | 198 | + | Unidentified beta-proteobacterium (AB015567) | 99 | beta-Proteobacteria, Comamonadaceae |
| B23 | + | 61 | − | Ochrobactrum anthropi (OAN242576) | 100 | alpha-Proteobacteria, Rhizobiaceae |
| B24 | + | 235 | − | Uncultured bacterium IAFBD8 (AF286174) | 98 | Firmicutes, Bacillus-Staphylococcus group |
| B25c | + | 39 | − | Enterobacter cloacae (ECU65720) | 98 | gamma-Proteobacteria, Enterobacteriaceae |
−, isolate derived from a control plant; +, isolate derived from a plant that was infected with the pathogen.
NCBI, National Center for Biotechnology Information.
Endophyte which was found in Agria plants as well as in Bionta plants.
CFB, Cytophaga-Flexibacter-Bacteroides.
Based on partial 16S rDNA gene analysis, 30 genetically different strains showed sequence similarities of more than 96% to the closest GenBank entry. Therefore, most endophytes could be assigned to certain bacterial species or genera. However, three bacteria isolated from Agria stems (A11, A12, and A13) and five bacteria isolated from Bionta stems (B4, B5, B9, B14, and B18) appeared to be less homologous to already known sequences.
A total of 23 theoretical T-RF sizes were found among the endophytes isolated, and 10 of them were also detected in total community fingerprints (Table 1). However, the remaining isolates could not be detected by cultivation-independent analysis. More T-RF sizes were obtained from pathogen-treated plants than from control plants. The sizes of two T-RFLP peaks (200 and 224 bp) of community fingerprints found specifically in infected plants matched the theoretical T-RF sizes of strains showing biocontrol activity.
Monitoring the biocontrol activity of the isolated endophytes.
A total of 16 strains were able to prevent disease in plants; 7 of them (A1, A12, A14, A15, A16, A18, and A21) were derived from cultivar Agria (Table 2), and 9 (B1, B2, B3, B4, B9, B10, B20, B21, and B22) were derived from Bionta (Table 3). Figure 2 shows plantlets after 2 weeks of incubation with the blackleg pathogen and endophytic strains B2 and B3. In addition, we found equal amounts of gram-positive and gram-negative strains that exhibited antagonistic activity against the blackleg pathogen. Some isolates, such as A9 and B17 (both showing 99% similarity to P. fluorescens), produced disease symptoms before exposure to E. carotovora subsp. atroseptica.
FIG. 2.
Tissue culture plants after 2 weeks of incubation with E. carotovora subsp. atroseptica (tube 1) or with the blackleg pathogen in combination with endophytic strains B2 (tube 3) and B3 (tube 4). Plants treated with 0.9% sterile NaCl (tube 2) were used as a growth control.
Strains that demonstrated antagonistic activity towards E. carotovora subsp. atroseptica were tested for production of antibiotics that inhibit growth of the pathogen. However, antibiotic production could not be demonstrated for any of the isolates tested.
DISCUSSION
Microbial community structure in healthy and infected potato plants.
Cultivation-independent population analysis of endophytes inhabiting potato stems revealed distinct populations of endophytes in healthy and pathogen-infected plants. Shannon-Weaver indices clearly indicated that there was increased structural diversity in plants that were inoculated with E. carotovora subsp. atroseptica. In general, pathogens induce a cascade of reactions in plants leading to the synthesis of stress metabolites, including H2O2, phytoalexins, and stress signals such as abscicic acid, jasmonic acid, and salicylic acid (18). However, in this study plants were hardly affected by the presence of the pathogen. Mobilization of nutrients may have taken place, causing improved conditions for growth of endophytes, possibly explaining the increased diversity in pathogen-treated plants. Similar findings were obtained with rhizosphere bacteria of avocado trees that were infected with the pathogen Phytophthora cinnamomi (45). In that study, infected roots which did not show visible symptoms of disease were colonized by much more variable bacterial communities and had significantly different community structures than healthy roots (45).
The higher diversity of endophytes in infected but healthy plants suggests that endoplant bacteria may be involved in pathogen defense. The role of endophytes in protecting plants against phytopathogens has been emphasized by several authors. Clay (8) proposed that the interaction between a host plant and its endophytic microflora is one mechanism by which disease resistance operates. Later, Sturz et al. (37) suggested that functioning communities of microbial endophytes in potato plants contribute to resistance to bacterial soft rot. The mechanisms by which pathogen invasion is prevented may be similar to those exhibited by rhizosphere microorganisms and may range from outcompetition of the pathogen to production of antibiotics to induction of systemic resistance. Furthermore, it has been proposed that the target plant benefits or suffers from bacterial metabolites produced as a response to the competing microflora (36). The pathogenicity of E. carotovora is regulated by quorum sensing, which is a population density-dependent modulation of the bacterial phenotype (38). Therefore, disease may be inhibited by a diverse endophyte community maintaining the pathogen population below the level that is required for expression of pathogenicity.
Characterization and biocontrol activity of isolated endophytes.
Cultivation on tryptic soy agar yielded bacterial endophytes that belonged to 20 different genera. Some genera were represented by several species, and some endophytes proved to be different strains of the same species. Our results indicated that a major portion of the endophyte community was not culturable on tryptic soy agar; however, several isolates were also detected by whole-population analysis. Some bacteria were not represented in community fingerprints. This may have been due to enrichment of particular strains in the growth medium that were not highly abundant in planta. Alternatively, as most cultured endophytes that were not found in community patterns were derived from spore-forming members of the Bacillus-Staphylococcus group, it may also be that these isolates inhabited plants as spores that were not lysed during the DNA isolation procedure.
By cultivation different endophyte populations were obtained from control and infected plants. The fact that a higher number of morphologically and genetically different strains were isolated from plants inoculated with E. carotovora subsp. atroseptica indicates that the diversity of culturable endophytes increased in the presence of the pathogen. Screening potato-inhabiting bacteria for putative antagonistic activity against E. carotovora subsp. atroseptica revealed that a high percentage (38%) of the isolates protected tissue culture plants from blackleg infection. These results are consistent with findings of van Buren and colleagues (40), who demonstrated that 32% of 192 endophytic bacterial strains isolated from potato stems exhibited biocontrol activity against the bacterial ring rot pathogen. Our results did not indicate a greater presence of potential biocontrol strains in plants inoculated with E. carotovora subsp. atroseptica. This might have been due to the limitations of bacterial cultivation. The contribution of uncultured endophytes to pathogen control merits further research.
Few differences between potato varieties were found by whole-community profiling; however, the culturable endophyte communities were very different. Similar phylogenetic distributions of endophytic isolates were found in the two cultivars used, except that more low-G+C-content gram-positive bacteria were isolated from Bionta plants. Our results suggest that similar phylogenetic groups and genera, but different species and strains, were present in the different plant varieties. Striking was the fact that only four isolates colonized both cultivars. The reason for this observation may be that tubers were used as the starting material for this experiment and the tubers were produced in different soils and therefore probably contained different endophytic strains. Most isolates could be clearly assigned by using the 16S rRNA gene sequence information. However, we found strains that showed the highest levels of homology to bacteria that have not been cultured or characterized yet. One group of strains (B4, B5, and B9) showed only 89% sequence similarity to its closest known relative, an uncultured Paenibacillus sp. These isolates were detected in Bionta plants but not in Agria plants, and one isolate (B9) exhibited antagonistic activity against E. carotovora subsp. atroseptica. Further analysis of this bacterial group should elucidate its phylogenetic position and function.
Several potato endophytes showed high 16S rDNA sequence homology to potential human pathogens, such as Enterobacter amnigenus, E. cloacae, Stenotrophomonas maltophilia, Staphylococcus xylosus, and Ochrobactrum anthropi (5, 7, 20, 22, 27). Although potato is a plant that is cooked before consumption, the potential occurrence of human pathogens in plants may merit further investigation. In addition, several potential phytopathogens were identified. A. tumefaciens, which is able to induce crown gall tumors in plants (44), as well as Clavibacter michiganensis (9), the bacterium that causes potato ring rot disease, were detected. Interestingly, potential phytopathogens were found to exhibit antagonistic activity against E. carotovora subsp. atroseptica, indicating that the distinction between plant-benefiting endophytes and pathogens can be unclear.
Conclusions.
Our results clearly demonstrated that the presence of a pathogen such as E. carotovora subsp. atroseptica has a significant effect on the endophytic bacterial community. Striking differences were found by a cultivation-independent approach, as well as by isolation and subsequent characterization of endophytes. Furthermore, we showed that endophytes represent a promising source of biocontrol strains and that their use may be more successful than that of rhizosphere bacteria due to less competition with other bacteria in the apoplast. Improved cultivation-independent community analysis targeting particular bacterial groups will continue to improve our understanding of the interactions among plants, pathogens, and bacterial endophytic communities.
Acknowledgments
This project was financed by the Austrian Science Foundation (Fonds zur Förderung der wissenschaftlichen Forschung). A. Sessitsch received an APART fellowship funded by the Austrian Academy of Sciences.
Potato tubers were obtained from the Niederösterreichische Saatbaugenossenschaft Windigsteig. We are grateful to B. Hyman of the Scottish Crop Research Institute (Invergowrie, Dundee, Scotland), who kindly provided the E. carotovora subsp. atroseptica strain used in this study.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäfer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, C. R., G. A. Dickie, W. L. G. Harvey, and J. W. Y. F. Chan. 1995. Endophytic bacteria in grapevine. Can. J. Microbiol. 41:46-53. [Google Scholar]
- 4.Blumer, C., and D. Haas. 2000. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173:170-177. [DOI] [PubMed] [Google Scholar]
- 5.Bollet, C., A. Elkouby, P. Pietri, and P. de Micco. 1991. Isolation of Enterobacter amnigenus from a heart transplant recipient. Eur. J. Clin. Microbiol. Infect. Dis. 10:1071-1073. [DOI] [PubMed] [Google Scholar]
- 6.Brown, C. M., and M. J. Dilworth. 1975. Ammonia assimilation by Rhizobium cultures and bacteroids. J. Gen. Microbiol. 86:39-48. [DOI] [PubMed] [Google Scholar]
- 7.Campos-Herrero, M. I., M. J. Pena, M. C. Perez, A. Bordes, and M. Mosquera. 1997. Bacteremia caused by Steonotrophomonas (Xanthomonas) maltophila. Enferm. Infect. Microbiol. Clin. 15:223-225. [PubMed] [Google Scholar]
- 8.Clay, K. 1988. Fungal endophytes of grasses: a defensive mutualism between plants and fungi. Ecology 69:10-16. [Google Scholar]
- 9.Davis, M. J., A. G. Gillaspie, A. K. Vidaver, and R. W. Harris. 1984. Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Bacteriol. 34:107-117. [Google Scholar]
- 10.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garbeva, P., L. S. van Overbeek, J. W. L. van Vuurde, and J. D. van Elsas. 2001. Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA based PCR fragments. Microb. Ecol. 41:369-383. [DOI] [PubMed] [Google Scholar]
- 13.Hallmann, J., A. Quadt-Hallmann, W. F. Mahaffee, and J. W. Kloepper. 1997. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43:895-914. [Google Scholar]
- 14.Kirchhof, G., B. Eckert, M. Stoffels, J. I. Baldani, V. M. Reis, and A. Hartmann. 2001. Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fibre plants. Int. J. Syst. E vol. Microbiol. 51:157-168. [DOI] [PubMed] [Google Scholar]
- 15.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1991. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA-specific and specific DNA probes. Appl. Environ. Microbiol. 57:3390-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloepper, J. W. 1983. Effect of seed piece inoculation with plant-growth promoting rhizobacteria on populations of Erwinia carotovora on potato roots and daughter tubers. Phytopathology 73:217-219. [Google Scholar]
- 17.Leong, J. 1986. Siderophores: their biochemistry and possible role in the biocontrol of plant pathogens. Annu. Rev. Phytopathol. 24:187-209. [Google Scholar]
- 18.Lichtenthaler, H. K. 1998. The stress concept in plants: an introduction. Ann. N. Y. Acad. Sci. 851:187-198. [DOI] [PubMed] [Google Scholar]
- 19.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahood, M. S., A. R. Sarwari, M. A. Khan, Z. Sophie, E. Khan, and S. Sami. 2000. Infective endocarditis and septic embolization with Ochrobactrum anthropi: case report and review of literature. J. Infect. 40:287-290. [DOI] [PubMed] [Google Scholar]
- 21.Massol-Deya, A. A., D. A. Odelson, R. F. Hickey, and J. M. Tiedje. 1995. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA), p. 1-8. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dortrecht, The Netherlands.
- 22.Mastroianni, A., O. Coronado, A. Nanetti, and F. Chiodo. 1994. Staphylococcus xylosus isolated from a pancreatic pseudocyst in a patient infected with the human immunodeficiency virus. Clin. Infect. Dis. 19:1173-1174. [DOI] [PubMed] [Google Scholar]
- 23.Murashige, T., and F. Skoog. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15:473-497. [Google Scholar]
- 24.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nei, M., and W.-H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowak, J. 1998. Benefits of in vitro “biotization” of plant tissue cultures with microbial inoculants. In Vitro Cell. Dev. Biol. Plant 34:122-130. [Google Scholar]
- 27.Oana, S., J. Aizawa, S. Tatebe, N. Ishiwada, H. Kuroki, and Y. Kohno. 2000. A case of persistent Enterobacter cloacae bacteremia. Pediatr. Int. 42:702-704. [DOI] [PubMed] [Google Scholar]
- 28.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with the chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiessendoppler, E., and P. Cate. 1996. Schwarzbeinigkeit, bakterielle Stengelfäule und Naßfäule der Knolle, p. 48-51. In B. Zwatz and G. Bedlan (ed.), Wichtige Krankheiten und Schädlinge der Kartoffel. Institut für Phytomedizin im Bundesamt und Forschungszentrum für Landwirtschaft, Vienna, Austria.
- 30.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sessitsch, A., B. Reiter, U. Pfeifer, and E. Wilhelm. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39:23-32. [DOI] [PubMed]
- 32.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication. University of Illinois Press, Urbana.
- 33.Sharma, V. K., and J. Novak. 1998. Enhancement of verticillium wilt resistance in tomato transplants by in vitro co-culture of seedlings with a plant growth promoting rhizobacterium (Pseudomonas sp. strain PsJN). Can. J. Microbiol. 44:528-536. [Google Scholar]
- 34.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoltzfus, J. R., R. So, P. P. Malarvithi, J. K. Ladha, and F. J. de Bruijn. 1998. Isolation of endophytic bacteria from rice and assessment of their potential for supplying rice with biologically fixed nitrogen. Plant Soil 194:25-36. [Google Scholar]
- 36.Sturz, A. V., and B. G. Christie. 1996. Endophytic bacteria of red clover as agent of allelopathic clover-maize syndromes. Soil Biol. Biochem. 28:583-588. [Google Scholar]
- 37.Sturz, A. V., B. R. Christie, B. G. Matheson, W. J. Arsenault, and N. A. Buchanan. 1999. Endophytic bacterial communities in the periderm of potato tubers and their potential to improve resistance to soil-borne plant pathogens. Plant Pathol. 48:360-369. [Google Scholar]
- 38.Swift, S., J. P. Throup, P. Williams, G. P. Salmond, and G. S. Stewart. 1996. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem. Sci. 21:214-219. [PubMed] [Google Scholar]
- 39.Tholozan, J. L., J. M. Cappelier, J. P. Tissier, G. Delattre, and M. Federighi. 1999. Physiological characterization of viable-but-nonculturable Campylobacter jejuni cells. Appl. Environ. Microbiol. 65:1110-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Buren, A. M., C. Andre, and C. A. Ishimaru. 1993. Biological control of the bacterial ring rot pathogen by endophytic bacteria isolated from potato. Phytopathology 83:1406. [Google Scholar]
- 41.van de Peer, Y., and R. de Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 42.van Wees, S. C., M. Luijendijk, I. Smoorenburg, L. C. van Loon, and C. M. Pieterse. 1999. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 41:537-549. [DOI] [PubMed] [Google Scholar]
- 43.Wilhelm, E., W. Arthofer, and R. Schafleitner. 1997. Bacillus subtilis, an endophyte of chestnut (Castanea sativa), as antagonist against chestnut blight (Cryphonectria parasitica), p. 331-337. In A. C. Cassells (ed.), Pathogen and microbial contamination management in micropropagation. Kluwer Academic Publishers, Dortrecht, The Netherlands.
- 44.Winans, S. C. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56:12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, C., D. E. Crowley, and J. A. Menge. 2001. 16S rDNA fingerprinting of rhizosphere bacterial communities associated with healthy and Phytophthora infected avocado roots. FEMS Microbiol Ecol. 35:129-136. [DOI] [PubMed] [Google Scholar]