Abstract
Spores of Bacillus species are being used commercially as probiotics and competitive exclusion agents. Unlike the more commonly used Lactobacillus-type probiotics, spores are dormant life forms. To address how spore probiotics might function we have investigated whether spores can germinate in the gastrointestinal tract by using a murine model. Using a genetically engineered chimeric gene, ftsH-lacZ, which is strongly expressed only in vegetative cells, we have developed a sensitive competitive reverse transcription-PCR assay which has enabled detection of as few as 102 vegetative bacteria in the mouse gut. Using this method we have administered doses of ftsH-lacZ spores to groups of mice and shown that spores can germinate in significant numbers in the jejunum and ileum. The levels of detection we obtained suggest that spores may colonize the small intestine, albeit briefly.
Bacterial spores are dormant life forms which can exist in a desiccated and dehydrated state indefinitely. The process of spore formation has been extensively studied as a simple model for understanding cellular differentiation and is one of the primary reasons for the interest in spores and spore formation (7). Intriguingly though, spores of Bacillus subtilis are being used as probiotics and competitive exclusion (CE) agents for both human and animal consumption (18). For humans they are available either as over-the-counter prophylactics for mild gastrointestinal disorders such as diarrhea or as health foods or nutritional supplements. In some countries though (e.g., Vietnam), bacterial spores are being used for oral bacteriotherapy of gastrointestinal disorders often under clinical supervision. In the agricultural industry spores are also receiving increasing attention as potential alternatives to antibiotics as growth promoters. The use of probiotics and/or CE agents seems likely to increase as public awareness of their potential benefits increases.
While spores are being sold as probiotics, an important question is that of how spores act to enhance the normal microbial flora of the gastrointestinal tract. This question must be addressed, because the majority of probiotics currently available are bacteria which are non-spore formers: i.e., they are given as vegetative cells (usually as lyophilized preparations). The best-known examples of these probiotic bacteria are the lactobacilli and bifidobacteria (2, 8, 9). If probiotic bacteria are to be taken seriously then we would assume that they would share a common mechanism for enhancing the normal well-being of the gut microflora or for CE of potential pathogens. If there is no common mechanism shared between conventional Lactobacillus-type bacteria and the spore probiotics, then the question must be asked as to whether there is any credibility to some of the claims made about the positive benefits of probiotic bacteria (2, 12).
Probiotics and CE agents are thought to enhance the gut microflora by preventing the colonization of the gastrointestinal tract by pathogenic bacteria. There are three basic ways (8, 9, 26) in which this might be achieved: (i) immune exclusion of a pathogenic bacterium, (ii) exclusion of a pathogen by competitive adhesion, and (iii) synthesis of antimicrobial substances which impair colonization of the gastrointestinal tract by a pathogen. In recent work we have shown that spores of B. subtilis do appear to have the potential to suppress all aspects of Escherichia coli 078:K80 infection in a 1-day-old-chick model (16). By analyzing spore counts in the feces of mice administered spore suspensions, we have also shown that it is possible that spores could germinate in the gastrointestinal tract (15). If our hypothesis is correct then spores, by germinating, could function as a probiotic in the same way as the conventional probiotic or CE bacteria. This has prompted the work described here, in which we have developed a molecular method to detect the germination of spores in the gastrointestinal tract of mice.
MATERIALS AND METHODS
Animals.
Six-week-old, female, BALB/c mice were obtained from Harlan UK (Oxon, United Kingdom) and maintained in animal facilities for the duration of the experiment.
Strains.
SC2288 (ftsH-lacZ) carries a transcriptional fusion of the 5′ segment of the ftsH gene fused to the E. coli lacZ gene. This genetically engineered gene carries resistance to chloramphenicol (final concentration of 5 μg/ml) and has been described elsewhere (17).
General methods.
Bacillus and general molecular biology methods were as described elsewhere (13, 22). DNA sequencing was performed using a commercial automated DNA sequencing facility (MWG Biotech, Ltd., Milton Keynes, United Kingdom).
Preparation of spore or vegetative cells. (i) Spores.
Spores were prepared from large culture volumes (200 ml) using Difco sporulation medium (without antibiotics). This was the exhaustion method for induction of sporulation, and the methodology has been reported in detail elsewhere (21). Spores were harvested 24 h after the estimated start of sporulation and treated with lysozyme to destroy any residual vegetative cells as described (21). Spores were then washed repeatedly with water, concentrated by centrifugation, and heat treated at 65°C (45 min) to kill any residual vegetative cells or germinated spores. Aliquots were frozen at known concentrations (CFU per milliliter) at −80°C till use.
(ii) Vegetative cells.
To prepare vegetative cells of SC2288 a single colony from a Difco sporulation medium agar plate containing chloramphenicol at 5 μg/ml was used to inoculate 5 ml of Luria-Bertani (LB) broth containing 0.2% l-glutamine and 5% d-glucose. l-Glutamine and d-glucose were incorporated to inhibit spore formation in LB growth (23). The culture was incubated at 30°C for 18 h in a roller drum. Three milliliters of this culture was used to inoculate 300 ml of LB broth (in a 2-liter flask) containing 0.2% l-glutamine and 5% d-glucose, and the culture was incubated at 37°C. When the culture had reached an optical density at 600 nm (OD600) of approximately 0.6, cells were harvested and suspended in 0.1 volume of sterile 12% glycerol. The number of CFU per milliliter of the suspension was then determined, and aliquots were frozen at −80°C.
Intragastric dosing and dissections.
Groups of mice were inoculated intragastrically with 0.2-ml suspensions of spores or vegetative cells using a ball-ended feeding needle. Mice were dissected following sacrifice, and sections of the small intestine (duodenum, jejunum, and ileum) or individual sections were excised and processed immediately.
Analysis of spore shedding.
Analysis of spore shedding was done as described previously (15) using mice housed in individual cages with gridded floors to prevent coprophagia.
Total RNA isolation.
Total RNA from gut sections was isolated using TRIzol based on the method of Chomczynski and Sacchi (4) as follows. Excised intestinal sections were homogenized in TRIzol (Life Technologies). Tissue was weighed and immersed in TRIzol (3 ml) immediately following dissection. Acid-washed glass beads (1.5 ml; bead diameter, 0.5 mm) were added, and the samples were frozen at −80°C until the RNA isolation step. Complete lysis of the gut tissue and disruption of the bacterial cells were achieved by vortexing the sample containing the glass beads for 3 min followed first by sonication for 20 s (on ice) and then by three cycles of freeze-thaw on dry ice. Homogenates were then clarified by centrifugation (12, 000 × g, 10 min, 4°C), and the supernatant was decanted and extracted with 0.9 ml of chloroform to destroy residual protein. RNA was precipitated with ethanol following the manufacturer's protocol, and total RNAs were dissolved in 100 μl of RNase-free water containing recombinant RNasin (1 U/μl; Promega) and stored at −80°C until reverse transcription (RT)-PCR analysis. Total RNAs were quantified spectrophotometrically by OD260 (GeneQuant II; Pharmacia). Prior to RT-PCR total RNAs were treated with RQ1 RNase-free DNase (1 U/μg of total RNA; Promega) for 45 min at 37°C, and the reaction was stopped by addition of 1 μl of 20 mM EGTA and DNase heat inactivated for 10 min at 65°C.
Primers.
Detection of the ftsH-lacZ transcript was carried out using the primers FtsHdc (5′ CGCAAGAAACAAGCGGATGGG 3′; anneals to nucleotides [nt] 307 to 326 of the ftsH open reading frame [ORF]) and RlacZdc (5′-ATCAACATTAAATGTGAGCGAG-3′; anneals to nt 366 to 386 of the lacZ ORF) to amplify a product of 509 bp. For synthesis of the competitor template Ft2dc was used as the 5′ primer (5′ CGCAAGAAACAAGCGGATGGGCCTGCTCAATCAGGCTCAAGGC 3′) together with RlacZdc to amplify a product of 453 bp using SC2288 chromosomal DNA as a template. The underlined sequence is the 5′ sequence also present in FtsHdc. For establishing the quality of total mouse RNA, mouse-specific β-actin primers βAct-F (5′ GCTGGTCGTCGACAACGGCTCC 3′; anneals to nt 101 to 122 of the β-actin ORF [accession no. X03672]) and βAct-R (5′ GAGGATGCGGCAGTGGCCATCT 3′; anneals to nt 757 to 779 of the β-actin ORF) were used to amplify a sequence of 679 bases.
Competitive RT-PCR assay.
Our assay was based on the studies of Shin et al. (24) and used the chimeric ftsH-lacZ gene containing the 5′ region (734 bp) of ftsH fused to the lacZ gene of E. coli (Fig. 1). This hybrid gene has been described previously (17) and is only expressed during vegetative cell growth controlled from the σA-recognized ftsH promoter. Spores of SC2288 (ftsH-lacZ) were prepared as described above and used to dose multiple groups of four mice orally (0.2-ml suspension containing 2 × 108 CFU/ml). At appropriate time points thereafter one group was sacrificed, gut sections were dissected, and total RNA (mouse and bacterial) was extracted.
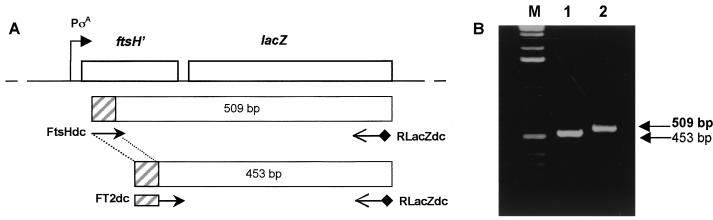
FIG. 1.
ftsH-lacZ chimera and PCR amplification products. (A) Physical map of the ftsH-lacZ chromosome of strain SC2288 as described by Lysenko et al. (17). The FtsHdc and RlacZdc primers used to amplify a 509-base segment across the ftsH-lacZ fusion junction and FT2dc and RlacZdc used to amplify a 453-base competitor template are shown together with the predicted amplification products. The hatched boxes indicate the sequence included in the 5′ end of the FT2dc primer that is recognized by the FtsHdc primer. (B) PCR amplifications with these primer sets using SC2288 chromosomal DNA. Lane M, 1-kb ladder.
For the assay a standard RT-PCR was performed on the total RNA samples obtained from mouse sections using the two primers, FtsHdc and RlacZdc (Fig. 1), to amplify the 509-base product extending from ftsH to lacZ. A product would only be detected if vegetative bacteria were present.
Next, to generate a competitive template we used the FT2dc and RlacZdc primers (Fig. 1) to generate a smaller amplification product of 453 bases using SC2288 chromosomal DNA as a template. FT2dc included a 5′ tail to which FtsHdc could anneal. The competitive PCR product was gel purified, denatured, and quantified (on the basis of OD260 determined using a Pharmacia Gene Quant II device). A competitive RT-PCR was then performed as follows. First, the competitive template was serially diluted and dilutions were mixed with a known concentration of total RNA extract (1 μg). Each PCR mixture therefore consisted of two templates, various concentrations of competitive DNA, and a fixed concentration of RNA. Next, in each PCR mixture, the primers FtsHdc and RlacZdc were used to generate a PCR product of which two species should be produced, a 453-base species amplified from the competitive template and a 509-base species amplified from the RNA template. The product of each PCR was size fractionated by agarose gel electrophoresis (2% [wt/vol] acrylamide). The dilution at which the 509- and 453-base PCR products were equivalent in density enabled extrapolation of the number of moles of ftsH-lacZ mRNA by regression analysis.
RT-PCR conditions.
RT-PCRs were performed using the Calypso One-Step RT-PCR system incorporating avian myeloblastosis virus (AMV) reverse transcriptase (DNAmp Ltd). The program conditions were 1 cycle at 50°C for 30 min followed by 1 cycle at 94°C for 2 min (for inactivation of the reverse transcriptase); 10 cycles with annealing at 58°C for 30 s, extension at 68°C for 45 s, and denaturation at 94°C for 20 s; a further 25 cycles with an extra cycle extension of 5 s per cycle; and a final extension at 68°C for 10 min.
To establish whether any DNA contamination had occurred samples were also analyzed in parallel using heat inactivation of the reverse transcriptase at 95°C for 5 min prior to amplification. As an internal control for RNA amplification, positive control reagents in the Calypso RT-PCR kit, consisting of MS2 bacteriophage RNA and two primers, were used to obtain an amplification product of 1,054 bp.
Competitive RT-PCR regression analysis.
Gel images were acquired using a UVP imaging system and then analyzed by densitometry using Sigma Scan software (Jandel Scientific). Densitometry data were subjected to regression analysis, and the log number of the moles of competitor template was plotted against the log ratio between the target product and the competitor band intensity. From the equimolar point of target and competitor product, the number of moles of vegetative SC2288 mRNA can be calculated.
RESULTS
RT-PCR primers.
We have used RT-PCR to develop a molecular method for the detection of spore germination in the mouse gastrointestinal tract. Our work was based on the studies of Shin et al., who used competitive RT-PCR to quantify the levels of respiratory syndrome virus in semen (24). In our work we exploited a chimeric gene in which the ftsH gene of B. subtilis has been fused to the lacZ gene of E. coli (17). The ftsH gene is transcribed exclusively during vegetative cell growth by RNA polymerase associated with σA. ftsH was chosen since this gene is expressed at high levels (6) which would increase the probability for detection by RT-PCR. Cells carrying this chimeric gene (strain SC2288) would produce a unique mRNA transcript which could be detected by RT-PCR. In the first instance we designed primer sets to amplify a defined segment of the ftsH-lacZ transcript across the fusion junction of ftsH and lacZ as shown in Fig. 1A. The first set of conventional PCR primers (5′-FtsHdc and 3′-RlacZdc) was designed to amplify a 509-base segment. The second set used a modified 5′ primer (5′-FT2dc) together with the same 3′ primer (RlacZdc) to amplify a 453-base PCR product. The FT2dc primer carried a 5′ sequence to which FtsHdc could anneal and that would enable the 453-base PCR product to be used as a competitive template in our assay. As shown in Fig. 1B both primer sets enabled the amplification of unique PCR products that could be separated by agarose gel electrophoresis.
Transit of SC2288 spores in the mouse gut.
To evaluate the transit of SC2288 spores in the mouse gut we prepared spores and dosed 5 mice with 6 × 108 spores. As well as lysozyme-treating spore suspensions we also heat treated them (65°C, 45 min) to ensure the inactivation of all contaminating vegetative cells. At 6-, 12-, 18- and 24-h intervals thereafter we collected the total feces from individual mice and determined the number of spores present. As shown in Table 1 we found that for two mice the total (cumulative) number of spores excreted was greater than the original inoculum. We have observed this previously and take this as evidence, although not proof, that B. subtilis can colonize, albeit briefly, the mouse gut (15). Our reasoning was that to account for the increased counts, spores would have germinated, replicated and, for some, entered the developmental pathway leading to the formation of spores. Our data also showed that spore counts gradually dropped over time yet considerable numbers of spores were detectable at each time point. Accordingly, 6, 12, 18, and 24 h were chosen as time points for sampling for RT-PCR analysis.
TABLE 1.
Shedding of B. subtilis spores in mouse fecesa
| Time (h) | Spore count in feces from mouse:
|
Mean spore count (SD) | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| 6 | 5.05 × 107 | 4.4 × 108 | 6.5 × 107 | 2.3 × 108 | 7.3 × 108 | 3.03 × 108 (2.86 × 108) |
| 12 | 5.1 × 107 | 7.0 × 107 | 1.2 × 108 | 3.6 × 107 | 1.3 × 108 | 8.14 × 107 (4.17 × 107) |
| 18 | 1.8 × 107 | 1.0 × 108 | 6.2 × 106 | 3.6 × 105 | 9.0 × 106 | 2.67 × 107 (4.15 × 107) |
| 24 | 4.35 × 107 | 2.36 × 106 | 1.45 × 106 | 6.36 × 105 | 2.5 × 106 | 1.10 × 107 (1.87 × 107) |
| Total (% of inoculum) | 1.63 × 108 (27) | 6.12 × 108 (102) | 1.93 × 108 (32) | 2.67 × 108 (44.5) | 8.72 × 108 (145) | |
Individually housed mice (BALB/c, female, 6 weeks old) were orally given suspensions with 6 × 108 spores of SC2288 containing no viable vegetative bacteria. Total feces were collected at 6-h time points using cages with gridded floors. Feces were then homogenized, heat treated at 65°C (45 min) to kill all vegetative bacteria, and plated in serial dilution onto drug-resistant plates to estimate spore counts as described before (15). Individual spore counts for each mouse and time point are shown together with the mean and standard deviation.
Optimization of RNA recovery from mouse gut tissue.
Before attempting to evaluate the fate of spores administered orally to mice we first established the parameters required to recover high-quality RNA from the gastrointestinal tract and then to establish the sensitivity of the RT-PCR using gut tissues spiked with vegetative B. subtilis. In the first instance we evaluated a number of techniques as well as commercial kits for isolating total RNA from excised sections of the gastrointestinal tract. The technique which we found reproducibly provided complete lysis and recovery of both mouse and bacterial RNA was emulsion of tissues that had been spiked with vegetative B. subtilis cells in guanidinium thiocyanate (TRIzol) followed by physical disruption with glass beads, sonication, and repeated freeze-thaw cycles as described in Materials and Methods. To determine the integrity of the extracted mRNA we used RT-PCR primers that recognized the β-actin gene. Successful amplification of β-actin produced a 679-base PCR product which was found to be consistent between samples (data not shown). Although not shown, formaldehyde agarose gel electrophoresis was first used to determine the quality of RNA prior to a RT-PCR.
Next, we performed experiments in which sections of the small intestine were mixed with various amounts of vegetative B. subtilis bacteria. Total RNA was then extracted, and RT-PCR was used to amplify a cDNA product. This technique enabled us to estimate the maximum sensitivity of our RT-PCR method, i.e., the minimum number of SC2288 cells that could be detected using RT-PCR. Figure 2 shows one experiment in which sections of jejunum have been spiked with different amounts of SC2288 vegetative cells. Total RNA was then prepared and RT-PCR was performed using the FtsHdc and RlacZdc primers to amplify a 509-base cDNA product. This experiment revealed a lower limit of detection of 104 cells/section (however, for the RT-PCR assay 0.01 volume was used; therefore, the actual detection limit was as low as 102 cells). We have been unable to reliably detect a lower number of bacteria using our methodology. These experiments have also been repeated using spiked duodenum sections (data not shown). As an important control we have found (data not shown) that if mouse tissues are spiked with SC2288 spores no RT-PCR product can be detected.
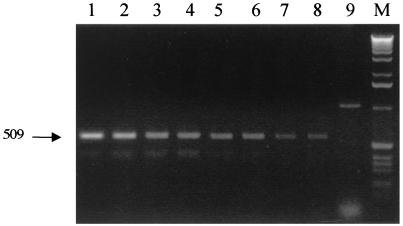
FIG. 2.
Sensitivity of RT-PCRs. To approximate the sensitivity of the RT-PCR small intestine sections excised from a mouse were spiked with different amounts of vegetative cells of SC2288 (ftsH-lacZ) (see Materials and Methods). Total RNA (1 μg) was then prepared, and RT-PCR was performed using FtsHdc and RlacZdc to amplify the 509-base ftsH-lacZ product. A positive control was also used to spike mouse sections with MS2 phage and to amplify a 1,054-base MS2-specific RT-PCR product of 1,054 bases. Lane 1, 5 × 107 SC2288 cells; lane 2, 1 × 107 SC2288 cells; lane 3, 5 × 106 SC2288 cells; lane 4, 1 × 106 SC2288 cells; lane 5, 5 × 105 SC2288 cells; lane 6, 1 × 105 SC2288 cells; lane 7, 5 × 104; lane 8, 1 × 104 SC2288 cells; lane 9, MS2 phage (positive PCR control); lane M, 1-kb ladder.
One further evaluation we performed was to use competitive RT-PCR to quantify the number of ftsH-lacZ mRNAs in spiked tissues (see Materials and Methods). This would, in turn, enable an extrapolation of the number of vegetative cells. Individual duodenum sections from six mice were spiked with 6 × 108 vegetative SC2288 cells. We used a competitive RT-PCR assay (Fig. 3 shows a representative example from one sample) followed by regression analysis (shown as an average for the group of six mice) to evaluate the number of moles of ftsH-lacZ transcripts. The number of ftsH-lacZ mRNA moles in 6 × 108 vegetative cells (i.e., equal to the intragastric inoculum) was found to correspond to 2.465 × 10−14 mol of RT-PCR product, corresponding to 2.81 μg (1.484 × 1010 copies) of ftsH-lacZ mRNA.
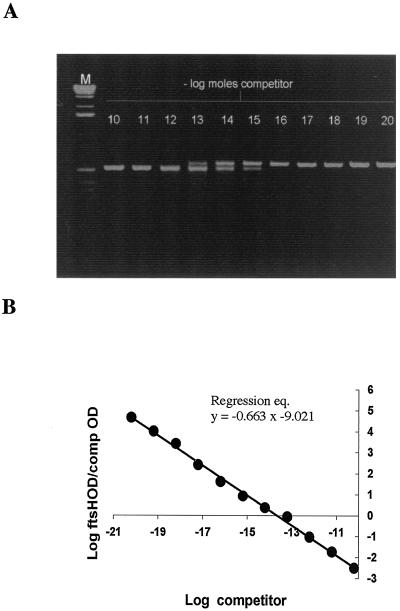
FIG. 3.
Competitive RT-PCR assay. Determination of the number of moles of ftsH-lacZ mRNA using a competitive RT-PCR assay. Jejunum sections prepared from six mice were spiked with 6 × 108 vegetative cells of SC2288, and total RNA was prepared. RT-PCR was used with two primers (FT2dc and RlacZdc) to generate a competitive template of 453 bases. This competitor template was purified, and dilutions were mixed with total RNA samples prepared from each mouse. These were then used in individual RT-PCRs using primers FtsHdc and RlacZdc to generate a 509-base product (from the RNA) and a 453-base product (from the competitor). (A) Representative titration from one mouse sample. As the concentration of competitor template decreases so the production of the 509-base product increases (left to right on the gel). Relative absorbance values from densitometric analysis of the 509- and 453-base PCR products were determined and normalized for size. (B) Least-squares regression analysis of average densitometric data from the group of six mice. The log moles of competitor RT-PCR product is plotted against the logarithmic ratio of absorbance of the 509-base species to absorbance of the 453-base product.
Detection of germination in the small intestine.
Groups of mice were administered 6 × 108 spores of SC2288. Groups were sacrificed at 6, 12, 18, and 24 h; sections of the small intestine (duodenum and jejunum) were excised; and total RNA was extracted. We used RT-PCR to first establish whether ftsH-lacZ mRNA was present in samples from these time points and from naïve mice. We failed to detect any signal in duodenum samples at any time point but could readily identify a PCR product in jejunum samples. The jejunum results are shown in Fig. 4 and reveal the strongest signals at the 18-h time point and weaker signals at 24 h. No signal could be detected in samples from the 6- and 12-h time points. To demonstrate that the signals we saw were due to ftsH-lacZ mRNA and were not from DNA, Fig. 4 also shows the absence of a signal in the 18- and 24-h samples in which the AMV reverse transcriptase was heat inactivated prior to initiation of cDNA synthesis. The only explanation for these results was that spores of SC2288 had germinated in the small intestine. To confirm that the RT-PCR products were indeed the correct product we sequenced the amplification products in their entirety on both strands.
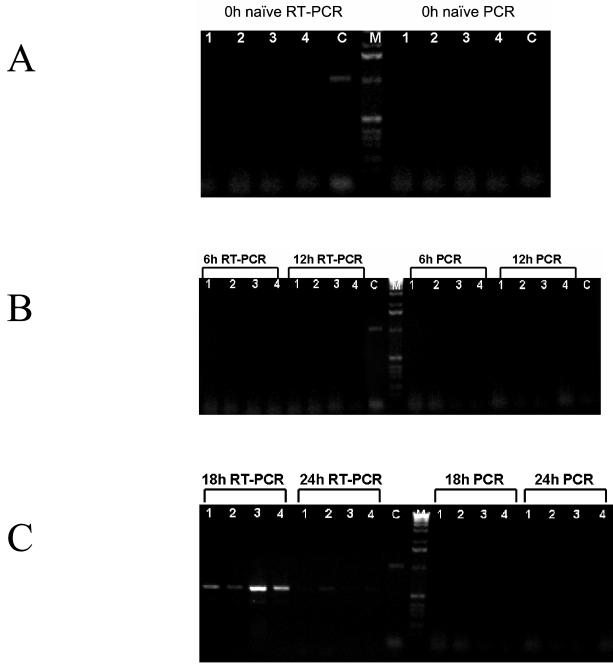
FIG. 4.
RT-PCR analysis of jejunum sections. Groups of four mice (lanes 1, 2, 3, and 4) were administered 6 × 108 spores of strain SC2288. Mice were sacrificed at 6, 12, 18, and 24 h (B and C) and dissected, the jejunum was excised, and total RNA was extracted. (A) In addition naïve, untreated mice and mice just before dosing (0 h) were also examined. Total RNA was subjected to RT-PCR analysis using two primers (FtsHdc and RlacZdc) which generate a 509-base cDNA product if more than 104 SC2288 vegetative cells are present. Samples to the right of the 1-kb ladder are identical RT-PCRs in which the AMV reverse transcriptase has been inactivated by heat treatment. (C) Control RT-PCR using MS2 bacteriophage as supplied with the Calypso RT-PCR kit.
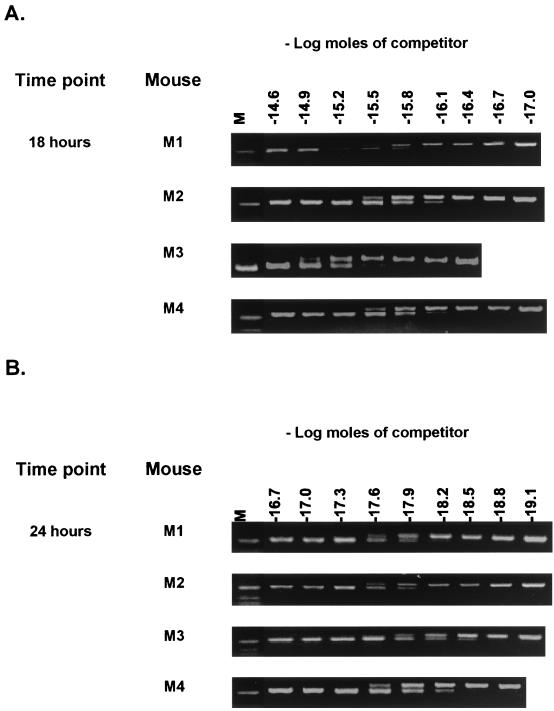
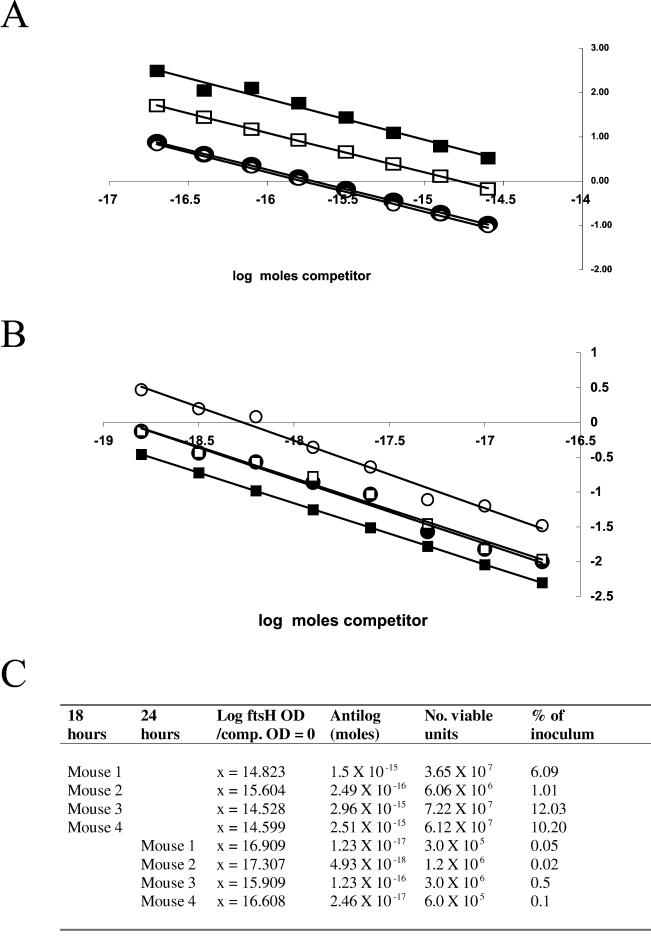
To quantify the signal strength we used a competitive RT-PCR assay to establish the number of vegetative cells that could produce this signal. The quantifications of the 18- and 24-h samples are shown in Fig. 5, and regression analysis is shown in Fig. 6A and 6B. The results of the regression analysis (Fig. 6C) revealed that in samples from the 18-h time point the signals detected corresponded to 1 to 12% of the original inoculum of viable units (6 × 108) and in those from the 24-h time point there were much lower levels (0.02 to 0.5%).
FIG. 5.
Competitive RT-PCR analysis. Total RNA extracts from 18 -h (A) and 24 -h (B) jejunum samples of mice dosed with SC2288 (ftsH-lacZ) cells were mixed with dilutions of a competitive template (as described in Materials and Methods and Fig. 3). Samples correspond to those of Fig. 4.
FIG. 6.
Quantitation of ftsH-lacZ transcripts. Densitometric analysis of the 453- and 509-base products from competitive RT-PCR assays shown in Fig. 5 were used for regression analysis. Shown are results of regression analysis of the 18-h jejunum samples for the four mice sampled (A) and the 24-h samples (B). (A) The equations for the regression lines relative to the 18-h samples were the following: mouse 1, y = −0.8833x − 13.093 (R2 = 0.99); mouse 2, y = −0.8961x − 13.982 (R2 = 0.99); mouse 3, y = −0.9286x − 13.491 (R2 = 0.98); mouse 4, y = −0.8897x − 12.989 (R2 = 0.99). (B) The equations for the regression lines for the 24-h samples were the following: mouse 1, y = −0.919x − 15.539 (R2= 0.98); mouse 2, y = −0.9712x − 17.820 (R2= 0.98); mouse 3, y = −0.8786x − 118.107 (R2 = 1); mouse 4, y = −0.9008x − 18.437 (R2 = 0.98). (C) Tabulation of the quantification of vegetative bacteria (viable units) using the extrapolated regression data.
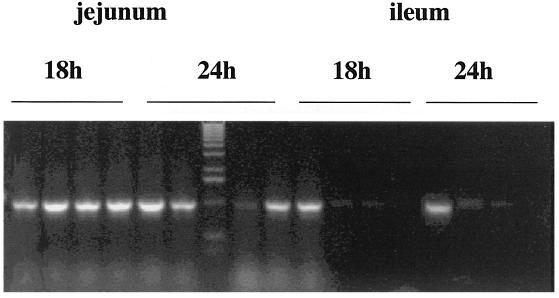
To add further support for these results we have repeated this analysis using a different chimeric gene, rrn0-lacZ. Strain SL6913 (obtained from P. Piggot) carries a transcriptional fusion of the rRNA gene, rrn0, to lacZ. This vegetative gene is strongly expressed. We administered SL6913 spores (6 × 108) to groups of four mice and used RT-PCR with appropriate primer sets to amplify a 1-kb PCR product in selected sections of the small intestine. As shown in Fig. 7, we could obtain signals from the 18- and 24-h samples of jejunum and from the ileum sections from three mice as well.
FIG. 7.
RT-PCR analysis using rrnO-lacZ. Groups of four mice were administered 6 × 108 spores of strain SL6913. Mice were sacrificed at 18 and 24 h and dissected, the jejunum and ileum were excised, and total RNA was extracted. Total RNA was subjected to RT-PCR analysis using two primers which generate a 1-kb cDNA product when SL6913 vegetative cells are present.
DISCUSSION
We have developed a molecular method to demonstrate that bacterial spores can germinate in the small intestine of mice. Our reason for attempting this is to address the question of how spore probiotics and CE agents currently consumed for both human and animal use function. Our results show that using a sensitive RT-PCR assay we can detect as few as 102 vegetative cells in the small intestine. Since our inoculum consisted only of spores, the one, and only, explanation for this results is that spores had germinated. Considering that our assay requires excision of tissues as well as breakage of bacteria and mammalian tissues we believe this to be a significant achievement. Spore germination was not detected in the duodenum but was readily detectable in the jejunum and in one experiment was detected in the ileum as well. The sensitivity of our method means that we cannot exclude the possible germination of spores in the duodenum, but this must be at very low levels. The small intestine contains regions of different physiochemical conditions, and obviously the duodenum would carry a high content of stomach acids as well as bile salts. These may inhibit spore germination as predicted before (25) or contaminate our RT-PCR assay. We have also found that we could detect the strongest germination signal in the sample from the 18-h time point. Analysis of the transit of spores through the gut, in this work and elsewhere (14), has shown that the majority of spores are shed between 6 and 24 h in mice. It therefore seems likely that germination is occurring during the first 24 h. In previous work we have shown that, following dosing of mice with spores, counts can be detected in the feces as early as 3 h (15).
We believe that our competitive RT-PCR assay is of sufficient accuracy to provide an estimate of the number of vegetative cells. However, we also realize that at a 10% level (the level detected in one 18-h mouse section) when the original inoculum was 6 × 108 cells, this would mean that in one section of the gut, and for one window of time, 6 × 107 vegetative cells are present. This is a remarkably high number considering the original dose. We are sufficiently cautious about these quantifications not to try to make any definitive conclusions about population size. However, if this estimate is correct, then a significant number of vegetative cells must be present in the small intestine, and we believe this would be more than could be accounted for simply by germination of the original inoculum. A reasonable explanation then is that a percentage of spores germinate and these then undergo limited rounds of growth. This does not seem improbable, since B. subtilis has been shown to grow under anaerobic conditions provided that a suitable nutritional environment is available (20) (it is also questionable whether the small intestine is completely anaerobic). Spores would be expected to germinate whether the environment is aerobic or anaerobic, and the only necessary condition is that suitable nutritional germinants (e.g., fructose, l-alanine [19]) be present, and these should be present in the small intestine. Supporting this, some spore-forming bacteria have been shown to carry out their entire life cycle of sporulation and germination in the gastrointestinal tract of the guinea pig (1), so there is no clear precedent for assuming that spore formation or germination cannot occur in the intestinal tract. If vegetative cells of B. subtilis can exist in the small intestine, then an interesting question is for how long and whether they can colonize. As has been proposed previously B. subtilis is sensitive to bile salts (25), and perhaps adverse conditions found in the small intestine eventually inhibit long-term colonization. Interestingly though, studies using ligated ileum loops from rabbits have suggested that spores might germinate in the gastrointestinal tract (14).
If the small intestine can be colonized even briefly by vegetative B. subtilis, then this may provide a partial explanation for how spores can be used, like the Lactobacillus species, as probiotics. That is, they would exert their beneficial effect as vegetative cells, and the fact that they are given as spores is simply an effective means for delivering large quantities of bacteria to the small intestine since the spore can survive transit across the stomach. Interestingly, the Lactobacillus-type probiotics are given as lyophilized preparations, and although this genus is normally resident in the gut we would expect the majority of these bacteria to be destroyed upon entry into the stomach. The few that can survive would populate the small intestine and somehow prevent colonization of the intestine by harmful bacteria (i.e., by serving as a CE agent) and/or by enhancing the gut microflora (as a probiotic). By analogy, delivery of a high concentration of spores would enable almost 100% survival through the stomach, where a small population would germinate, and the vegetative cells would briefly colonize one or more sections of the small intestine. This is the model we propose that could explain the apparent paradoxical use of spores and Lactobacillus-type probiotics. For obvious reasons we have used a murine model for these studies. It goes without saying, then, that we cannot state that in a human, spores might follow a similar pattern of germination and limited colonization. However, since B. subtilis is not a normal resident of either the mouse gut or the human gut and spores are being used as a probiotic or CE agent, it seems probable that delivery of large numbers of this foreign bacterium will lead to germination and limited colonization.
Our work suggests that the small intestine is briefly colonized by B. subtilis. However, it is also possible that the spore itself exerts an immunostimulatory effect which serves to exclude the colonization of the gut by harmful pathogens. A number of reports have shown that the spore is immunostimulatory and can elicit a number of cellular immune responses in the gastrointestinal tract (3, 5, 10, 11). One final question our work raises is that of the fate of vegetative B. subtilis. As observed before (15) and also here (Table 1), on some occasions the number of excreted spores we can detect exceeds that given in the original inoculum. Experimental error is of course the immediate explanation, yet we have repeated these experiments many times and sometimes observe this increase. The errors that would decrease the counts are the most probable, and this then only supports the increase observed as being real. We wonder, then, whether B. subtilis cells can sporulate in the gut. The hostile conditions encountered, particularly as the germinated cells enter the lower regions of the gastrointestinal tract, make spore formation seem a likely escape route. Our analysis of spore shedding here and in other work (15) has shown that a small increase in spore numbers is found in the feces 18 to 24 h after dosing. Taking into account our RT-PCR analysis performed here, we propose that after 18 h significant numbers of spores are germinating but the majority are shed in the feces. Germinated spores will either be killed, perhaps by the action of bile salts, or, after a few, limited rounds of growth and division, form spores to escape the increasingly hostile conditions found in the gut. Since spore formation in the laboratory takes a minimum of 7 to 8 h, we reason that spore formation must be occurring simultaneously with spore germination at about 18 h and thereafter.
Acknowledgments
We thank Dana Cohen for her assistance in the early part of this study and Pat Piggot for the gift of SL6913.
This work was supported by grants from the Wellcome Trust and the EU Vth Framework to S.M.C.
REFERENCES
- 1.Angert, E. R., and R. M. Losick. 1998. Propagation by sporulation in the guinea pig symbiont Metabacterium polyspora. Proc. Natl. Acad. Sci. USA 95:10218-10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1999. Probiotics: snake oil for the new millenium? Environ. Microbiol. 1:375-382. [DOI] [PubMed] [Google Scholar]
- 3.Caruso, A., G. Flamminio, S. Folghera, L. Peroni, I. Foresti, A. Balsari, and A. Turano. 1993. Expression of activation markers on peripheral-blood lymphocytes following oral administration of Bacillus subtilis spores. Int. J. Immunopharmacol. 15:87-92. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Ciprandi, G., A. Scordamaglia, D. Venuti, M. Caria, and G. W. Canonica. 1986. In vitro effects of Bacillus subtilis on the immune response. Chemioterapia 5:404-407. [PubMed] [Google Scholar]
- 6.Deuerling, E., B. Paeslack, and W. Schumann. 1995. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J. Bacteriol. 177:4105-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller, R. 1991. Probiotics in human medicine. Gut 32:439-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 10.Gialdroni-Grassi, G., and C. Grassi. 1985. Bacterial products as immunomodulating agents. Int. Arch. Allergy Appl. Immunol. 76:119-127. [DOI] [PubMed] [Google Scholar]
- 11.Grasso, G., P. Migliaccio, C. Tanganelli, M. A. Brugo, and M. Muscettola. 1994. Restorative effect of Bacillus subtilis spores on interferon production in aged mice. Ann. N. Y. Acad. Sci. 717:198-208. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton-Miller, J. M. T., and G. R. Gibson. 1999. Efficacy studies of probiotics: a call for guidelines. Br. J. Nutr. 82:73-75. [DOI] [PubMed] [Google Scholar]
- 13.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 14.Hisanga, S. 1980. Studies on the germination of genus Bacillus spores in rabbit and canine intestines. J. Nagoya City Med. Assoc. 30:456-469. [Google Scholar]
- 15.Hoa, T. T., L. H. Duc, R. Isticato, L. Baccigalupi, E. Ricca, P. H. Van, and S. M. Cutting. 2001. The fate and dissemination of Bacillus subtilis spores in a murine model. Appl. Environ. Microbiol. 67:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Ragione, R. M., G. Casula, S. M. Cutting, and S. M. Woodward. 2001. Bacillus subtilis spores competitively exclude Escherichia coli 070:K80 in poultry. Vet. Microbiol. 2062:133-142. [DOI] [PubMed] [Google Scholar]
- 17.Lysenko, E., T. Ogura, and S. Cutting. 1997. Characterization of the ftsH gene of Bacillus subtilis. Microbiology 143:971-978. [DOI] [PubMed] [Google Scholar]
- 18.Mazza, P. 1994. The use of Bacillus subtilis as an antidiarrhoeal microorganism. Boll. Chim. Farm. 133:3-18. [PubMed] [Google Scholar]
- 19.Moir, A., and D. A. Smith. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44:531-553. [DOI] [PubMed] [Google Scholar]
- 20.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Schaeffer, P., J. Millet, and J. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin, J., E. M. Bautista, Y.-B. Kang, and T. W. Molitor. 1998. Quantitation of porcine reproductive and respiratory syndrome virus RNA in semen by single-tube reverse transcription-nested polymerase chain reaction. J. Virol. Methods 72:67-79. [DOI] [PubMed] [Google Scholar]
- 25.Spinosa, M. R., T. Braccini, E. Ricca, M. De Felice, L. Morelli, G. Pozzi, and M. R. Oggioni. 2000. On the fate of ingested Bacillus spores. Res. Microbiol. 151:361-368. [DOI] [PubMed] [Google Scholar]
- 26.Tannock, G. W. (ed.). 1999. Probiotics: a critical review. Horizon Scientific Press, Norfolk, United Kingdom.