Abstract
Bioregenerative life support systems may be necessary for long-term space missions due to the high cost of lifting supplies and equipment into orbit. In this study, we investigated two biological wastewater treatment reactors designed to recover potable water for a spacefaring crew being tested at Johnson Space Center. The experiment (Lunar-Mars Life Support Test Project—Phase III) consisted of four crew members confined in a test chamber for 91 days. In order to recycle all water during the experiment, an immobilized cell bioreactor (ICB) was employed for organic carbon removal and a trickling filter bioreactor (TFB) was utilized for ammonia removal, followed by physical-chemical treatment. In this study, the spatial distribution of various microorganisms within each bioreactor was analyzed by using biofilm samples taken from four locations in the ICB and three locations in the TFB. Three target genes were used for characterization of bacteria: the 16S rRNA gene for the total bacterial community, the ammonia monooxygenase (amoA) gene for ammonia-oxidizing bacteria, and the nitrous oxide reductase (nosZ) gene for denitrifying bacteria. A combination of terminal restriction fragment length polymorphism (T-RFLP), sequence, and phylogenetic analyses indicated that the microbial community composition in the ICB and the TFB consisted mainly of Proteobacteria, low-G+C gram-positive bacteria, and a Cytophaga-Flexibacter-Bacteroides group. Fifty-seven novel 16S rRNA genes, 8 novel amoA genes, and 12 new nosZ genes were identified in this study. Temporal shifts in the species composition of total bacteria in both the ICB and the TFB and ammonia-oxidizing and denitrifying bacteria in the TFB were also detected when the biofilms were compared with the inocula after 91 days. This result suggests that specific microbial populations were either brought in by the crew or enriched in the reactors during the course of operation.
A mission of the Advanced Life Support (ALS) Project is to develop bioregenerative life support systems to enhance the International Space Station and ultimately sustain a lunar base or a voyage to Mars. This necessitates creating a combination of physicochemical and biological systems that can regenerate resources (or materials) needed to sustain life for long time periods, possibly without the potential of resupply. Since bacteria perform recycling of various elements on Earth, it is reasonable to assume that a bioregenerative ALS system for a space vehicle or colony may also rely on bacterial activity. Understanding the microbial ecology in a biological treatment system is important for designing and operating it at optimal efficiency and to ensure reliability. In 1997, two fixed-film biological wastewater treatment reactors were tested by the National Aeronautics and Space Administration (NASA) at the Johnson Space Center (JSC) during a 91-day simulated long-term manned space mission. This experiment (Lunar-Mars Life Support Test Project—Phase III) consisted of four crew members confined in a test chamber. The system utilized an immobilized cell bioreactor (ICB) for organic carbon removal and a trickling filter bioreactor (TFB) for ammonia removal, supplemented by additional physicochemical devices for reclaiming potable water.
In this report, we characterize the microbial community structures in the ICB and the TFB. Previously, many studies have demonstrated the existence of diverse groups of microorganisms in activated sludge and in fixed-film treatment systems (7, 8, 14, 20, 22, 23, 28, 31). However, the Phase III test at JSC represents a unique opportunity in which the microbial populations responsible for treating the wastewater (and a human crew) were allowed to develop in isolation for a 3-month period. The major questions addressed in this research include (i) which microorganisms were present in the JSC bioreactors and whether they differ from those previously reported and (ii) which microorganisms, if any, were brought in by the crew or enriched during the course of the Phase III test.
To address these questions, we characterized the total, ammonia-oxidizing, and denitrifying bacterial communities in the system by using three target genes, i.e., the 16S rRNA, ammonia monooxygenase (amoA), and nitrous oxide reductase (nosZ) genes, respectively. The amoA gene was chosen to identify bacteria responsible for ammonia removal (24, 25), and the nosZ gene (26, 27) was used to detect bacteria responsible for loss of fixed nitrogen from the wastewater treatment systems. These two functional groups (nitrifiers and denitrifiers) are essential for waste processing and resource recovery of nitrogen. A functional gene approach was selected for the N-cycling groups because of concerns regarding specificity when a 16S rRNA gene approach is used for nitrifiers (24) or because the ability to denitrify is not limited to specific taxonomic groups (32). This analysis, in conjunction with physical and chemical data obtained during the Phase III test, may help improve the design and performance of biological wastewater treatment reactors for the ALS Project.
MATERIALS AND METHODS
The wastewater treatment system consisted of six subsystems, including two fixed-film bioreactors (Fig. 1), which are the target of this study, and four physicochemical reactors. The reactor structures and operating conditions are shown in Fig. 1 and Tables 1 and 2 (see reference 19 for additional information). The system was designed to process 110.6 kg of wastewater generated by the crew per day. The average total organic carbon (TOC) and ammonia removal percentages were 94 and 48%, respectively, during the manned portion of the test. The water supply was recycled 10 times during the test without any additional resupply.
FIG. 1.
Schematic of biological wastewater treatment reactors for Phase III test. Table 2 contains the wastewater characteristics at various places within the reactors indicated by the numbers. P/C, physicochemical treatment.
TABLE 1.
Reactor specifications and operating conditions of the ICB and TFB
| System | Vol (m3) | Diam (m) | Ht (m) | Feed flow rate (ml/min) | Recycling flow rate (ml/min)a | Hydraulic retention time (min) | TOC loading rate (mg/liter · h) |
|---|---|---|---|---|---|---|---|
| ICB | 0.11 | 0.254 | 2.18 | 75 | 150 | 1,467 (489)b | 675 |
| TFB | 0.07 | 0.254 | 1.45 | 75 | 10,000 | 933 (6.9) | 3.39 |
From TFB effluent. The high rate in the TFB was used to minimize gradients and ensure aerobic conditions within the reactor.
Nominal, based on empty bed volume; time for a single pass is in parentheses.
TABLE 2.
Average wastewater characteristics of the ICB and TFBa
| Stage | TOC (mg/liter) | NH4+ N (mg/liter) | NO2− N (mg/liter) | NO3− N (mg/liter) | pH | Alkalinity (meq/liter) |
|---|---|---|---|---|---|---|
| 1. Wastewater | 247 | 150 | Trace | Trace | 8.44 | 11.7 |
| 2. ICB influent | 150 | 153 | 14 | 11 | 8.58 | 10.0 |
| 3. ICB postaeration | 93 | 143 | 5 | 6 | 8.57 | 10.3 |
| 4. ICB effluent/TFB influent | 19 | 143 | 12 | 12 | 7.66 | 8.2 |
| 5. TFB effluent | 12 | 90 | 35 | 32 | 6.27 | 1.3 |
ICB Influent is a mixture of TFB effluent, recycled to the front of the process, and wastewater. The number for each stage (superscript) corresponds to that in Fig. 1.
ICB.
An ICB was constructed by using an acrylic cylinder (25.4-cm inside diameter, 218-cm height) filled with 60 acrylic plates held 2.54 cm apart by spacers (Tables 1 and 2). Each plate was covered on both sides by a porous polymer support for microbial colonization. Wastewater entered the bottom of the reactor, and air was injected at a rate of 1.5 liters/min at a distance 16.5 cm from the bottom in an upflow cocurrent manner. This bottom layer was designed to promote denitrification, followed by aerobic carbon oxidation in the remainder of the column. The influent of the ICB was the mixture of wastewater from a feed tank and a portion of the TFB effluent (see below).
TFB.
A TFB was also constructed by using an acrylic cylinder (25.4-cm inside diameter, 145-cm height) packed with alternating layers of 2.54-cm polypropylene Pall rings and 6-mm ceramic saddles to support microbial growth (Tables 1 and 2). The influent to the TFB consisted of the ICB effluent and recycled water from the TFB itself. A total of 2 liters of air per min was distributed throughout the TFB by injection at three locations: an aeration cup, the bottom of the reactor, and an aeration tank (Fig. 1). The effluent from the TFB was recycled into the TFB and ICB or processed in a series of physicochemical treatments and stored for human consumption.
Bioreactor inoculation, operation, and sampling.
The ICB for organic carbon removal was inoculated with a commercial microbial consortium (DBC Plus Type ABH; Enviroflow, Inc., Manassas, Va.) on 23 August 1997 and acclimatized for 4 days. On 27 August, the ICB began receiving wastewater (shower, handwash, and urine) donated by personnel at JSC, as well as simulated humidity condensate. From 19 September to 19 December (91 days), the ICB reactor received wastewater generated by the four crew members enclosed in a sealed test chamber. The sources of wastewater were laundry, shower, handwash, oral hygiene, urine, and humidity condensate. The reactor was continuously operated throughout the phase III test at a nominal influent flow rate of 75 ml of wastewater per min and 150 ml of recycled water per min from the TFB. The average TOC concentration measured was 294 mg/liter in the feed tank, 180 mg/liter at the inlet to the ICB, and 17 mg/liter in the ICB effluent during the manned portion of the test. The concentration of organic carbon at the inlet to the ICB was lower than that in the wastewater collection tank due to the effect of dilution by the recycled water.
The TFB for ammonium removal was inoculated with a commercial nitrifying bacterial consortium (DBC Plus NitroTreat 20; Enviroflow) and acclimatized for 10 days. The influent was a mixture of the effluent from the ICB at 225 ml/min with the recycled flow from the aeration tank below the TFB outlet at 10 liters/min. (The high recycling rate provided a uniform distribution of wastewater throughout the reactor.) The ammonium N concentration ranged from 59 to 370 mg/liter in the influent mixture with an average concentration of 87 mg/liter in the TFB effluent.
DNA extraction and PCR amplification.
Genomic DNA was extracted from biofilm samples by a phenol-chloroform procedure described in reference 25 with eight freeze-thaw cycles. Between 100 and 800 mg of biofilm or commercial inoculum was used to extract genomic DNA. For 16S rRNA gene amplification, genomic DNA (<10 ng) was amplified with eubacterial universal primers 27F and 1525R (13) as described previously (25). For ammonia monooxygenase (amoA) genes, amplification was done with primers A189 and A682 under the conditions described in reference 9. For the nitrous oxide reductase (nosZ) genes, primers Nos661F and Nos1773R were used as described previously (26, 27).
Cloning and sequence analysis.
Cloning of the various target genes was done with the CLONEAMP pAMP1 system (Life Technologies, Gaithersburg, Md.) in accordance with the manufacturer's instructions. A total of 480 clones for 16S rRNA genes (four libraries from the ICB and one library from the TFB), 288 clones for amoA genes (two libraries from the ICB and one library from the TFB), and 288 clones for nosZ genes (two libraries from the ICB and one library from the TFB) were analyzed in this study. Clonal libraries were not created for the inocula. Plasmid purification of recombinant clones for sequence analysis was performed with a FlexiPrep Kit (Amersham Pharmacia Biotech, Piscatway, N.J.), and sequence data were generated with BigDye chemistry on an ABI 310 genetic analyzer (Perkin-Elmer, Foster City, Calif.). Identification of 16S sequence similarities of the small subunit (SSU) of rRNA utilized the Ribosomal Database Project (16). The BLASTN (1) and FASTA (21) programs were used for preliminary amoA and nosZ gene sequence similarity analyses, and a distance matrix with Jukes-Cantor correction was generated to examine sequence similarity among the pure cultures and uncultured microorganisms at the nucleic acid and amino acid levels. Phylogenetic tree reconstruction utilized both the maximum-likelihood (5) and neighbor-joining methods in the genetic data environment (30).
T-RFLP fingerprinting.
For terminal restriction fragment length polymorphism (T-RFLP) analysis, a fluorescent dye was incorporated on the 5′ primer for each target gene (2, 4, 15, 17, 18). The fluorescently labeled product (ca. 50 ng) was digested with MnlI (New England Biolabs, Beverly, Mass.) for 16S rRNA as described previously (6, 12) with TaiI (MBI Fermantas, Amherst, N.Y.) for amoA and with HinpI (New England Biolabs) for nosZ fingerprints (27). (The restriction endonucleases were selected to give maximal resolution of terminally labeled fragments to known target sequences in the database.) All digestions were done in accordance with the manufacturer's instructions for 12 h. After ethanol precipitation, T-RFLP analysis was also carried out on an ABI 310 genetic analyzer with Genescan software and an internal size standard. The comparative T-RFLP data analyses were carried out by using the Combinatorial Polythetic Agglomerative Hierarchical clustering package (3; http://www.es.umb.edu/edgwebp.htm).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been deposited in the GenBank database and assigned accession numbers AF390906 to AF390962 and AF390116 to AF390135.
RESULTS
T-RFLP analysis of the reactors.
Initial characterization of the microbial communities at various places in the reactors relied on T-RFLP fingerprinting. Fluorescent amplifications were successful for nearly all of the samples, with the exception of amoA and nosZ for the ICB inoculum. It may be that ammonia-oxidizing and denitrifying bacteria were a small portion of the total community in this sample and were below the detection limit of the PCR assay (17, 18). An alternative explanation (PCR inhibitors) was not deemed likely given the 16S rRNA amplifications from the ICB inoculum did not demonstrate appreciable product suppression, and numerous attempts to eliminate any such inhibitors were unsuccessful.
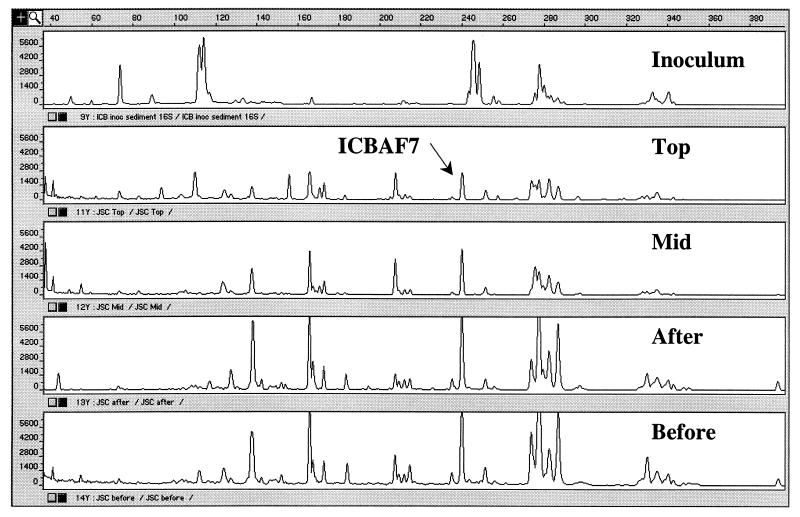
In the T-RFLP fingerprints of the microbial communities, total peak numbers varied by a factor of 5 (range, 28 to 129) for SSU genes and by a factor of 3 for the functional genes (ranges, 3 to 8 for amoA and 19 to 57 for nosZ) (Table 3). The middle of the ICB contained the lowest number of T-RFLP peaks for the SSU and nosZ genes, while the anaerobic part of the ICB (before) harbored the lowest number of amoA peaks. In the TFB, the bottom and middle regions also enclosed the lowest numbers of SSU genes and amoA and nosZ genes, respectively. Not only were changes in absolute numbers of peaks discernible, but marked differences in profiles could also be seen between the inoculum and the various biofilm samples taken throughout the reactor at the end of the test (Fig. 2). Here, a dominant T-RFLP peak is shown that corresponds to a clone (ICBAF7) not found in the inoculum (Table 4; see below). For the nitrifying population, the observed peaks in the inoculum were only of the lengths associated with Nitrosomonas-like amoA genes (177 and 282 bp), while those in the reactor represented both Nitrosomonas- and Nitrosospira-like amoA gene peaks (453 bp) (data not shown). Differences in the nosZ fingerprints between inocula and samples were observed. Some nosZ peaks matched the following sequences in the database: Pseudomonas sp.-Sinorhizobium meliloti (152 bp), Achromobacter cycloclastes-Bradyrhizobium japonicum (199 bp), Paracoccus denitrificans (153 bp), and Rhodobacter sphaeroides (203 bp).
TABLE 3.
Number of terminal restriction fragment peaks of three target genes in the two bioreactors
| Bioreactor layer | No. of terminal restriction fragment peaks
|
||
|---|---|---|---|
| 16S rRNA genes | amoA genes | nosZ genes | |
| ICB inoculum | 129 | NAa | NA |
| Top | 75 | 8 | 56 |
| Middle | 62 | 7 | 43 |
| After air injection | 110 | 5 | 50 |
| Before air injection | 111 | 4 | 57 |
| TFB inoculum | 43 | 3 | 38 |
| Top | 35 | 7 | 30 |
| Middle | 35 | 5 | 19 |
| Bottom | 28 | 5 | 34 |
NA, not available due to the unsuccessful PCR amplification.
FIG. 2.
T-RFLP profiles of 16S rRNA genes from biofilm samples of the ICB after digestion with MnlI. A unique peak (clone ICBAF7) not detectable in the inoculum but found in the final samples is indicated.
TABLE 4.
Comparison of the 16S rRNA genes from the clonal libraries in the ICB to the T-RFLP fingerprint of the inoculum and to sequences in the Ribosomal Database Project
| Taxon and clone | Peak length (bp)a | Most similar organism | Sabb | Ic | Td | Md | Ad | Bd |
|---|---|---|---|---|---|---|---|---|
| Proteobacteria | ||||||||
| Alpha group | ||||||||
| ICBTD9 | 124 | Holospora obtusa | 0.716 | + | ++ | −+ | −+ | −+ |
| ICBTE8 | 211 | Sphingomonas sp. strain BRW2 | 0.773 | + | ++ | −+ | −+ | −+ |
| ICBMF2 | 288 | Marine snow-associated bacterium Adriatic83 | 0.712 | + | −+ | ++ | −+ | −+ |
| Beta group | ||||||||
| ICBBA5 | 132 | Diazotroph strain BA27 | 0.938 | + | −− | −− | −+ | +− |
| ICBTA10 | 164 | Thauera terpenica strain 58 | 0.918 | + | ++ | ++ | −+ | ++ |
| ICBTC9 | 215 | Brachymonas denitrificans strain AS-P1 | 0.748 | + | ++ | −+ | −+ | −+ |
| ICBBD2 | 214 | B. denitrificans strain AS-P1 | 0.704 | + | −+ | −+ | −+ | ++ |
| ICBBB10 | 208 | Unidentified beta proteobacterium sewCLONE2 | 0.836 | + | −+ | −+ | −+ | ++ |
| ICBBD7 | 277 | Type 0803 filamentous bacterium strain Ben04Be | 0.799 | + | −+ | −+ | −+ | ++ |
| ICBTF8 | 276 | Unidentified bacterium strain rA1e | 0.824 | + | ++ | −+ | −+ | −+ |
| ICBTF7 | 276 | Uncultured bacterium clone KC208 | 0.784 | + | ++ | −+ | −+ | −+ |
| ICBBC5 | 279 | Unidentified bacterium strain rA1e | 0.789 | + | −+ | −+ | −+ | ++ |
| ICBBA10 | 279 | Uncultured bacterium clone KC208 | 0.792 | + | −+ | −+ | −+ | ++ |
| ICBMD10 | 278 | Strain SBR 2090e | 0.864 | + | −+ | ++ | −+ | −+ |
| ICBAA12 | 278 | Uncultured bacterium SJA-10 | 0.717 | + | ++ | −+ | ++ | −+ |
| Gamma group | ||||||||
| ICBMF3 | 54 | Acinetobacter lwoffii strain A391 | 0.837 | + | ++ | ++ | −+ | −+ |
| ICBTE5 | 90 | Iron-oxidizing lithotroph ES-1 | 0.664 | + | ++ | −− | −+ | −+ |
| ICBBE7 | 100 | Denitrifying Fe(II)-oxidizing bacterium strain BrG3 | 0.766 | + | ++ | −− | −+ | ++ |
| ICBTB11 | 126 | Serratia fonticola strain DSM4576 | 0.900 | + | ++ | ++ | −+ | −+ |
| Epsilon group | ||||||||
| ICBBA6 | 136 | Arcobacter cryaerophilus ATCC 49615 | 0.839 | + | −+ | −+ | −+ | ++ |
| ICBAA7 | 139 | Unidentified bacterium clone T55a | 0.890 | + | ++ | −+ | ++ | ++ |
| ICBAF5 | 141 | A. cryaerophilus ATCC 49615 | 0.908 | + | −+ | −+ | ++ | −+ |
| ICBTE4 | 142 | A. cryaerophilus ATCC 49615 | 0.878 | + | ++ | −+ | ++ | −+ |
| ICBTA1 | 142 | A. cryaerophilus ATCC 49615 | 0.941 | + | ++ | ++ | ++ | ++ |
| ICBTG2 | 289 | Unidentified bacterium clone T55e | 0.804 | + | ++ | ++ | ++ | ++ |
| ICBTC5 | 288 | Arcobacter butzleri CCUG 10373 | 0.742 | + | ++ | −+ | −+ | −+ |
| ICBTD7 | 289 | A. butzleri CCUG 10373 | 0.803 | + | ++ | −+ | −+ | ++ |
| ICBTC7 | 289 | A. butzleri CCUG 10373 | 0.724 | + | ++ | −+ | −+ | ++ |
| Cytophagales | ||||||||
| ICBTA3 | 44 | Uncultured eubacterium WCHB1-29 | 0.621 | + | ++ | −+ | −+ | −+ |
| ICBAF7 | 241 | Uncultured eubacterium WCHB1-69 | 0.639 | − | −+ | −+ | ++ | −+ |
| Low-G+C gram-positive bacteria | ||||||||
| ICBBB9 | 125 | Clostridium hastiforme DSM 5675 | 0.731 | + | −+ | −+ | −+ | ++ |
| ICBBB7 | 133 | Lactosphaera pasteurii strain KoTa2 | 0.856 | + | −− | −− | −− | +− |
| ICBAI4 | 155 | Acetonema longum strain AP0-1 | 0.638 | + | −+ | −+ | ++ | −+ |
| ICBBC4 | 170 | Clostridium hastiforme DSM 5675 | 0.774 | + | −+ | −+ | −+ | ++ |
| ICBTA2 | 252 | Clostridium viride strain T2-7 | 0.531 | + | ++ | −+ | −+ | −+ |
| ICBTG1 | 297 | Bacterium ASF500 | 0.650 | − | ++ | −+ | −+ | −+ |
| Verrucomicrobia, ICBTG4 | 187 | Verrucomicrobiales PB90-1 | 0.635 | + | +− | +− | −− | −+ |
| Planctomycetes, ICBMG6 | 384 | Isosphaera sp. strain Schlesner 666 | 0.646 | − | −− | +− | −− | −− |
| Unclassified, ICBMG3 | 187 | Unidentified eubacterium | 0.801 | + | −− | +− | −− | −+ |
Base pair length of terminal restriction fragment peak expected based on sequence.
Sab value in the Ribosomal Database Project database; a value of 0.9 or greater indicates a close match to a sequence in the database.
I, inoculum. No clonal library was generated. A plus or minus indicates the presence or absence of a terminal restriction fragment peak at this length.
T: top; M: middle; A, after air injection; B, before air injection (layers in the ICB). Within each pair, plus or minus on the left means that sequence is present or absent in the clonal library; a plus or minus on the right indicates the presence or absence of a terminal restriction fragment peak at this length.
Sequence from activated sludge reported by other researchers.
Cluster analysis of the various T-RFLP fingerprints verified much of this visual inspection. The inocula always grouped outside the samples from the various places within the reactor (Fig 3) for all three target genes. This result suggests that many members of the community are lost during reactor operation or that most of the microbial inhabitants of the reactors are induced from undetectable levels in the inocula or introduced in the wastewater feed. Total communities based on 16S T-RFLP patterns demonstrated similarities ranging from 35 to 75% (Fig. 3) for the ICB and from 28 to 67% for the TFB, with much of the difference in similarity based on very small peaks (data not shown). The clustered profiles of the 16S rRNA genes demonstrated a separation between the bacterial communities in the ICB and the TFB, implying that different microbial populations had developed despite the recycling of effluent between the reactors.
FIG. 3.
Cluster analyses of the T-RFLP profiles in the ICB and TFB based on the 16S rRNA, amoA, and nosZ genes. The samples from the ICB are indicated by the following suffixes: I, inoculum; T, top layer; M, middle layer; A, after-air injection layer; B, before-air injection layer. For the TFB, the following similar suffixes are used: T, top layer; M, middle layer; B, bottom layer. The cluster analyses were performed with the Combinatorial Polythetic Agglomerative Hierarchical clustering package (COMPAH96; http://www.es.umb.edu/edgwebp.htm).
The distributions of the ammonia-oxidizing bacteria based on the amoA profiles also showed a clustering pattern similar to that of the 16S rRNA genes. For the ICB, the after- and before-air injection layers clustered together (89%) and the top and middle layers clustered together (80%), with the three TFB samples also clustering (83%) (Fig. 3). In the before- and after-air injection zones of the ICB and all three zones of the TFB, the peaks of both Nitrosomonas-like and Nitrosospira-like amoA genes were identified; however, in the top and middle parts of the ICB, only Nitrosomonas-like amoA genes were found (data not shown). The similarity values of nosZ were lower than those of the other two target genes, ranging from 47 to 58% for the ICB and from 27 to 34% for the TFB. In addition, the T-RF peak profiles clustered differently from the other two genes. In the ICB, the middle-layer nosZ populations grouped with the before and after samples rather than with the top layer sample. In the TFB, the top and bottom samples grouped together and the middle sample was more distant.
Clonal analysis of 16S rRNA genes.
To gain a better understanding of the microbial populations in the reactors, clonal libraries of 16S rRNA genes were constructed from all four layers of the ICB and from the middle layer of the TFB as described previously (12, 25). Due to the similarities of the T-RFLP profiles, only two samples from the ICB and the middle sample from the TFB were analyzed for amoA or nosZ genes. Of the 384 16S rRNA clones obtained from the ICB, 39 unique partial 16S rRNA gene sequences (541 bp) were identified. Comparison of these ICB SSU sequences to the Ribosomal Database Project database indicated low homology, with similarity (Sab) values ranging from 0.621 to 0.941 (Table 4), suggesting that many of the SSU genes found in the ICB have not been described in previous studies of waste treatment systems. Only five sequences showed high levels of homology (Sab values of >0.9) to sequences in the database. A total of 28 SSU clones were affiliated with the Proteobacteria. Low-G+C gram-positive and Cytophagales-like 16S rRNA gene sequences were also detected in many of the libraries. No distinct spatial distribution patterns were discerned based on the SSU genes in the different layers of the ICB.
In the TFB middle layer, analysis of 96 clones revealed 18 unique 16S rRNA gene sequences (Table 5). A total of 14 proteobacterium-like 16S rRNA gene sequences were detected (Sab values of 0.791 to 0.973), with 12 demonstrating high levels of similarity (Sab values of >0.9) to sequences in the Ribosomal Database Project. The remaining six clonal sequences were loosely related to the Cytophaga-Flexibacter-Bacteroides group, Planctomycetales, and Nitrospira.
TABLE 5.
Comparison of the 16S rRNA genes from the middle layer of the TFB to the T-RFLP fingerprints from the inoculum and other parts of the reactor and sequences in the Ribosomal Database Project
| Taxon and clone | Peak length (bp)a | Most similar organism | Sabb | Ic | Tc | Mc | Bc |
|---|---|---|---|---|---|---|---|
| Proteobacteria | |||||||
| Alpha group TFBMC2 | 252 | Ochrobactrum tritici strain SCII24 | 0.973 | +d | − | + | + |
| Beta group | |||||||
| TFBMD9 | 208 | Dechlorisoma suila strain PS | 0.935 | + | + | + | + |
| TFBMF4 | 165 | Unidentified bacterium strain rJ13e | 0.833 | + | + | + | + |
| TFBME9 | 164 | Ralstonia eutropha strain CH34 | 0.931 | + | + | + | + |
| TFBMA4 | 219 | Alcaligenes sp. strain NKNTAU | 0.904 | − | − | − | − |
| TFBMA5 | 132 | Diazotroph strain BA27 | 0.938 | − | − | − | − |
| Gamma group | |||||||
| TFBMH1 | 126 | Enterobacter agglomerans strain AH16 | 0.964 | − | − | + | + |
| TFBMF5 | 126 | Enterobacter sp. | 0.933 | − | − | + | + |
| TFBMB11 | 126 | Enterobacter agglomerans strain AH16 | 0.791 | − | − | + | + |
| TFBMH12 | 126 | Enterobacter agglomerans strain AH16 | 0.907 | − | − | + | + |
| TFBMH2 | 101 | Stenotrophomonas nitritireducens strain L2 | 0.925 | − | − | − | − |
| TFBME10 | 237 | Unidentified gamma proteobacterium strain R2A30 | 0.962 | − | − | − | − |
| TFBME4 | 279 | Pseudomonas sp. strain CRE 12 | 0.937 | − | − | − | − |
| TFBMC1 | 279 | Pseudomonas sp. strain CRE12 | 0.905 | − | − | − | − |
| Cytophagales, TFBMF3 | 97 | Unidentified bacterium Riz1026 | 0.708 | − | − | − | − |
| Nitrospira, TFBMG11 | 50 | Unidentified eubacterium | 0.802 | + | + | + | + |
| Planctomycetes | |||||||
| TFBME6 | 111 | Gemmata obscuriglobus UQM446 | 0.554 | − | + | − | − |
| TFBMI5 | 386 | Isosphaera sp. strain Schlesner 666 | 0.659 | − | − | − | − |
Length of terminal restriction fragment peak.
Sab value in the Ribosomal Database Project; a value of 0.9 or greater indicates a close match to a sequence in the database.
I, inoculum; T, top; M, middle; A, after air injection; B, before air injection (layers in the ICB).
A plus indicates that a terminal restriction fragment peak was found at this length; a minus indicates that none found.
Sequence from activated sludge reported by other researchers.
Ammonia-oxidizing bacteria in the ICB and TFB based on the amoA genes.
Among the 288 clones, a total of 27 different amoA genes (14 in the ICB and 13 in the TFB) were observed. Most shared high levels of sequence similarity to sequences in the database, ranging from 99.1% for Nitrosospira sp. strain Np39-19 to 100% for N. europaea and were considered proof that these nitrifying microorganisms inhabited the bioreactors. However, four novel amoA genes were obtained that were more distantly related to amoA genes in the database. Two sequences (clones ICBTG2 and ICBBA2) appeared to be affiliated with the N. europaea cluster as defined in references 22 and 29 (Fig. 4), showing 93.5 and 93.9% similarity at the nucleotide level and 98.8 and 98.2% similarity at the amino acid level to N. europaea, respectively. Two other novel sequences (clones ICBBC11 and ICBBG2) grouped with the Nitrosospira multiformis lineage, with 92.7 and 97.1% similarity at the nucleotide level and 97.1% similarity for both at the amino acid level to N. multiformis, respectively. No clones were affiliated with the Nitrosomonas eutropha or other Nitrosospira branches (e.g., N. briensis and N. tenuis).
FIG. 4.
Phylogenetic tree based on the partial amoA genes (513-bp alignment) in the ICB and TFB reconstructed by the neighbor-joining method. Nodes with more than 50 bootstrap iterations (of 100) are highlighted by circles. Clonal sequences obtained in this study are highlighted. The accession numbers of the amoA genes are as follows: Nitrosomonas europaea, L08050; Nitrosomonas eutropha, U51630; Nitrosomonas sp., AB031869; Nitrosospira briensis, U76553; Nitrosospira multiformis, AF042171; Nitrosospira multiformis, U89833; Nitrosospira sp. strain AHB1, X90821; Nitrosospira sp. strain Np39-19, AF006692; Nitrosospira sp. strain NpAV, AF016003; Nitrosospira tenuis, U76552; Nitrosococcus oceani, U96611; bioreactor samples with an ABIA prefix, AF070983 to AF070987.
In the TFB, a total of four unique amoA sequences (TFBMH12, TFBME3, TFBMH4, and TFBMB10) were identified (Fig. 4). The clone TFBMH12 sequence had 94.5 and 99.4% identity with the N. europaea amoA gene at the nucleotide and amino acid levels, respectively. The other three sequences were close to the N. multiformis amoA gene, with similarities ranging from 89.9 to 92.1% and from 96.4 to 97.7% at the nucleotide and amino acid levels, respectively. The clone TFBMB10 sequence, however, appeared to be more closely related to the clone ABIAC5 sequence (97.3% identity at the nucleotide level and 98.8% identity at the amino acid level), which was present in an ammonia biofilter reported previously (25). Many of the amoA genes found in these bioreactors clustered at the periphery of the known groups of nitrifying bacteria.
Denitrifying bacteria in the ICB and TFB based on the nosZ genes.
In the ICB, nine novel partial nosZ genes (489 bp) were obtained from the clonal libraries generated from the top and before-air injection layers, and their affiliation is shown in a reconstructed phylogenetic tree in Fig. 5. Various degrees of similarity were observed between the clonal nosZ gene sequences from the bioreactors and known cultured denitrifying bacteria (Pseudomonas sp., Achromobacter sp., and Sinorhizobium sp.) and environmental nosZ sequences in the GenBank database. The similarities ranged from 76.7 to 94.2% at the nucleotide level and from 68.0 to 94.2% at the amino acid level. One nosZ sequence, ICBBE12, was nearly identical to an environmental clone from marine sediments (696O; 99.7% identity at the nucleotide level and 100% identity at the amino acid level) (26). In the TFB, three unique nosZ genes were identified in the middle-layer clonal library (Fig. 5) and the clone TFBB2 sequence appeared to be related to the P. stutzeri nosZ gene, with similarities of 89.6% (nucleotide level) and 88.9% (amino acid level). Clone TFBD2 showed sequence similarities to the P. fluorescens nosZ gene of 86.5% (nucleotide level) and 88.9% (amino acid level). Clone TFBC2 was affiliated with the P. denitrificans clade at 94.7% (nucleotide level) and 94.2% (amino acid level) similarity.
FIG. 5.
Phylogenetic tree based on the partial nosZ genes (489-bp alignment) in the ICB and TFB reconstructed by the neighbor-joining method. Nodes with more than 50 bootstrap iterations (of 100) are highlighted by circles. Designations of clonal sequences obtained in this study start with ICB or TFB. The accession numbers of the nosZ genes are as follows: Achromobacter cycloclastes, AF047429; Bradyrhizobium japonicum, AJ002531; Paracoccus denitrificans, X74792; Pseudomonas aeruginosa, X65277; Pseudomonas denitrificans, AF016059; Pseudomonas fluorescens, AF056319; Pseudomonas stutzeri, M22628; Ralstonia eutropha, X65278; Rhodobacter sphaeroides, AF125260; Sinorhizobium meliloti, U47133; 696C, AF119951; 696E, AF119949; 696L, AF119927; 696H, AF119947; ProJ, AF119934; ProO, AF119933; ProR, AF119937.
DISCUSSION
The Phase III test at JSC demonstrated an integrated biologically-physicochemically based system capable of treating and recycling wastewater for consumption by a human crew (19). This analysis was carried out to obtain baseline information on the microorganisms in the reactors associated with organic carbon degradation, nitrification (ammonia oxidation), and denitrification being developed at JSC and to test whether these microbial populations are similar to those of other wastewater treatment facilities. In this study, three eubacterial divisions were found in both bioreactors: Proteobacteria (α, β, and, γ groups), Cytophagales, and Planctomycetales. The taxonomic divisions observed in this study concur with the findings of other studies associated with wastewater (reviewed in reference 11) in finding Proteobacteria (α, β, and γ groups), Cytophagales, Planctomycetales, Verrucomicrobium, and low-G+C gram-positive bacteria. However, sequences affiliated with high-G+C gram-positive bacteria and the δ group of Proteobacteria reported in other wastewater studies were not detected in the JSC bioreactors. Conversely, sequences belonging to the ɛ group of Proteobacteria were found in the ICB in our study but not in the previous studies.
With respect to the spatial distribution of ammonia-oxidizing bacterial populations, the ICB top and middle layers contained only Nitrosomonas-like amoA genes and the before- and after-air injection sections harbored both Nitrosomonas- and Nitrosospira-like amoA genes, consistent with other reports for a bioreactor (25) and the environment (10, 24, 22, 29). The detection of Nitrosospira-like amoA may not be a surprise, in contrast to its “sporadic” detection in other wastewater treatment systems, as noted in reference 22, since biofilms may provide a greater diversity of niches compared with activated sludge. However, questions have been raised about whether a T-RFLP approach can adequately resolve amoA sequences, especially when using the Holmes priming set as we have in this study (22).
For this fingerprinting exercise, we used the enzyme TaiI, in contrast to previous work using TaqI, to distinguish amoA genes (10). Although no restriction enzyme is able to resolve all of the possible target genes for T-RFLP analysis, the TaiI enzyme can give peaks to diagnostic groups such as the Nitrosomonas-like (177 and 282 bp) and Nitrosospira-like (483 bp) groups. Improved resolution with T-RFLP approaches will require multiple digestions to fully resolve the various components of the different clusters. As for the Holmes priming set, in reference 22 it is reported that there exist up to six mismatches for members of the N. europaea-like cluster, which implies that no PCR product will be obtained when this priming set is used. However, in this and a previous study of biofilters (25), N. europaea-like genes have been amplified and characterized. Although these primers contain mismatches, these do not appear to significantly inhibit the detection of N. europaea-like amoA genes from these bioreactor samples.
Additionally, the question arises of whether these amoA genes belong to different species of nitrifying bacteria inhabiting these bioreactors or represent divergent copies of amoA genes from previously described nitrifying bacteria. Purkhold et al. suggested a threshold value of 80% similarity for amoA genes from different species of nitrifiers based on an extensive comparison of known strains using DNA-DNA hybridization, 16S rRNA similarity, and amoA similarity. None of the amoA genes described in this study meet this threshold criterion for novel nitrifying species (similarities in these data ranged from 90 to 97%). Although we cannot unequivocally rule out the possibility that these amoA genes represent novel nitrifying bacteria, it is more likely that the bioreactors harbor nitrifying strains closely related to Nitrosomonas and Nitrosospira cultures that have been characterized.
As for the denitrifying populations, the spatial distribution profiles of nosZ genes in the TFB differed slightly from those of the 16S rRNA and amoA genes, with the top and bottom portions of the reactor clustering together. Based on the nosZ T-RFLP profiles, the ICB appeared to harbor a more diverse group of denitrifying bacteria than the TFB. As with the 16S rRNA T-RFLP profiles, this finding is expected because of the higher total organic carbon concentrations (180 mg/liter on average, ranging from 74 to 500 mg/liter) and pH values (7.37 to 8.48) in the ICB influent than in the TFB influent (TOC of 16.6 mg/liter on average, ranging from 9 to 56 mg/liter, and pHs of 6.07 to 7.37) (19). The clonal analysis of nosZ genes suggests that the denitrifying bacteria that inhabit the bioreactors are generally related to cultured denitrifiers (Pseudomonas-Paracoccus-Achromobacter group); however (as with the amoA genes), the nosZ sequences all cluster at the periphery of the various known denitrifying microorganisms. This is in contrast to an environmental study in which few of the nosZ genes discovered were associated with any cultured denitrifying bacteria (26). In spite of this, three sequences were identified that were not closely affiliated with any known groups of nosZ genes. More attempts should be made to identify those microorganisms possessing unique nosZ genes from bioreactors and other samples to gain a better understanding of denitrifying bacteria in these systems.
In conclusion, the bioreactors at JSC were found to harbor unique heterotrophic (ɛ-Proteobacteria, Cytophagales, and Planctomycetales) and denitrifying populations of bacteria, but the ammonia-oxidizing populations were generally found to be closely related to known strains of nitrifiers from other wastewater treatment systems (22). This information on microbial populations is important to establish and enhance the reliability of biological water recovery systems for long-duration space flight missions. The next phase of research should focus on measuring gene expression of total and specific microorganisms, such as ammonia-oxidizing and denitrifying bacteria, to provide information about active populations in a given system (e.g., a wastewater treatment reactor). Combination of these data and quantitative data (e.g., fluorescence in situ hybridization) for these organisms with physical and chemical measurements of processes within the reactors may begin to describe the workings of what has been, until now, a “black box” system and eventually improve the design and operation of these biological systems.
Acknowledgments
This work was supported in part by the New Jersey-NASA Center for Research and Training (NJ-NSCORT) and by grants from the National Science Foundation (NSF 982024) and NASA (NAG 9-1283) to L.J.K.
We thank the anonymous reviewers whose comments significantly improved the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. L. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Avaniss-Aghajani, E., K. Jones, D. Chapman, and C. Brunk. 1994. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques 17:144-149. [PubMed] [Google Scholar]
- 3.Berardesco, G., S. Dyhrman, E. Gallagher, and M. P. Shiaris. 1998. Spatial and temporal variation of phenanthrene-degrading bacteria in intertidal sediments. Appl. Environ. Microbiol. 64:2560-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruce, K. D. 1997. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR-restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 63:4914-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 6.Garland, J. L., K. L. Cook1, J. L. Adams, and L. Kerkhof. 2001. Culturability as an indicator of succession in microbial communities. Microb. Ecol. 42:150-158. [DOI] [PubMed] [Google Scholar]
- 7.Hallin, S., and P.-E. Lindgren. 1999. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65:1652-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmer, C., S. Kunst, S. Juretschko, M. C. Schmid, K. H. Schleifer, and M. Wagner. 1999. Nitrogen loss in a nitrifying biofilm system. Water Sci. Technol. 39:13-21. [Google Scholar]
- 9.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 10.Hortz, H. P., J. H. Rotthauwe, T. Lukow, and W. Liesack. 2000. Identification of major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J. Microbiol. Methods 39:197-204. [DOI] [PubMed] [Google Scholar]
- 11.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerkhof, L., M. Santoro, and J. Garland. 2000. Response of soybean rhizosphere communities to human hygiene water addition as determined by community-level physiological profiling (CLPP) and terminal restriction fragment length polymorphism (T-RFLP). FEMS Microbiol. Lett. 184:95-101. [DOI] [PubMed] [Google Scholar]
- 13.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Son, Ltd., Chichester, England.
- 14.Lazarova, V., D. Bellahcen, J. Manem, D. A. Stahl, and B. E. Rittmann. 1999. Influence of operating conditions on population dynamics in nitrifying biofilms. Water Sci. Technol. 39:5-11. [Google Scholar]
- 15.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1995. The Ribosomal Database Project (RDP). Nucleic Acids Res. 24:82-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 18.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Aeronautics and Space Administration. 2000. Lunar-Mars life support test project: phase III final report. CTSD-ADV-341. Lyndon B. Johnson Space Center, Houston, Tex.
- 20.Neef, A., A. Zalglauer, H. Meier, R. Amann, H. Lemmer, and K. H. Schleifer. 1996. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl. Environ. Microbiol. 62:4329-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson, W., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rittmann, B. E., C. S. Laspidou, J. Flax, D. A. Stahl, V. Urbain, H. Hardiun, J. J. van der Waarde, B. Geurkink, M. J. Henssen, H. Brouwe, A. Klapwijk, and M. Wetterauw. 1999. Molecular and modeling analyses of the structure and function of nitrifying activated sludge. Water Sci. Technol. 39:51-59. [Google Scholar]
- 24.Rotthauwe, J.-H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakano, Y., and L. Kerkhof. 1998. Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl. Environ. Microbiol. 64:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scala, D. J., and L. J. Kerhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schramm, A., C. M. Santegoeds, H. K. Nielsen, H. Ploug, M. Wagner, M. Pribyl, J. Wanner, R. Amann, and D. de Beer. 1999. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl. Environ. Microbiol. 65:4189-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, W. W., R. Overbeek, C. R. Woese, W. Gilbert, and P. M. Gillevet. 1994. The genetic data environment: an expandable GUI for multiple sequence analysis. Comput. Appl. Biosci. 10:671-675. [DOI] [PubMed] [Google Scholar]
- 31.Surampalli, R. Y., O. K. Scheible, and S. K. Banerji. 1995. Nitrification in single-stage trickling filters. Environ. Prog. 14:164-171. [Google Scholar]
- 32.Zumft, W. G. 1997. Cell biology and the molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]