Abstract
The pyruvate dehydrogenase multienzyme complex (PDHC) was found to be upregulated by osmotic stress in the osmotolerant pathogen Staphylococcus aureus. Upregulation was detectable in the levels of both activity and protein and was judged to be about fourfold when sodium chloride was used to adjust the water activity (aw) of the growth medium to 0.94. The upregulation of the PDHC was also found to be humectant dependent and was greatest when impermeant, nonmetabolizable humectants were used to adjust aw. Further experiments provided evidence that in addition to osmotic upregulation, the PDHC complex is also subject to catabolite repression, thus providing a possible explanation for the observation that high concentrations of carbohydrates are generally more inhibitory to the growth of this bacterial pathogen than are high concentrations of salts.
Staphylococcus aureus, an important human pathogen, is considered a highly osmotolerant bacterium and is able to grow at a water activity (aw) of as low as 0.86 (33). Its osmotolerance may be an important determinant of its ability to grow in foods and on human skin, from where it can spread and/or form toxins and cause disease. Although osmoadaptation of S. aureus has been studied fairly extensively in the past (8, 9, 29, 30, 38, 42, 43, 46), it remains unclear why this organism is considerably more osmotolerant than other bacterial pathogens. In fact, thus far, studies have revealed an osmoadaptive process apparently typical for bacteria in general: namely, the accumulation of glycine betaine, proline, or other common compatible solutes through transport from the growth medium (25, 29, 39) and increased synthesis of stress proteins such as chaperones (31) and alkyl hydroperoxide reductase C (8). However, recent studies have given indications that the regulation of stress gene expression may be somewhat unusual in the staphylococci. For example, expression of the alternative sigma factor σB is repressed during osmotic stress in S. aureus (13) while it is strongly induced in Bacillus subtilis under those conditions (11, 45). Although a few studies have attempted to identify osmotically upregulated proteins in S. aureus (8, 31, 44), this field still remains relatively unexplored.
A major goal of the present study was to investigate changes in protein composition of this osmotolerant organism in response to osmotic stress and to identify proteins important for its osmoadaptation. Herein, we present evidence that the pyruvate dehydrogenase multienzyme complex (PDHC) of S. aureus, an enzyme of central importance in this organism's primary metabolism, is upregulated by osmotic stress. The implications of this finding with respect to the osmoadaptation of S. aureus and the regulation of its metabolic activity are discussed.
MATERIALS AND METHODS
Strains and media.
S. aureus strain ATCC 12600 was obtained from the American Type Culture Collection (Manassas, Va.) and grown in Trypticase soy broth (TSB; Difco Laboratories, Detroit, Mich.) at 37°C. The aw values of media were measured in duplicate at room temperature in an Aqua Lab CX-2 water activity meter (Decagon, Pullman, Wash.). For testing growth on different carbon sources and measuring glucose consumption, the defined medium (DFM) described previously (44a) was used, except proline levels were set at 5 mM and glucose was replaced, where appropriate, with other potential carbon sources (d-fructose, d-arabinose, l-arabinose, d-sucrose, d-mannose, d-xylose, glycine betaine, glycerol) at 0.5% (wt/vol).
Isolation and two-dimensional (2D) electrophoresis of membrane proteins.
Since osmoadaptation is a response to changing environmental conditions, it was decided to focus on the part of the cellular protein profile most exposed to the environment, namely plasma membrane-associated proteins. Cells were grown in approximately 1 liter of TSB or TSB adjusted to an aw of 0.95, 0.92, or 0.89 with glycerol, NaCl, or sucrose; cells were harvested at the late exponential or stationary growth phase. Harvesting (by centrifugation), lysis (by osmotic disruption of protoplasts), and isolation of plasma membranes (by centrifugation) were performed essentially as described by Sprott et al. (36), except lysostaphin (1.0 U ml of culture−1) was used instead of lysozyme and sorbitol (50% [wt/vol]) was used instead of sucrose to control osmotic pressure. Membrane proteins were solubilized according to the method of Ames and Nikaido (7). An aliquot containing approximately 200 μg of membrane protein, as determined by a modified Lowry method (23), was diluted with 2 volumes of an isoelectrofocusing buffer described by Ames and Nikaido (7) containing 3% (vol/vol) IGEPAL CA-630 (Sigma Chemical Company, St. Louis, Mo.) instead of NP-40. The samples were electrophoresed in the first dimension on 4% polyacrylamide (PA) tube gels (1-mm diameter) for 16,500 V · h. The second dimension was done on a 4% PA stacking gel and a 10% PA running gel at 35 mA per gel. The gels were stained with 0.2% Coomassie brilliant blue R-250 in 5:4:1 distilled water:methanol:acetic acid and destained in 0.5 M NaCl (37), except when the gels were to be used for sequencing, in which case the gels were stained with 0.05% Coomassie brilliant blue R-250 in 20% methanol (sequencing grade) and 0.5% acetic acid. Destaining was performed in 30% sequencing grade methanol, and the gels were stored in distilled water. The gels were scanned with a Bio-Rad (Hercules, Calif.) model GS-670 imaging densitometer and analyzed with a Molecular Analyst 2DFull software package (Bio-Rad).
Spot elution and N-terminal sequencing.
Selected spots were excised from four 2D gels, homogenized in a 1.5-ml-capacity tissue grinder, and extracted by passive diffusion as described by Hames (18). The extracts were electrophoresed on a 4% PA stacking gel and a 10% PA running gel with 7 μl of thioglycolic acid liter−1 in the upper running buffer. The samples were then electroblotted onto a sequencing grade polyvinylidene difluoride membrane (Bio-Rad) according to protocols supplied by the manufacturer. N-terminal sequencing by Edman degradation was performed at the Macromolecular Core Facility at the Hershey Medical Center, Hershey, Pa.
Preparation of cell extracts and PDHC activity assays.
Cultures (100 ml) were grown in TSB or TSB adjusted to a set aw value (usually 0.94 or 0.96) with a humectant (d-sucrose, d-glucose, d-fructose, d-xylose, d-arabinose, l-arabinose, d-mannose, glycine betaine, sodium chloride, potassium chloride, sodium sulfate, glycerol, or polyethylene glycol with an average molecular weight of 8,000 [PEG 8000]). Cells were harvested by centrifugation (10,000 × g for 10 min), washed with 10 ml of a 100 mM sodium phosphate buffer (pH 6.6), and lysed by treatment with lysostaphin (100 U ml−1) at 37°C for 60 min. After centrifugation (10,000 × g for 10 min), the pellet was resuspended in 150 μl of 100 mM sodium phosphate buffer (pH 6.6) containing 0.1% Triton X-100, 2 U of DNase, and 2 U of RNase. Protease inhibitor cocktail (catalogue no. P 8465; Sigma) was added according to the manufacturer's instructions, and the resulting suspension was incubated for 20 min at 37°C. The remaining cell debris was removed by centrifugation (10,000 × g for 10 min). Protein concentration in the extract was determined by a modified Lowry method (23) suitable for membrane protein preparations.
PDHC activity was assayed according to a procedure based on that of Seals and Jarett (34). Briefly, an aliquot containing a fixed amount (usually 100 μg) of protein was added to a 20-ml-capacity serum vial that had been equipped with an elevated CO2-trapping cup holding a piece of Whatman no. 1 filter paper. To the sample, 155 μl of 5× preincubation buffer (100 μM EDTA, 10 mM dithiothreitol, 5 mM ADP, 1% Triton X-100, 250 mM HEPES; pH 7.7) was added, and the resulting mixture was diluted to 775 μl with water and incubated at 37°C for 10 min. After incubation, 125 μl of assay buffer (10 mM NAD [Sigma], 2 mM coenzyme A [ICN Biomedicals, Aurora, Ohio], 2.5 mM thiamine pyrophosphate, 5 mM dithiothreitol, 5 mM MgCl2, 0.5% Triton X-100, 100 mM Tris-HCl; pH 8.0) was added, and the vials were sealed and incubated at 37°C for a further 5 min. After the second incubation, 100 μl of 1 mM (2 Mcpm ml−1) [1-14C]pyruvate (Amersham-Pharmacia, Piscataway, N.J.) (27 mCi mmol−1) was injected into the solution and the vials were incubated at 30°C for 10 min, after which time the reaction was terminated by placing the vials on ice and adding 1 ml of ice-cold 3 M sulfuric acid to the solution. Finally, 150 μl of 1 M NaOH was injected onto the filters, and CO2 was collected at room temperature with gentle shaking for 3 h. The filters were suspended in 10 ml of Ecoscint H scintillation fluid (National Diagnostics, Atlanta, Ga.) and counted for 10 min in a Beckman (Palo Alto, Calif.) LS1701 scintillation counter. To eliminate the possibility that the measured activity resulted from the action of pyruvate-metabolizing enzymes other than the PDHC (e.g., pyruvate decarboxylase) and/or of pyruvate dehydrogenase alone (i.e., in isolation from the other PDHC components), assays were also performed in the absence of NAD, an essential cofactor for the PDHC whose turnover is mediated by the dehydrolipoamide dehydrogenase component of the PDHC (10).
Preparation of genomic DNA.
Cells grown to late exponential growth phase in 6 ml of TSB were harvested by centrifugation at 10,000 × g for 2 min. The cell pellet was resuspended in 10 mM Tris-HCl (pH 7.5) containing 1 mM EDTA, 1 U of lysostaphin μl−1, 1 U of RNase A μl−1, 100 ng of proteinase K μl−1, and 0.5% sodium dodecyl sulfate and incubated for 1 h at 37°C. Genomic DNA was extracted essentially as described by Moore (26).
PCR and sequencing.
Preliminary sequence data were obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org) and used to design oligonucleotide primers for amplification of a 1,129-bp fragment containing the S. aureus pdhB gene. Primers were synthesized by Integrated DNA Technologies (Coralville, Iowa) and were as follows: left primer, 5′-TGCCTCAAAACTTAGCAGAACA-3′ (binds the sequence 59 to 38 bases upstream of the pdhB initiation codon); right primer, 5′-CACGTTTTTGCCCTCCTAAG-3′ (binds the sequence 74 to 93 bases downstream of the pdhB termination codon). PCR was carried out in a GeneAmp 9600 thermocycler (Perkin-Elmer, Norwalk, Conn.). Initial denaturation was at 96°C for 5 min and was followed by 30 cycles of denaturation at 94°C for 2 min, annealing at 55°C for 2 min, and extension at 72°C for 1 min, which in turn was followed by final extension at 72°C for 15 min and indefinite holding at 4°C. Taq polymerase (Promega, Madison, Wis.) was added after initial denaturation. PCR products were purified with a QIAquick kit (Qiagen, Valencia, Calif.) according to a protocol supplied by the manufacturer. The purified products were sequenced at the Nucleic Acid Facility at The Pennsylvania State University, University Park, Pa. The pdhB sequence obtained from S. aureus ATCC 12600 has been deposited in GenBank (accession no. AF235026).
Carbon source consumption per unit of biomass as a function of growth medium aw.
Cultures (100 ml) were grown at 37°C in DFM and in DFM containing NaCl at aw values of 0.945 and 0.910. Glucose was the only utilizable carbon source and was present at a concentration of 0.5% (wt/vol). Aliquots (0.5 ml) were removed at various time points between 10 and 175 h of incubation, and cells were harvested by centrifugation (10,000 × g for 10 min). Glucose concentrations in the spent medium as well as in uninoculated media were measured with a Sigma Diagnostics glucose oxidase kit according to the manufacturer's instructions. The cell pellet was washed with 0.5 ml of 100 mM sodium phosphate buffer (pH 6.6) and subsequently digested in 1 M NaOH at 90°C for 10 min. The protein content of the digested cell pellets was determined according to the modified Lowry method of Daniels et al. (15), which is suitable for alkaline cell digests. The decrease in glucose concentration in the spent media compared to that in uninoculated medium was expressed on a per-milligram-of-cellular-protein basis to give an estimate of glucose consumed per unit of biomass produced.
RESULTS AND DISCUSSION
Synthesis of several membrane-associated proteins of S. aureus is induced when cells are grown in the presence of high concentrations of NaCl.
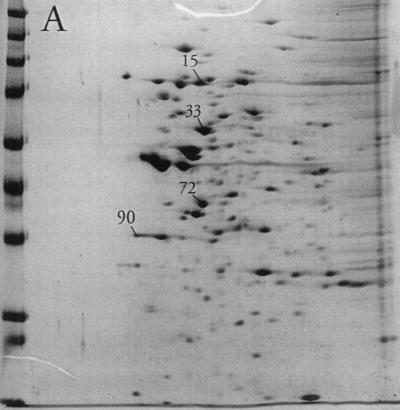
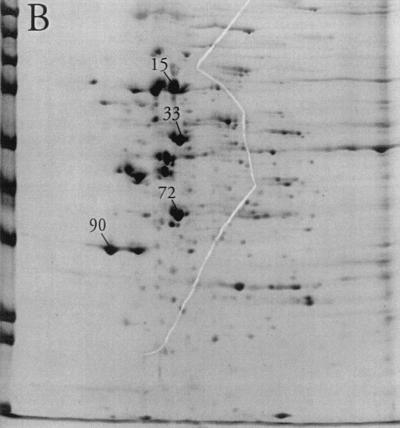
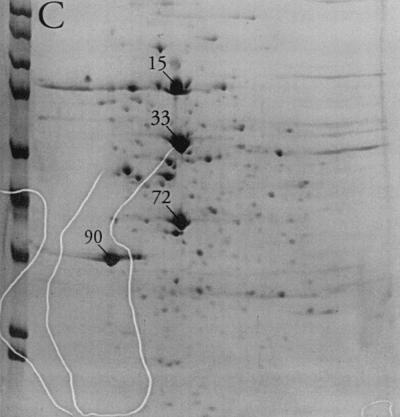
Substantial changes were observed in the membrane-associated protein profile of S. aureus when the aw of the growth medium was reduced by the addition of NaCl. For most proteins where a change could be observed, the protein level decreased with decreasing aw, as judged by gel spot density. In a few cases, however, a dramatic increase in spot density could be observed (Fig. 1). Indeed, the relative abundance of four spots (no. 15, 33, 72, and 90) increased from a total of 9% in the absence of osmotic stress to more than half (51%) of the total membrane-associated protein visible on the gels at an aw of 0.89 (Table 1). Of these, proteins 72 and 90 were the most clearly separated from neighboring spots and could be isolated from the gels with relative ease. They were therefore selected for further analysis. Additional gels (not shown) from cultures grown in the presence of high levels of sucrose or glycerol indicated that the increase in the levels of proteins 72 and 90 in response to osmotic stress was humectant specific, as their relative abundance was essentially unaltered compared to that of nonstress controls (data not shown).
FIG. 1.
2D polyacrylamide gel electrophoresis analysis of the membrane protein fraction of cells grown in TSB (A), TSB with the aw adjusted to 0.950 with NaCl (B), or TSB with the aw adjusted to 0.890 with NaCl (C). Spots 15, 33, 72, and 90 are labeled. The molecular mass markers from top to bottom at left are E. coli β-galactosidase (116 kDa), rabbit phosphorylase b (97 kDa), rabbit fructose-6-phosphate kinase (84 kDa), bovine serum albumin (66 kDa), bovine glutamic dehydrogenase (55 kDa), chicken egg ovalbumin (45 kDa), rabbit glyceraldehyde-3-phosphate dehydrogenase (36 kDa), bovine carbonic anhydrase (29 kDa), and bovine trypsinogen (24 kDa).
TABLE 1.
Relative increase in levels of protein spots 72, 90, 15, and 33 as a function of aw
| Spot | Relative abundancea at aw of 0.996, 0.95, 0.89 | Apparent Mw (in thousands)b | Apparent pIb |
|---|---|---|---|
| 72 | 2, 4, 11 | 40 | 5.5 |
| 90 | 1, 4, 11 | 36 | 5.0 |
| 15 | 2, 4, 13 | 70 | 5.5 |
| 33 | 4, 6, 16 | 57 | 5.5 |
The relative abundance of each protein spot is expressed as a percentage of the total protein detected on a representative 2D PA gel by scanning densitometry as described in Materials and Methods. Cells were grown in TSB medium adjusted to an aw value of 0.996, 0.95, or 0.89 by the addition of NaCl.
The apparent Mw and pI for each protein spot were estimated by comparing mobilities in 2D gels to a variety of protein standards.
Proteins 72 and 90 share homology with the PdhA and PdhB subunits, respectively, of the PDHC.
N-terminal sequencing of protein 90 yielded the sequence QMTMVQAIN starting at amino acid 2. A search in GenBank (12) with BLASTp (6) indicated a high degree of similarity to the N-terminal region of the PdhB proteins (the β subunit of pyruvate dehydrogenase) from B. subtilis, Bacillus stearothermophilus, and Mycobacterium tuberculosis (Table 2). In light of this result, the putative pdhB gene of S. aureus was PCR amplified and sequenced. The predicted amino acid sequence (GenBank accession no. AF235026) corresponded to a polypeptide of 35.2 kDa with a pI of 4.65, consistent with the mobility of protein 90 on 2D PA gels (Table 1). Furthermore, the predicted N-terminal sequence of the S. aureus PdhB protein was found to be identical to that determined by N-terminal sequencing of spot 90 beginning at amino acid 3, providing further evidence that spot 90 does indeed correspond to the S. aureus PdhB protein. The fact that amino acid identity began at residue 3 also indicates that the N-terminal methionine of PdhB is cleaved off posttranslationally.
TABLE 2.
Homology of bacterial protein sequences to the protein spot 90 N-terminal sequence
| Organism | Polypeptide | N positiona | Sequenceb | GenBank accession no. |
|---|---|---|---|---|
| Staphylococcus aureus | Protein 90 | 2 | QMTMVQAIN | AF235026 |
| Bacillus stearothermophilus | PDHC-E1βc | 3 | QMTMVQAI | X53560 |
| Bacillus subtilis | PDHC-E1β | 3 | QMTMIQAI | M57435 |
| Mycobacterium tuberculosis | PDHC-E1β | 26 | LTMVQAIN | Z95556 |
| Edwardsiella tarda | Unknown | 73 | TMVEAIN | L43071 |
| Bacillus cereus | Hemolysin BL | 137 | TMVEAIN | L20441 |
| Helicobacter pylori | Unknown | 244 | EMTMVQKI | AE000542 |
| Pseudomonas putida | BCKDHC-E1βd | 6 | MTMIQA | M57613 |
The position of the sequence shown with respect to the protein N terminus (S. aureus) or to the putative translation start site (all others).
Obtained by N-terminal amino acid sequencing (S. aureus) or deduced from gene sequence (all others).
PDHC-E1β, PDHC, enzyme 1, subunit β.
BCKDHC-E1β, branched-chain keto acid dehydrogenase multienzyme complex, enzyme 1, subunit β.
PdhB is one of four subunits of the PDHC. Although neither PdhB nor any other PDHC component is an integral membrane protein, the PDHC has previously been reported to be isolated from the membrane fractions of S. aureus and B. subtilis (1, 2, 3, 19, 20), consistent with our finding in the present paper. Thus, it was reasonable to expect that the levels of the other subunits of this complex also should increase within cells grown at elevated osmolarity and be visualized on 2D gels of membrane protein fractions. Indeed, we were able to obtain the N-terminal sequence for spot 72 (PKLQAQFDA) and found it to exactly match the predicted S. aureus PdhA sequence (beginning with amino acid 3) identified from genome sequence data provided by the TIGR website. The relative migration of spot 72 on 2D gels is also consistent with the theoretical Mw (41,400) and theoretical pI (4.9) for the S. aureus PdhA protein as predicted from the TIGR sequence data. Due to a lack of sufficient purified protein, we were unable to determine the N-terminal sequences for protein spots 15 and 33; however, the possibility remains that spots 15 and 33 may correspond to the PdhC and PdhD subunits of the PDHC (note that the relative migration of PdhC and PdhB on 2D gels has previously been shown to be strongly influenced by the presence of lipoamide moieties on these proteins [5, 32]). In order to obtain further evidence that the levels of the PDHC are elevated when S. aureus is grown in the presence of high NaCl concentrations, functional activity assays were performed on cell extracts as described below.
Activity assays confirm that pyruvate dehydrogenase activity is elevated in cells grown in low-aw media.
The finding that levels of the PDHC may be osmotically regulated in S. aureus was somewhat surprising, because to our knowledge, enzymes of primary metabolism have not previously been implicated in the osmotic stress response in any bacterial species. Thus, in order to obtain more definitive evidence, pyruvate dehydrogenase activity assays were performed on cell extracts derived from cultures grown in TSB media at different aw values. Results were in good agreement with the 2D gel results reported above and indicated that levels of PdhB were approximately three to four times higher in cultures grown in media adjusted to an aw of 0.95 with NaCl than in nonstress controls (Table 3). It is noted that pyruvate dehydrogenase activity levels within extracts from cultures grown in the presence of Na2SO4 or KCl were not significantly different from levels found within extracts derived from cultures grown in the presence of NaCl at the same aw value (data not shown), suggesting that the increased level of enzyme activity occurred in response to osmotic stress and was not due to a specific response to the Na+ or Cl− ion
TABLE 3.
PDHC activity and protein spot 90 levels in cell extracts from cultures grown at different aw values
| awa | Protein 90 levelb | PDHC activityc |
|---|---|---|
| 0.996 | 1.67 | 1.6 ± 0.3 |
| 0.97 | NDd | 3.3 ± 0.7 |
| 0.95 | 6.23 | 4.9 ± 1.0 |
| 0.94 | ND | 6.8 ± 1.0 |
| 0.89 | 9.17 | ND |
Cells were grown to the exponential phase in TSB medium containing various amounts of added NaCl to bring the aw of the medium to the indicated value.
The relative level of protein spot 90 on representative 2D PA gels was determined by scanning densitometry and is expressed in arbitrary volume units. Each gel was loaded with 150 μg of total membrane-associated protein.
The measured 14CO2-generating activity (nanomoles of CO2 per milligram of protein per minute) was found to be dependent on the presence of NAD in the assay buffer (data not shown), eliminating the possibility that other pyruvate-metabolizing enzymes (such as pyruvate decarboxylase) were responsible for all or part of the measured activity and indicating that the activity was carried out by an intact PDHC (since the NAD-consuming reaction is performed by PdhD). Values shown are averages obtained from duplicate measurements from at least two replicate experiments ± standard deviations.
ND, not determined.
Increased pyruvate dehydrogenase activity may provide S. aureus with a strategy to meet increased energy demands resulting from osmotic stress.
Pyruvate dehydrogenase is a central enzyme of the primary metabolism of S. aureus. Therefore, its apparent induction by osmotic stress brought to mind the common but, with two notable exceptions (16, 21), largely untested assumption that osmoadaptation is an energy-consuming process (17, 27). The rationale behind this assumption is that the maintenance of very steep humectant and compatible-solute concentration gradients across the cytoplasmic membrane (which is necessary if the cell is to maintain its volume and turgor without accumulating high intracellular concentrations of a humectant that may be disruptive to the cell's metabolism) requires substantial activity of the cell's transport systems, which will, directly or indirectly, consume large quantities of ATP. The findings of the present study provided a compelling reason to test this hypothesis. Indeed, we observed that S. aureus consumes greater amounts of its carbon and energy source per unit of biomass produced when it is growing under conditions of osmotic stress. Cells growing in DFM with NaCl-adjusted aw values of 0.996, 0.945, and 0.910 consumed 1.8 ± 1.6, 5.4 ± 1.1, and 12.9 ± 2.6 mg of glucose per mg of protein, respectively. This effect appears quite substantial, with consumption of glucose per unit of biomass produced increasing approximately sevenfold when cells are grown at an aw of 0.910 compared to when they are grown with no stress (aw, 0.996). The only other studies of which we are aware that directly address energy requirements in relation to osmotic stress were conducted on the relatively nonosmotolerant bacteria Escherichia coli (21) and Klebsiella pneumoniae (16). While the former study concluded that low water activity did not impose a “large energetic burden” on E. coli, the latter study revealed an approximately 10-fold-higher level of carbon source consumption by K. pneumoniae per unit of biomass when cells were grown at an aw of 0.970 (compared to when cells were grown in the absence of osmotic stress). It may be worth noting that an aw of 0.970 is very restrictive to the growth of K. pneumoniae, yielding a growth rate of less than 10% of that observed at an aw of 0.996 (16), whereas an aw of 0.910 is only moderately inhibitory to S. aureus, yielding a growth rate of approximately 20 to 25% of that observed at an aw of 0.996 in complex media at 37°C (O. Vilhelmsson and K. J. Miller, unpublished data). It is also worth noting that in addition to this increased carbon source consumption, S. aureus has been shown to decrease production of most extracellular proteins in response to osmotic stress (40), which should further increase the energy available for growth and maintenance. Furthermore, extracellular proteins were not included in our biomass estimates, and therefore the glucose consumption per unit of biomass, particularly at high aw values, is probably overestimated. Consequently, the increase in consumption at low aw values reported herein is probably an underestimate.
Presumably, the apparent increase in energy requirements reported above reflects the cell's efforts to maintain very steep humectant and compatible-solute concentration gradients across the cell membrane. It would therefore be expected that a humectant that is permeant and compatible with cellular metabolism would not lead to increased energy requirements and, hence, increased levels of the PDHC (since other compatible solutes would not be accumulated). Glycerol has just such properties (4), and additional experiments revealed that growth in the presence of high glycerol concentrations (aw, 0.95) did not lead to increased levels of the PDHC in S. aureus (Table 4). Glycerol is utilizable as a carbon source by S. aureus (47), and thus, it could be argued (especially in light of our results with sugars as humectants, discussed below) that the low PDHC activity in the presence of glycerol is the result of catabolite repression rather than low energy demands. However, glycerol is highly permeant towards biological membranes (4) and thus probably enters the cells via passive and/or facilitated diffusion. It is therefore unlikely to induce expression of any phosphotransferase system which was shown by Smith et al. (35) to be required for catabolite repression of enterotoxin A production in this organism. However, based on these results, we were prompted to examine the effects of other carbon sources on PDHC activity.
TABLE 4.
PDHC activity in cell extracts from exponentially growing cultures in media with aw adjusted with various humectantsa
| Humectant | aw | PDHC activityb | C sourcec |
|---|---|---|---|
| Sucrose | 0.94 | 0.86 ± 0.4 | Yes |
| Glucose | 0.94 | <0.1 | Yes |
| Mannose | 0.95 | 0.77 ± 0.07 | Yes |
| Fructose | 0.94 | <0.1 | Yes |
| Glycerol | 0.95 | 1.1 ± 0.2 | Yes |
| PEG 8000 | 0.96 | 2.6 ± 0.4 | No |
| Glycine betaine | 0.94 | 4.7 ± 0.5 | No |
Cells were grown in TSB media.
PDHC activity is defined in footnote c of Table 3, where the activity value (1.6 ± 0.3) for non-osmotically stressed cultures (aw, 0.996) is also given. Values shown are averages obtained from duplicate measurements from at least two replicate experiments ± standard deviations.
Experimentally determined as the presence (yes) or absence (no) of growth of S. aureus in DFM with the indicated humectant added as the only available carbon source at a concentration of 0.5% (wt/vol).
PDHC activity may be subject to catabolite repression.
The effects of a variety of nonionic humectants on pyruvate dehydrogenase activity were examined. For these experiments, cells were grown in the presence of high concentrations (aw adjusted to between 0.94 and 0.96) of several metabolizable carbon sources (sucrose, glucose, fructose, and mannose) as well as two nonmetabolizable humectants (PEG 8000 and glycine betaine). Consistent with an osmotic effect, pyruvate dehydrogenase activity was found to be elevated within extracts derived from cells grown in the presence of high concentrations of PEG 8000 and glycine betaine (Table 4). However, this was not found to be the case when cells were grown in the presence of high concentrations of metabolizable carbon sources. Indeed, levels of pyruvate dehydrogenase activity found within cells grown in the presence of high concentrations of sucrose, glucose, fructose, and mannose were even lower than levels present within nonstress control cultures. This reduction in activity was particularly dramatic in the case of glucose and fructose, suggesting that the levels of the PDHC may be regulated through catabolite repression. Attempts to use nonmetabolizable sugars (d-xylose, d-arabinose, and l-arabinose) as humectants were unsuccessful, as no growth occurred in media adjusted to aw values of 0.94 or 0.96 with these sugars. To our knowledge, this study is the first to show a link between osmotic stress responses and catabolite repression in S. aureus. In light of the effect of osmotic stress on cellular energy demands, discussed above, such a link would seem likely to exist. Interestingly, it may be noted that Ueguchi et al. (41) have provided evidence for a link between stress responses and catabolite repression in E. coli. Specifically, these researchers have shown that catabolite repression has a role in the regulation of the E. coli RpoS sigma factor.
Concluding remarks.
Although the effects of low aw on the growth of S. aureus have been studied for decades, the literature is surprisingly scant on the effects of different humectants on the rate of growth of this organism. Indeed, most growth studies utilizing more than one humectant have primarily focused only on identifying the minimum aw required for growth (14, 24, 28). However, available data reveal that the growth rate of this food-borne pathogen is, in fact, lower when carbohydrates are used as humectants than when salts are used in this capacity (22, 33, 44a). This effect becomes particularly pronounced at very low aw values (44a). To our knowledge, there have been no prior attempts to explain this observation, but the results of the present study may provide important insight in this regard. Specifically, increased levels of PDHC may represent an important osmotic stress response for this organism and provide it with a strategy for meeting the increased energy demands associated with maintaining high levels of intracellular compatible solutes. If, however, the osmotic upregulation of the PDHC is counteracted by catabolite repression when metabolizable carbohydrates are used as humectants, it may be predicted that the growth rate of S. aureus should be substantially reduced.
Acknowledgments
This work was supported in part by a Grant for Study Abroad (Fulbright-Hays Program) from the Iceland-United States Educational Commission, a NATO Science Fellowship, and a special USDA grant on milk safety.
Preliminary sequence data were obtained from The Institute for Genomic Research website (http://www.tigr.org). We thank Anne Stanley at the Macromolecular Core Facility, Hershey Medical Center, for performing the N-terminal sequencing.
REFERENCES
- 1.Adler, L.-Å., and S. Arvidson. 1984. Detection of a membrane-associated protein on detached membrane ribosomes in Staphylococcus aureus. J. Gen. Microbiol. 130:1673-1682. [DOI] [PubMed] [Google Scholar]
- 2.Adler, L.-Å., and S. Arvidson. 1987. Correlation between the rate of exoprotein synthesis and the amount of the multiprotein complex on membrane-bound ribosomes (MBRP) in Staphylococcus aureus. J. Gen. Microbiol. 133:803-813. [DOI] [PubMed] [Google Scholar]
- 3.Adler, L.-Å., and S. Arvidson. 1988. Cloning and expression in Escherichia coli of genes encoding a multiprotein complex involved in excretion of proteins from Staphylococcus aureus. J. Bacteriol. 170:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alemohammad, M. M., and C. J. Knowles. 1974. Osmotically induced volume and turbidity changes of Escherichia coli due to salts, sucrose and glycerol, with particular reference to the rapid permeation of glycerol into the cell. J. Gen. Microbiol. 82:125-142. [DOI] [PubMed] [Google Scholar]
- 5.Allen, A. G., and R. N. Perham. 1991. Two lipoyl domains in the dihydrolipoamide acyltransferase chain of the pyruvate dehydrogenase multienzyme complex of Streptococcus faecalis. FEBS Lett. 287:206-210. [DOI] [PubMed] [Google Scholar]
- 6.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ames, G. F.-L., and K. Nikaido. 1976. Two-dimensional gel electrophoresis of membrane proteins. Biochemistry 15:616-623. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong-Buisseret, L., M. B. Cole, and G. S. A. B. Stewart. 1995. A homologue to the Escherichia coli alkyl hydroperoxide reductase AhpC is induced by osmotic upshock in Staphylococcus aureus. Microbiology 141:1655-1661. [DOI] [PubMed] [Google Scholar]
- 9.Bae, J.-H., S. H. Anderson, and K. J. Miller. 1993. Identification of a high-affinity glycine betaine transport system in Staphylococcus aureus. Appl. Environ. Microbiol. 59:2734-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates, D. L., M. J. Danson, G. Hale, E. A. Hooper, and R. N. Perham. 1977. Self-assembly and catalytic activity of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Nature 268:313-316. [DOI] [PubMed] [Google Scholar]
- 11.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson, D. A., M. S. Boguski, D. J. Lipman, J. Ostell, B. F. F. Oulette, B. A. Rapp, and D. L. Wheeler. 1999. GenBank. Nucleic Acids Res. 27:12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christian, J. H. B. 1981. Specific solute effects on microbial water relations, p. 825-854. In L. B. Rockland and G. F. Stewart (ed.), Water activity: influences on food quality. Academic Press, New York, N.Y.
- 15.Daniels, L., R. S. Hanson, and J. A. Phillips. 1994. Chemical analysis, p. 512-554. In P. Gerhardt, R. G. A. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 16.Esener, A., G. Bol, N. Kossen, and J. A. Roels. 1981. Effect of water activity on microbial growth, p. 339-344. In M. Moo-Young, C. W. Robinson, and C. Vezina (ed.), Advances in biotechnology, vol. 1. Pergamon Press, Toronto, Canada. [Google Scholar]
- 17.Gould, G. W. 1985. Present state of knowledge of aw effects on microorganisms, p. 229-245. In D. Simatos and J. L. Multon (ed.), Properties of water in foods in relation to quality and stability. Martinus Nijhoff Publishers, Dordrecht, The Netherlands.
- 18.Hames, B. D. 1981. An introduction to polyacrylamide gel electrophoresis, p. 1-91. In B. D. Hames and D. Rickwood (ed.), Gel electrophoresis of proteins. A practical approach. IRL Press, Oxford, United Kingdom.
- 19.Hemilä, H., A. Palva, L. Paulin, L.-Å. Adler, S. Arvidson, and I. Palva. 1991. The secretory S complex in Bacillus subtilis is identified as pyruvate dehydrogenase. Res. Microbiol. 142:779-785. [DOI] [PubMed] [Google Scholar]
- 20.Hemilä, H., A. Palva, L. Paulin, S. Arvidson, and I. Palva. 1990. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J. Bacteriol. 172:779-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krist, K. A., T. Ross, and T. A. McMeekin. 1998. Final optical density and growth rate; effects of temperature and NaCl differ from acidity. Int. J. Food Microbiol. 43:195-203. [DOI] [PubMed] [Google Scholar]
- 22.Li, K.-Y., and J. A. Torres. 1993. Water activity relationships for selected mesophiles and psychrotrophs at refrigeration temperature. J. Food Prot. 56:612-615. [DOI] [PubMed] [Google Scholar]
- 23.Markwell, M. A., S. M. Haas, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 24.Marshall, B. J., D. F. Ohye, and J. H. B. Christian. 1971. Tolerance of bacteria to high concentrations of NaCl and glycerol in the growth medium. Appl. Microbiol. 21:363-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, K. J., S. C. Zelt, and J.-H. Bae. 1991. Glycine betaine and proline are the principal compatible solutes of Staphylococcus aureus. Curr. Microbiol. 23:131-137. [Google Scholar]
- 26.Moore, D. 1992. Preparation and analysis of DNA, p. 2-1-2-38. In E. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology, 2nd ed. John Wiley & Sons, New York, N.Y.
- 27.Oren, A. 1999. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63:334-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plitman, M., Y. Park, R. Gomez, and A. J. Sinskey. 1973. Viability of Staphylococcus aureus in the intermediate moisture range. J. Food Sci. 38:1004-1008. [Google Scholar]
- 29.Pourkomailian, B., and I. R. Booth. 1992. Glycine betaine transport by Staphylococcus aureus: evidence for two transport systems and for their possible roles in osmoregulation. J. Gen. Microbiol. 138:2515-2518. [DOI] [PubMed] [Google Scholar]
- 30.Pourkomailian, B., and I. R. Booth. 1994. Glycine betaine transport by Staphylococcus aureus: evidence for feedback regulation of the activity of the two transport systems. Microbiology 140:3131-3138. [DOI] [PubMed] [Google Scholar]
- 31.Qoronfleh, M. W., U. N. Streips, and B. J. Wilkinson. 1990. Basic features of the staphylococcal heat shock response. Antonie Leeuwenhoek 58:79-86. [DOI] [PubMed] [Google Scholar]
- 32.Schmid, R., J. Bernhardt, H. Antelmann, A. Völker, H. Mach, U. Völker, and M. Hecker. 1997. Identification of vegetative proteins for a two-dimensional protein index of Bacillus subtilis. Microbiology 143:991-998. [DOI] [PubMed] [Google Scholar]
- 33.Scott, W. J. 1953. Water relations of Staphylococcus aureus at 30°C. Aust. J. Biol. Sci. 6:549-564. [PubMed] [Google Scholar]
- 34.Seals, J. R., and L. Jarett. 1980. Activation of pyruvate dehydrogenase by direct addition of insulin to an isolated plasma membrane/mitochondria mixture: evidence for generation of insulin's second messenger in a subcellular system. Proc. Natl. Acad. Sci. USA 77:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, J. L., M. M. Bencivengo, and C. A. Kunsch. 1986. Enterotoxin A synthesis in Staphylococcus aureus: inhibition by glycerol and maltose. J. Gen. Microbiol. 132:3375-3380. [DOI] [PubMed] [Google Scholar]
- 36.Sprott, G. D., S. F. Koval, and C. A. Schnaitman. 1994. Cell fractionation, p. 72-103. In P. Gerhardt, R. G. A. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 37.Sreeramulu, G., and N. K. Singh. 1995. Destaining of Coomassie brilliant blue R-250-stained polyacrylamide gels with sodium chloride solutions. Electrophoresis 16:362-365. [DOI] [PubMed] [Google Scholar]
- 38.Stimeling, K. W., J. E. Graham, A. Kaenjak, and B. J. Wilkinson. 1994. Evidence for feedback (trans) regulation of, and two systems for, glycine betaine transport by Staphylococcus aureus. Microbiology 140:3139-3144. [DOI] [PubMed] [Google Scholar]
- 39.Townsend, D. E., and B. J. Wilkinson. 1992. Proline transport in Staphylococcus aureus: a high-affinity system and a low-affinity system involved in osmoregulation. J. Bacteriol. 174:2702-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Troller, J. A., and J. V. Stinson. 1978. Influence of water activity on the production of extracellular enzymes by Staphylococcus aureus. Appl. Environ. Microbiol. 35:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueguchi, C., N. Misonou, and T. Mizuno. 2001. Negative control of rpoS expression by phosphoenolpyruvate:carbohydrate phosphotransferase system in Escherichia coli. J. Bacteriol. 183:520-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijaranakul, U., M. J. Nadakavukaren, D. O. Bayles, B. J. Wilkinson, and R. K. Jayaswal. 1997. Characterization of an NaCl-sensitive Staphylococcus aureus mutant and rescue of the NaCl-sensitive phenotype by glycine betaine but not by other compatible solutes. Appl. Environ. Microbiol. 63:1889-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vijaranakul, U., M. J. Nadakavukaren, B. L. M. de Jonge, B. J. Wilkinson, and R. K. Jayaswal. 1995. Increased cell size and shortened peptidoglycan interpeptide bridge of NaCl-stressed Staphylococcus aureus and their reversal by glycine betaine. J. Bacteriol. 177:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vijaranakul, U., A. Xiong, K. Lockwood, and R. K. Jayaswal. 1998. Cloning and nucleotide sequencing of a Staphylococcus aureus gene encoding a branched-chain-amino-acid transporter. Appl. Environ. Microbiol. 64:763-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Vilhelmsson, O., and K. J. Miller. Humectant permeability influences growth and compatible solute uptake by Staphylococcus aureus subjected to osmotic stress J. Food Prot., in press. [DOI] [PubMed]
- 45.Völker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Völker, R. Schmid, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741-752. [DOI] [PubMed] [Google Scholar]
- 46.Wengender, P. A., and K. J. Miller. 1995. Identification of a PutP proline permease gene homolog from Staphylococcus aureus by expression cloning of the high-affinity proline system in Escherichia coli. Appl. Environ. Microbiol. 61:252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson, B. J. 1997. Biology, p. 1-38. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.