Abstract
To identify Shiga toxin-producing Escherichia coli genes associated with severe human disease, a genomic subtraction technique was used with hemolytic-uremic syndrome-associated O91:H21 strain CH014 and O6:H10 bovine strains. The method was adapted to the Shiga toxin-producing E. coli genome: three rounds of subtraction were used to isolate DNA fragments specific to strain CH014. The fragments were characterized by genetic support analysis, sequencing, and hybridization to the genome of a collection of Shiga toxin-producing E. coli strains. A total of 42 fragments were found, 19 of which correspond to previously identified unique DNA sequences in the enterohemorrhagic E. coli EDL933 reference strain, including 7 fragments corresponding to prophage sequences and others encoding candidate virulence factors, such a SepA homolog protein and a fimbrial usher protein. In addition, the subtraction procedure yielded plasmid-related sequences from Shigella flexneri and enteropathogenic and Shiga toxin-producing E. coli virulence plasmids. We found that lateral gene transfer is extensive in strain CH014, and we discuss the role of genomic mobile elements, especially bacteriophages, in the evolution and possible transfer of virulence determinants.
Shiga toxin-producing Escherichia coli (STEC) has the capacity to cause a variety of diseases ranging from uncomplicated diarrhea to hemorrhagic colitis and life-threatening systemic infections, such as the hemolytic-uremic syndrome. Foodborne STEC infections, either outbreaks or sporadic cases, appear worldwide. The major characteristic of STEC that has been linked to virulence is the production of Shiga toxins (Stx1 and/or Stx2) (25). Several other determinants have been implicated in virulence, such as intimin, which is involved in the binding of bacteria to target cells, and factors encoded by a large virulence plasmid. Among these are an enterohemolysin (E-hlyA), an extracellular serine protease (EspP), a catalase-peroxidase (KatP), and a type II secretory system. These factors are encoded by elements that have been acquired by horizontal transfer from an outside source, i.e., prophages, pathogenicity islands, and plasmids (12, 35, 46, 50).
Cattle appear to be the main reservoir of various STEC strains. Several studies have shown a high prevalence of STEC strains belonging to a wide range of serotypes in animals and food products (3, 6, 34, 48). However, only a limited number of serotypes have been associated with human disease, among which O157:H7 is predominant. Moreover, different combinations of potential virulence factors have been observed in STEC clinical isolates, in addition to the production of Shiga toxins. Thus, the known virulence factors do not allow differentiation of STEC strains with a high pathogenic potential from their counterparts of lesser clinical significance.
Between 1996 and 1997, six non-O157:H7 STEC strains were isolated from stool samples of adults with hemolytic-uremic syndrome in the teaching hospital of Clermont-Ferrand in France (9). Among them was strain CH014,which belongs to the O91:H21 serotype, which was previously associated with hemolytic-uremic syndrome cases in Finland and Canada (22, 26). Strain CH014 has the capacity to produce Stx2 and E-hlyA. In a prospective study done 1 year later in the same geographic area, STEC was isolated from bovine feces, food samples, and asymptomatic children (37). Among the strains found were eight belonging to the O6:H10 serotype that were isolated from both bovine and food samples. To our knowledge, strains of serotype O6:H10 have never been associated with human disease although they have the capacity to produce Stx2.
In a previous study, we have shown a high level of heterogeneity among STEC isolates from the same geographic area, even within strains of the same serotype. This heterogeneity seems to be due to mobile elements of the genome (36, 37). No characteristic has been found to be diagnostic for the pathogenic strains by comparison to their counterparts of cattle and food origin. Further studies are needed to identify special attributes, other than Stx production, necessary for the development of STEC pathogenesis in humans. The genomic subtraction technique has been previously used with success to identify specific DNA from several bacterial species (16, 19, 27). In this technique, an excess of sheared and denatured subtracter DNA is allowed to reassociate with enzyme-restricted and denatured DNA from the target bacterium. Nonspecific target sequences hybridize with complementary sequences of the subtracter DNA, leaving the preparation enriched for sequences unique to the target strain. The enriched sequences are amplified by PCR and cloned. They are then used as probes in Southern blot and colony blot assays to verify the specificity for the target DNA.
In the present study, a genomic subtractive hybridization procedure was used to identify CH014-specific DNA sequences that might encode factors involved in virulence. Several DNA fragments were identified that did not hybridize with DNA from the O6:H10 strains or with the E. coli K-12 laboratory strain. The data suggest that pathogenic STEC strains have been more extensively submitted to lateral gene transfer than have strains of lesser virulence. Some of the isolated fragments are good candidates for components of virulence determinants of STEC strains.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in the study are listed in Table 1. Pathogenic O91:H21 STEC strain CH014 was used for subtractive hybridization against strains NV110 and NV183 of serotype O6:H10. STEC strain CH014 was obtained from a patient with hemolytic-uremic syndrome in April 1997 (9). Strains NV110 and NV183 were collected in the feces of healthy cattle at the city slaughterhouse in October 1997 and March 1998, respectively (37). Eighteen strains were used in colony blot hybridization experiments. Five STEC strains of serotype O91:H21 and five of serotype O6:H10 were collected in the same geographic area (central France) between October 1997 and July 1998 from the feces of healthy cattle or from food products (beef or cheese); five human STEC isolates were obtained from sporadic cases of hemolytic-uremic syndrome in central France; and E. coli EDL933 (ATCC 43895) of serotype O157:H7, enteropathogenic E. coli (EPEC) E2348/69, and E. coli DH5α were used as reference strains. A collection of 220 STEC strains (designated NV) was used for screening with particular genomic fragments (37).
TABLE 1.
Characteristics of the bacterial strains used in this study
| Strain | Serotype | Characteristic(s)a | Source (reference)b |
|---|---|---|---|
| CH013 | O91:H10 | stx2stx2vhastx2vhb | HUS (9) |
| CH014 | O91:H21 | stx2stx2vhastx2vhbehxA | HUS (9) |
| CH015 | OR:H16 | stx2ehxA espP | HUS (9) |
| CH016 | Ox3:H− | stx2vha | HUS (9) |
| CH017 | Ont:H− | stx2vha | HUS (9) |
| CH087 | O103:H2 | stx1eae ehxA espP efa-1 | HUS (this study) |
| EDL933 | O157:H7 | stx1stx2eae ehxA espP efa-1 | STEC reference strain |
| E2348/69 | O127:H6 | eae ehxA espP efa-1 | EPEC reference strain |
| DH5α | K-12 | Laboratory strain | |
| JM109 | K-12 | Laboratory strain | |
| NV32 | O91:H21 | stx2vhbehxA | Beef (37) |
| NV74 | O91:H21 | stx2vhbehxA | Cheese (37) |
| NV127 | O91:H21 | stx2vhastx2vhbehxA | Cattle (37) |
| NV197 | O91:H21 | stx2, stx2vhbehxA | Cattle (37) |
| NV200 | O91:H21 | stx2stx2vhbehxA | Cattle (37) |
| NV106 | O6:H10 | stx2-NV206 | Cattle (37) |
| NV107 | O6:H10 | stx2-NV206 | Cattle (37) |
| NV110 | O6:H10 | stx2-NV206 | Cattle (37) |
| NV139 | O6:H10 | stx2-NV206 | Cattle (37) |
| NV183 | O6:H10 | stx2-NV206 | Cattle (37) |
| NV206 | O6:H10 | stx2-NV206 | Cattle (37) |
| NV268 | O6:H10 | stx2 | Cattle (37) |
stx1, stx2 variants, eae, ehxA, espP, and efa-1 genes were detected by PCR and specific hybridizations (unpublished data).
HUS, hemolytic-uremic syndrome.
Bacteria were grown at 37°C in Müller-Hinton broth (Biokar Diagnostics, Beauvais, France) or in Luria broth (Difco, Detroit, Mich.). As necessary, media were supplemented with 100 μg of ampicillin per ml. Bacteria were stored as frozen cultures at −80°C in either Müller-Hinton or Luria broth containing 15% glycerol.
DNA preparation for genomic subtraction experiments.
Total genomic DNA was extracted by the method of Picard-Pasquier et al. (33). DNA purity was verified by determining the UV absorption ratio. Target DNA was digested to completion with the Sau3A enzyme (Roche Molecular Biochemicals, Mannheim, Germany) in accordance with the supplier's instructions. After digestion, DNA was extracted once with phenol-chloroform, precipitated with ethanol, washed, and suspended at 0.25 μg/μl in 2.5× EE buffer (25 mM N-2-hydroxyethylpiperazine-N′-3-propanesulfonic acid, 2.5 mM EDTA, pH 8.0).
Five hundred micrograms of the DNA used to subtract nonspecific sequences (subtracter DNA) was fragmented by sonication in 0.5 ml of H2O in a Branson Sonifier 450 in the continuous mode and with an output setting of 2 for 30 s. The average size of DNA fragments was approximately 1 kb. Biotinylation of subtracter DNA was performed on ice in a dark room. One hundred microliters of sheared DNA at 1 μg/μl and 100 μl of photobiotin acetate (Sigma-Aldrich Chimie, St Quentin Fallavier, France) at 2 μg/μl in distilled water were added. The mixture was photoactivated three times by 10 min of illumination (365 nm) from a UV lamp (VL-6LC; Vilber-Lourmat, Marne-la-Vallée, France). After addition of 1 M Tris-HCl (pH 9.0) to a final concentration of 100 mM, biotinylated DNA was extracted four times with water-saturated 1-butanol, ethanol precipitated, washed, and suspended at 5 μg/μl in 2.5× EE buffer.
Genomic subtractive hybridization.
Ten micrograms of biotinylated subtracter strain DNA and 0.25 μg of target strain DNA were used for the first round of genomic subtraction by the protocol of Straus and Ausubel (44) with minor modifications. Briefly, the mixture of target and subtracter DNAs in 4 μl of 2.5× EE buffer, overlaid with mineral oil, was denatured at 100°C for 1 min and then mixed with 1 μl of 5 M NaCl. Hybridization was carried out in a 65°C air incubator in a 0.5 ml centrifuge tube. After 18 h at 65°C, 95 μl of EEN buffer (1× EE, 500 mM NaCl) was quickly and thoroughly mixed with the sample, which was then combined with 100 μl of a 2% suspension of streptavidin-coated polystyrene beads (Dynabeads M-280; Dynal A.S., Oslo, Norway) that had been washed in EEN. The sample was incubated at room temperature for 15 min and then placed on a magnet (Dynal MPC) for 2 min in accordance with the supplier's instructions. The beads, which were retained, were washed once with 200 μl of EEN and once with 100 μl of EEN. The unbound nucleic acid in the supernatant was precipitated at −20°C following the addition of 2 volumes of ethanol. The resulting pellet was washed with ethanol, dried, and resuspended in 5 μl of 1× EE buffer. A 0.5-μl volume was saved for analysis. The remaining sample was dried, resuspended in 2 μl of 2.5× EE buffer, and combined with 10 μg of biotinylated subtracter DNA. Two more rounds of denaturation, reassociation, and avidin selection were performed as described above. Aliquots (1/10 of each unbound fraction) were saved after each cycle.
PCR amplification of remaining subtracted DNA.
Double-stranded adapters for PCR amplification were prepared by complementary annealing of synthetic 24-mer oligonucleotide Sau1 (5′ GACACTCTCGAGACATCACCGTCC 3′) and 26-mer oligonucleotide Sau2 (5′P GATCGGACGGTGATGTCTCGAGAGTG 3′; Sau3A site underlined), which overlap at 22 nucleotide positions. Five-microgram samples of the two oligomers at 1 μg/μl (10 μl in total) were combined, and the mixture was heated to 100°C for 2 min in a 200-ml water bath that was then allowed to cool to room temperature. At each subtraction cycle, 0.5 μl of residual subtracted DNA was ligated for 5 min at room temperature with 150 ng of the resulting adapters by using a Rapid DNA Ligation Kit (Roche Molecular Biochemicals).
After ligation, the sample was purified by using a QIAquick PCR Purification Kit (QIAGEN S.A., Courtaboeuf, France) in accordance with the supplier's instructions. DNA capped with adapters was eluted with 30 μl of water, and 5 μl was used as a template for PCR amplification. Primer Sau1 was used at 1 μM with 200 μM each deoxynucleoside triphosphate (Roche Molecular Biochemicals), 1× reaction buffer, and 1 U of Taq DNA polymerase (Appligène-Oncor, Illkirch, France) in a 50-μl reaction volume. The PCR cycle included denaturation for 90 s at 94°C, primer annealing for 90 s at 65°C, and an extension step of 90 s at 72°C (30 cycles) on a Perkin-Elmer Cetus DNA thermal cycler 2400. Reaction products were then separated by electrophoresis on 3% agarose gel with 1 μg of ethidium bromide (ProLabo, Strasbourg, France) per ml at 100 V for 6 h in 1× Tris-acetate-EDTA buffer. DNA fragments separated by electrophoresis were cut on the gel, purified separately on 0.22-μm-pore-size filters (SPIN-X; Costar, Cambridge, Mass.), ethanol precipitated, and resuspended in 25 μl of water.
Cloning of fragments from the subtracted library.
Five microliters of each purified DNA fragment was used for a second round of amplification under the conditions described above. The reaction products were again purified on QIAGEN columns and eluted with 50 μl of water. One microliter of this solution was used for the ligation reaction with plasmid pCR2.1 (Original TA Cloning Kit; Invitrogen, Groningen, The Netherlands). The ligation protocol used was that described by the suppliers. E. coli strain JM109 (Promega, Charbonnieres, France) was transformed by electroporation (18) with selection for ampicillin (100 μg/ml) on Luria-Bertani agar (Difco) plates. Clones obtained were cultured individually in 10 ml of Luria broth (Difco) containing ampicillin (100 μg/ml). Lysates were prepared from 1.5 ml of an overnight Luria broth (Difco) culture by boiling at 100°C for 15 min. Following centrifugation of the lysate, 5 μl of the supernatant was tested by PCR with primer Sau1 as described above.
Probe preparation and hybridization experiments.
PCR products used as probes were purified on 0.22-μm-pore-size SPIN-X filters and radiolabeled with [α-32P]dATP (ICN Pharmaceuticals France S.A., Orsay, France) by using a random-primed DNA labeling kit (Roche Molecular Biochemicals) in accordance with the manufacturer's specifications. Southern blot assays were performed either with DNA transferred after gel electrophoresis of genomic DNA digested with the EcoRI or HindIII enzyme (Roche Molecular Biochemicals), with pulsed field gel electrophoresis (PFGE) patterns, with PCR products, or with plasmids obtained by the alkaline lysis method described by Kado and Liu (24). Smart Ladder (Eurogentec, Angers, France) was used as a digested genomic DNA size marker. After gel electrophoresis, DNA was transferred to Hybond N+ nylon membranes (Amersham) by standard methods. Colony blots were performed by following standard procedures (29). Hybridization was performed with a rapid hybridization buffer (Amersham Pharmacia Biotech, Orsay, France) as described by the manufacturer. Hybridized membranes were then washed successively at 65°C for 20 min, once with 0.1% sodium dodecyl sulfate-0.3 M NaCl-0.03 M sodium citrate and twice with 0.1% sodium dodecyl sulfate-75 mM NaCl-7.5 mM sodium citrate, and then exposed to Hyperfilm MP (Amersham) and processed in an automated film developer (Hyperprocessor; Amersham). The stx2-specific probe was prepared from the PCR product of strain EDL933 with primers LP43 and LP44, which were described by Cebula et al. (15).
Sequencing.
Plasmid DNA for sequencing was prepared with QIAGEN minicolumns (QIAGEN Plasmid Mini Kit) in accordance with the supplier's instructions. PCR products for sequencing were prepared by using the QIAquick PCR Purification Kit (QIAGEN S.A.). DNA sequencing was carried out on an automated sequencer by the fluorescent dye termination method, and the sequence was edited by using the manufacturer's software (GENOME Express, Grenoble, France). BLASTN and BLASTX sequence homology analyses were performed by using the National Center for Biotechnology Information BLAST network service.
PFGE analysis.
Genomic DNA was prepared by following the protocol described by Böhm and Karch (8). DNA digestion was performed with 50 U of XbaI (Life Technologies, Cergy Pontoise, France) for 18 h at 37°C. PFGE was performed in 1.2% agarose with a contour-clamped homogeneous electric field (CHEF)-PFGE Gene Navigator apparatus (Pharmacia, Uppsala, Sweden) in 0.5× Tris-borate-EDTA buffer at 200 V and 14°C. Pulse times were increased from 10 to 40 s over 24 h. A Lambda Ladder (Bio-Rad, Ivry, France) was used as molecular weight markers.
Nucleotide sequence accession numbers.
The sequences of the O91:H21 strain CH014 DNA fragments described here, which are absent in the O6:H10 STEC strains and in E. coli K-12, have been deposited in the GenBank database under accession no. AF467504 to AF467532.
RESULTS
Enrichment in sequences specific to pathogenic STEC strain CH014.
STEC strains of serotype O6:H10, although largely present in the bovine reservoir, have not been associated with pathogenicity in humans and thus might lack some of the attributes present in virulent STEC strains (37). To identify genomic sequences of O91:H21 strain CH014 that are absent from STEC strains of serotype O6:H10, genomic subtractive hybridization was used.
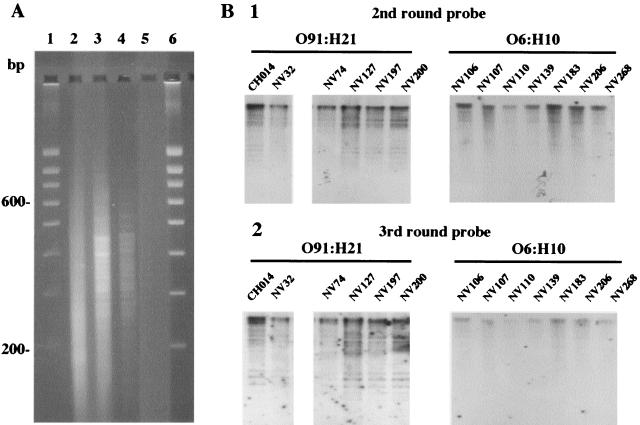
The DNA of pathogenic strain CH014 was subjected to subtraction by using mixed DNAs from two STEC strains (NV110 and NV183) of serotype O6:H10. The use of two subtracter strains ensured that sequences were not isolated due to their absence from one particular strain of serotype O6:H10. In this experiment, DNA fragments of CH014 that hybridize with the DNAs of strains NV110 and NV183 are selectively removed. The number of subtraction cycles was chosen to optimize enrichment in nonhomologous sequences. At each cycle, a fraction of subtracted DNA was amplified by PCR. Figure 1A shows the electrophoretic analysis of the amplified DNA derived from cycles 1 to 3. The product was 32P labeled and used to probe HindIII-digested genomic DNAs from the target and subtracter strains. For comparison, five isolates of serotype O91:H21 and five isolates of serotype O6:H10, obtained from cattle or food (Table 1), were included in the analysis. Figure 1B shows the results obtained. After hybridization with the probe derived from the third cycle, the prominent signals correspond to bands generated by the target genomic DNA and its O91:H21 counterparts but missing in O6:H10. The use of four cycles of subtraction resulted in DNA signal loss (data not shown). Thus, three rounds of genomic subtraction provided sufficient enrichment for accurate identification of sequences that are absent in the subtracter strains.
FIG. 1.
Analysis of the PCR products of genomic subtraction. (A) Gel electrophoresis of the PCR products in 3% agarose. Lanes: 1 and 6, Smart Ladder SF (Eurogentec); 2, amplified DNA of strain CH014 after a first cycle of genomic subtraction; 3, amplified DNA after a second round of genomic subtraction; 4, amplified DNA after a third cycle of genomic subtraction; 5, PCR-negative control. (B) Southern blots of HindIII-digested genomic DNAs of 13 STEC strains with the radioactively labeled PCR products of subtractive hybridization: 1, hybridization with the PCR product of the second round of subtraction; 2, hybridization with the PCR product of the third round of subtraction. Strains: CH014, target O91:H21 STEC strain; NV32, O91:H21 STEC strain isolated from beef; NV74, O91:H21 STEC strain isolated from cheese; NV127, NV197, and NV200, O91:H21 STEC strains isolated from cattle; NV110 and NV183, subtracter O6:H10 STEC strains isolated from cattle; NV106, NV107, NV139, NV206, and NV268, O6:H10 STEC strains isolated from cattle.
Molecular cloning of subtracted genomic sequences.
Following three rounds of subtraction, PCR amplification of CH014 DNA gave distinct products ranging in size from <0.2 to 1 kb, with 10 main fragments of approximately 0.3 to 0.6 kb (Fig. 1A). Each of the 10 fragments was submitted to a second round of PCR using the Sau1 oligonucleotide, corresponding to one of the adapter sequences adjacent to the CH014 DNA fragments. The amplified products were cloned, and inserts were individually amplified by PCR using primer Sau1. To eliminate clones carrying the same insert, an initial screening was performed by blotting individual amplified inserts from each clone onto a membrane and using the labeled PCR product corresponding to each insert as a probe. Of 110 inserts tested (11 each of 10 size classes), 62 corresponded to different sequences.
Probe specificity assessment and distribution among STEC strains.
To confirm the absence of the 62 fragments on the genomes of the two O6:H10 subtracter strains, colony blots and Southern blots of EcoRI- or HindIII-digested genomic DNAs from strains CH014, NV110, and NV183 were probed with the radioactively labeled PCR product of each insert. Of 62 inserts, 42 (68%) gave no signal with the NV110 and NV183 subtracter DNAs. The extent of hybridization of the 42 specific subtracted sequences to genomic DNAs from 18 strains was studied by colony blot experiments. As expected, no hybridization with genomic DNAs from five O6:H10 STEC strains was observed. Ten of the 42 inserts hybridized to DNA from DH5α, a standard laboratory E. coli K-12 strain. These 10 fragments were then eliminated for subsequent analyses. The hybridization profiles of the remaining 32 fragments are summarized in Table 2. Hybridization with DNAs from all five O91:H21 STEC strains obtained from cattle or food products was detected for the majority (25 of 32) of the fragments. Two fragments (S-16 and S-28) hybridized only with one of the O91:H21 strains (profiles K and S). In order to determine whether they were present only in rare STEC strains or whether they were found in varied STEC serotypes from diverse origins, the incidence of the two fragments was studied by colony blot assays of a large sample of STEC isolates (NV collection of 220 strains [37]). Their presence was not correlated with the origin of the isolates or with particular serotypes (data not shown).
TABLE 2.
Colony blot hybridization of 32 probes derived from subtractive hybridization with a panel of E. coli strains
| Straina | Serotype | Hybridizationb with colony blot pattern (fragment[s]):
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A (S-1) | B (S-2, S-10, S-30) | C (S-3, S-4, S-27, S-31) | D (S-5, S-25) | E (S-6) | F (S-7) | G (S-8) | H (S-9) | I (S-11) | J (S-12, S-13, S-14, S-15, S-32) | K (S-16) | L (S-17) | M (S-18) | N (S-19, S-20, S-21) | O (S-22) | P (S-23) | Q (S-24) | R (S-26) | S (S-28) | T (S-29) | ||
| NV32 | O91:H21 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| NV74 | O91:H21 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| NV127 | O91:H21 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NV197 | O91:H21 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| NV200 | O91:H21 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| CH014 | O91:H21 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| CH013 | O91:H10 | + | + | + | + | + | + | + | |||||||||||||
| CH015 | OR:H16 | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||
| CH016 | Ox3:H− | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||
| CH017 | Ont:H− | + | + | + | + | + | + | + | + | + | + | ||||||||||
| CH087 | O103:H2 | + | + | + | + | + | + | + | + | + | + | + | |||||||||
| EDL933 | O157:H7 | + | + | + | + | + | + | + | + | + | |||||||||||
| EPEC E2348/69 | O127:H6 | + | + | + | + | + | + | ||||||||||||||
The NV strains are from animal or food origins (Table 1), and the CH strains are of hemolytic-uremic syndrome origin.
No hybridization to DNA from five O6:H10 STEC strains or from strain DH5α was observed. Fragments are named S-1 to S-32. +, positive hybridization signal.
The abilities of the probes to hybridize with six hemolytic-uremic syndrome-associated STEC strains of different serotypes and to EPEC E2348/69 varied. Four fragments (S-2, S-10, S-24, and S-30) hybridized with the six pathogenic strains tested (profiles B and Q). One of them (S-24) also hybridized with the EPEC strain (profile Q). Ten other fragments hybridized with the E2348/69 EPEC reference strain but not with all of the pathogenic STEC strains (profiles A, D, G, H, and J). In contrast to the reactivity with all of the O91:H21 strains tested, four fragments (profile C) gave no signal with the other strains tested, suggesting that these fragments could be specific for the O91:H21 serotype.
Location on the CH014 genome.
The location of each fragment was analyzed by Southern hybridization on the >90-kb CH014 plasmid (named pO91:H21) (36). Nine of the 32 fragments were found to be plasmid located (Table 3). Five of them (S-1, S-2, S-6, S-7, and S-9) gave different hybridization profiles with six pathogenic STEC strains by colony blot assay, and the other four (S-3, S-4, S-27, and S-31) corresponded to the O91:H21-specific fragments (Table 2).
TABLE 3.
Sequence analysis of CH014-specific subtracted DNA fragments.
| Group and fragment name | Sequence size (bp) | Protein with similaritya | Organism | % Nucleo- tide identity | % Amino acid identity | Locationb | EcoRI- digested fragment size(s) (kb) | HindIII-digested fragment size(s) (kb)c |
|---|---|---|---|---|---|---|---|---|
| I | ||||||||
| S-1 | 327 | Transport protein E-HlyB | EDL933(pO157) | 96 | 98 | pl | NDd | ND |
| S-2 | 305 | Unknown protein | Shigella flexneri(pWR100) | 92 | 86 | pl | ND | >10 |
| Unknown protein L,7076 | EDL933(pO157) | 88 | 85 | |||||
| S-3 | 307 | Tissue invasion protein SepA | Shigella flexneri(pWR100) | 37 | pO91:H21 | 9 | 8 | |
| Serine protease EspP | EDL933(pO157) | 26 | ||||||
| S-4 | 160 | Probable IS transposase | Shigella flexneri(pWR100) | 81 | 57 | pO91:H21 | 10 | ND |
| S-5 | 336 | Copy number control protein CopB | EPEC(pB171) | 99 | 98 | chr | >10 | >10 |
| Copy number control protein CopB | EDL933(pO157) | 93 | 60 | |||||
| S-6 | 281 | Helicase Tral | E. coli(plasmid F) | 97 | 97 | pl | >10 | 10 |
| S-7 | 193 | Unknown protein ORF2 | E. coli(plasmid IncF) | 89 | 75 | pl | 9 | 8 |
| S-8 | 287 | Type I restriction enzyme EcoR124II R HsdR | E. coli(plasmid R124/3) | 95 | 95 | chr | >10 | 8 |
| II | ||||||||
| S-9 | 324 | Probable capsid portal protein GpQ | Phage P2 | 94 | 99 | pl | 5 | ND |
| S-10 | 327 | H tail component | Phage λ | 99 | 98 | chr | >10 | ND |
| CP-933X tail component | EDL933 | 97 | 97 | |||||
| S-11 | 234 | DNA-packaging protein | Phage λ | 95 | 89 | chr | 2.5 | ND |
| CP-933X DNA-packaging protein | EDL933 | 94 | 89 | |||||
| S-12 | 288 | CP-933X-V head-tail adapter | EDL933 | 98 | 98 | chr | >10, >10 | >10, >10 |
| S-13 | 302 | CP-933M, -N, -V, -X unknown protein | EDL933 | 99 | 97 | chr | >10, >10 | >10, >10 |
| S-14 | 123 | CP-933M, -N, -V portal protein | EDL933 | 95 | 68 | chr | >10, >10 | >10, >10 |
| S-15 | 277 | CP-933M, -N, -V unknown protein | EDL933 | 97 | 96 | chr | ND | >10, >10 |
| S-16 | 631 | CP-933M DicA similar protein | EDL933 | 50 | chr | <1 | ND | |
| S-17 | 224 | CP-933N, -O, -U unknown protein | EDL933 | 93 | 90 | chr | 2.5 | ND |
| S-18 | 295 | CP-933U unknown protein | EDL933 | 99 | 98 | chr | 10 | >10 |
| S-19 | 234 | CP-933C terminase | EDL933 | 86 | 94 | chr | >10, >10 | ND |
| S-20 | 299 | CP-933C terminase | EDL933 | 95 | 97 | chr | >10, >10 | ND |
| S-21 | 300 | CP-933C head maturation protease | EDL933 | 95 | 98 | chr | >10, >10 | ND |
| III | ||||||||
| S-22 | 1,342 | O island no. 154 fimbrial usher protein | EDL933 | 78 | 76 | chr | 5 | 10 |
| Fimbrial usher protein LpfC | Salmonella typhimurium | 50 | ||||||
| S-23 | 279 | O island no. 134 DNA processing protein | EDL933 | 98 | 97 | chr | 8 | ND |
| S-24 | 300 | O island no. 133 unknown protein | EDL933 | 91 | 93 | chr | 8 | ND |
| IV | ||||||||
| S-25 | 220 | General secretion pathway protein EpsK | Vibrio cholerae | 36 | chr | 8 | >10 | |
| S-26 | 340 | Peptide synthetase McyG | “Microcystis aeruginosa” | 40 | chr | 10, 8 | ND | |
| S-27 | 241 | pO91:H21 | 7 | >10 | ||||
| S-28 | 285 | chr | 10 | ND | ||||
| S-29 | 305 | chr | 10 | >10, 8 | ||||
| S-30 | ND | chr | 8 | 8 | ||||
| S-31 | ND | pO91:H21 | 10 | 5 | ||||
| S-32 | ND | chr | >10, >10 | >10, >10 |
Determined by comparison of sequences and coding regions with the EMBL and GenBank DNA databases. BLAST and BLASTX network services were used. IS, insertion sequence.
chr, chromosome located; pl, plasmid located; pO91:H21, O91:H21-specific plasmid located.
Two values mean two different-size fragments.
ND, not determined.
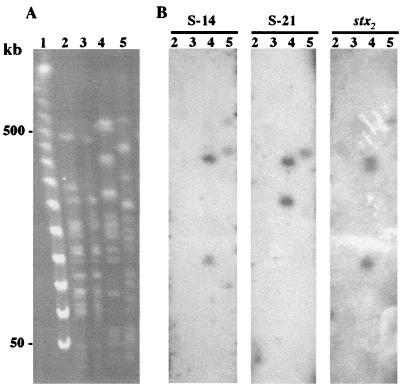
Southern blot analysis of EcoRI- or HindIII-digested CH014 genomic DNA with the 32 radioactively labeled subtracted fragments gave patterns of one or two bands varying in size from <1 to >10 kb (Table 3). Among them, eight probes (S-12, S-13, S-14, S-15, S-19, S-20, S-21, and S-32) gave the same pattern of two bands of >10 kb when hybridized with either EcoRI or HindIII restriction fragments. These eight chromosome-located fragments were grouped into two colony blot profiles, J and N (Table 2). In order to determine whether these fragments corresponded to contiguous sequences, genomic DNAs of the CH014, NV110, and NV183 strains were digested with XbaI, resolved by PFGE (Fig. 2A), and blotted for Southern hybridization with each probe. The eight probes hybridized with two bands of the CH014 PFGE profile (Fig. 2B). The patterns obtained with the S-12, S-13, S-15, and S-32 probes were identical to the pattern obtained with the S-14 probe. The patterns obtained with the S-19 and S-20 probes were identical to the pattern obtained with the S-21 probe. As indicated by their sequences (see below), the probes did not contain EcoRI, HindIII, or XbaI sites. Therefore, they should correspond to sequences present in distantly related duplicates on the genome.
FIG. 2.
Genomic XbaI restriction patterns of subtracter strain NV110 (lane 2), subtracter strain NV183 (lane 3), target strain CH014 (lane 4), and reference strain EDL933 of serotype O157:H7 (lane 5) obtained by PFGE (A) and Southern hybridization (B) with S-14, S-21, and stx2 as DNA probes. Lambda Ladder (Bio-Rad) was used as a DNA size marker (lane 1 in panel A).
DNA sequencing and sequence analysis.
The 32 fragments (named S-1 to S-32) that gave no signal with the O6:H10 STEC DNA or with the DH5α DNA were sequenced. The sequences of three fragments (S-30, S-31, and S-32) could not be determined. For each of the 29 remaining sequences, the GenBank (release 123.0), EMBL (release 66.0), and DDBJ (release 37.0) databases were screened at the National Center for Biotechnology Information for similarities. Table 3 summarizes the sequence analysis and possible similar proteins derived from six-frame translations of the DNA databases (see also http://www.u-clermont1.fr/pharma/recherche/bacterioviro/presentation.htm). The sequences fell into three major groups: (i) 8 fragments with similarities to sequences from plasmids of E. coli or from the pWR100 virulence plasmid of Shigella flexneri (13) (S-1 to S-8), (ii) 13 fragments with similarities to phage sequences (S-9 to S-21), and (iii) 3 fragments (S-22 to S-24) with similarities to specific regions of the EDL933 genome called O islands (32). A fourth group included two further sequences (S-25 and S-26) that presented only 54 and 58% similarity, at the amino acid level, to proteins described in other bacterial genera (31, 41). Furthermore, the nucleotide sequences and the deduced amino acid sequences of three fragments (S-27 to S-29) did not exhibit similarity to any sequences available in the current databases.
Among the sequences of interest, the S-1 fragment corresponded to the sequence of the ehxB gene, which is part of the ehxABCD operon located on STEC large virulence plasmids (42). The ehxA gene, which is part of the operon, was previously detected on the >90-kb CH014 plasmid (36) and was absent from the O6:H10 strains, possessing only a small 3.5-kb plasmid (data not shown). Thus, this result validates the experiments and confirms that the genomic subtraction technique has potential for pinpointing of regions that are most likely to be involved in the differential virulence of bacterial pathogens.
Two fragments (S-3 and S-22) presented similarities at the amino acid level to proteins whose functions have been established in the colonization and survival of pathogenic strains in the host, such as the SepA protein of S. flexneri, which is involved in tissue invasion (2), and the LpfC outer membrane usher protein of Salmonella enterica serovar Typhimurium, which is involved in the formation of long polar fimbriae that could play a role in host colonization (1). Such a fimbrial operon seems to be located on the EDL933 O#154 island (32). Fragment S-3 also presented similarities at the amino acid level, but to a lesser extent, to the pO157-located serine protease EspP. These two fragments are good candidates as components of new virulence determinants of STEC strains.
Overall, many sequences related to DNA rearrangements and to plasmid and phage sequences have been identified. They include those showing similarities at the amino acid level to a probable insertion sequence transposase of pWR100 (S-4); an EDL933-specific protein involved in DNA processing (S-23); sequences from the pO157, pWR100, pB171, F, IncF, and R124/3 plasmids; and sequences from the P2 and λ phages and seven CP-933 prophages. They suggest evolutionary events by which virulence genes may have been acquired and adaptation to the host could occur (28).
Comparison of the genomes of strains CH014 and EDL933.
As many as 19 of the 29 sequenced CH014 fragments, which are lacking on the O6:H10 STEC genome, had similarities to several regions identified on the EDL933 genome (Table 3). Fragments S-1, S-2, S-3, and S-5 presented similarities, at the amino acid level, to E-hlyB, the unknown protein L7076, EspP, and CopB, which are encoded on pO157, respectively (14).
Furthermore, as many as 15 fragments presented similarities to sequences of the EDL933 chromosome (32). Figure 3 shows the locations of the sequences on the diagrammatic EDL933 genome map. Of the 15 fragments, 12 (S-10 to S-21) corresponded to the sequences of seven prophages, namely, CP-933 C, CP-933 M, CP-933 N, CP-933 O, CP-933 U, CP-933 V, and CP-933 X (Table 3). By hybridization with EcoRI- and HindIII-digested genomic DNA, we found that fragments S-12, S-13, S-14, S-15, S-19, S-20, S-21, and S-32 were related and were present in two or more copies on the CH014 genome. Fragments S-19, S-20, and S-21 showed similarities to sequences of the CP-933C prophage, and four others showed similarities to sequences of prophages CP-933 M, CP-933 N, CP-933 V, and CP-933 X (Table 3). Differences between the two groups were correlated with those suggested by colony blot experiments (Table 2, profiles J and N) and with those obtained by hybridization on XbaI-PFGE profiles (Fig. 2B). Indeed, probes S-19, S-20, and S-21 hybridized with one band at 500 kb on the EDL933 PFGE profile. The three probes hybridized with two bands on the CH014 chromosome (at 450 and 350 kb), which suggests the presence of at least two prophages related to the CP-933 C prophage on the CH014 chromosome. Probes S-12, S-13, S-14, S-15, and S-32 hybridized with three bands (at 600, 500, and 150 kb) on the EDL933 PFGE profile and two bands on the CH014 profile (at 450 and 200 kb). One band was found, upon hybridization with each probe, on both the CH014 (at 450 kb) and EDL933 (at 500 kb) chromosomes, which suggests a relationship among the eight fragments. Sequencing confirmed that the fragments corresponded to several EDL933 prophages, some parts of which were present on the CH014 chromosome in at least two copies. In order to analyze if such prophage-like regions could correspond to stx2-encoding bacteriophages, XbaI-digested CH014 DNA was hybridized with an stx2-specific probe. Three bands at 450, 425, and 200 kb were probed (Fig. 2B). Two bands (at 450 and 200 kb) were common to fragments S-12, S-13, S-14, S-15, and S-32. Thus, the stx2 gene seems to be present in at least three copies on the CH014 genome. Two copies appear to be physically related to fragments showing similarities to several EDL933 prophages.
FIG. 3.
Representative locations of strain CH014 O91:H21-specific fragments on the E. coli EDL933 O157:H7 strain genome map. Open squares show the distribution of the 23 major EDL933-specific sequences compared to the E. coli K-12 genome that are missing in CH014. Filled squares show the distribution of the 10 additional CH014-specific sequences in the EDL933-specific regions. The size of each block is proportional to the size of the corresponding island described by Perna et al. (32).
Three further fragments corresponded to other EDL933-specific regions named O islands. They presented similarities to genes encoding a DNA-processing protein (S-23), an unknown protein (S-24), and a fimbrial usher protein (S-22) that could be an unidentified virulence factor.
DISCUSSION
STEC is an emerging enteric pathogen, yet neither its virulence factors nor its phylogeny has been completely characterized. Recently, Reid et al. (40) showed that the highly virulent pathogen E. coli O157:H7 separated 4.5 million years ago from an ancestor it has in common with E. coli K-12. Moreover, phylogenetic analysis of housekeeping genes suggested that separate old lineages of E. coli have acquired the same virulence factors in parallel (20). On the other hand, significant strain-to-strain heterogeneity has been shown by the prevalence of various virulence factors and by the fact that only a minority of STEC strains have caused diseases in humans or animals (4). A STEC strain (CH014) of serotype O91:H21 was isolated from a stool specimen from a patient with hemolytic-uremic syndrome at the Clermont-Ferrand hospital (9). STEC strains of serotype O91:H21 have been previously isolated from patients with hemolytic-uremic syndrome in Canada and Finland and from a patient with diarrhea in New Zealand (22, 26, 39). In a previous study, we showed that a large proportion of bovine feces and food samples was contaminated with STEC strains in the Clermont-Ferrand area (37). Among the major STEC isolates were strains of serotype O6:H10, which have never been associated with human disease. Therefore, we hypothesized that some CH014 genes that are absent in the O6:H10 genome might have a role in STEC virulence.
To identify genes that are lacking in O6:H10 STEC strains, a subtractive hybridization procedure was applied to STEC strains. Indeed, subtractive hybridization remains the method of choice for large-scale isolation of virulence genes. By genomic subtraction against the E. coli K-12 reference strain, an avian pathogenic strain was found to carry a total of 12 unique regions with an estimated 350 kb of unique DNA (10). Well-known EHEC clone O157:H7, for which the strain EDL933 genome has just been sequenced, has a genome 20% larger than that of E. coli K-12 (5,341 versus 4,639 kb) (32). This provides a hint of the patterns we can expect to see as we explore the global species genome.
We have adapted the subtractive hybridization technique to allow isolation of a large number of probes that are specific for pathogenic STEC strains but not for their closely related nonpathogenic counterparts. Modifications were made to the original procedure described by Straus and Ausubel (44) that are appropriate for the different application. Thirty-two specific fragments were isolated from pathogenic strain CH014. The fragments corresponded to sequences that were absent from the NV110 and NV183 DNAs and from E. coli K-12, as was shown by Southern and colony blot hybridizations. The approach was validated by the fact that among the CH014 subtracted fragments recovered, one was found to be part of the ehxABCD operon previously identified in serotype O91:H21 target strain CH014 but not in the serotype O6:H10 subtracter strains (9, 37).
A possible outcome of this work was to isolate sequences related to the pathogenicity of STEC strains. However, it is likely that certain sequences are related to other physiological characteristics specific to CH014 or to the strains of serotype O91:H21. The data showed no correlation between the presence of a particular sequence and the strains obtained from patients with hemolytic-uremic syndrome. The recovered sequences showed different colony blot hybridization profiles with the pathogenic strains tested, indicating a heterogeneity among STEC strains and supporting the previous hypothesis of the existence of multiple pathogenic lineages (3, 38, 43, 49, 50). On the other hand, colony blot experiments showed that the O91:H21 strains were fairly homogeneous although some heterogeneity was seen with probes S-16 and S-28. Interestingly, strains NV32 and NV74, isolated from food products, differed from their counterparts isolated from cattle in not being recognized by probes S-19, S-20, and S-21 (corresponding to sequences of the CP-933 C prophage) and probe S-26, respectively (Table 2).
One application of genomic subtraction is to isolate DNA probes for diagnosis, as previously shown for Rhizobium meliloti or Xanthomonas (5, 27). One such application could be envisaged with the S-3, S-4, S-27, or S-31 probe (Table 2, profile C). As the large virulence plasmids are highly stable, these sequences, even if plasmid located, could be further investigated for use in detecting the presence of STEC strains of serotype O91:H21 in patients or in food products.
Of particular interest is recovered fragment S-3, which shows 61% amino acid similarity to the SepA protein of S. flexneri and 42% similarity to the EspP serine protease of large virulence plasmid pO157 of strain EDL933 (Table 3). The S-3 fragment localized on the >90-kb plasmid of strain CH014 seems to be O91:H21 specific. This suggests that a gene similar to the espP gene of pO157, previously not detected by colony blot hybridization with the specific probe described by Brunder et al. (11), could be present on the CH014 virulence plasmid. Thus, pO157 and the CH014 plasmid could be less different in composition than at first expected. The espP gene was only recently reported for STEC strains (11, 14, 30). The proteolytic activity of EspP suggests that it could be an autotransporter protein such as the SepA protein of S. flexneri (2), which has C-terminal homology to EspP. In O26:NM and O157:H7 strains, the espP gene is located within remnants of different insertion sequences on the large virulence-associated plasmid. Analysis of the DNA region in the neighborhood of the gene similar to espP in strain CH014 could yield clues about the evolution of this particular gene. Moreover, the results of this report question the hypothesis that large plasmids of STEC are variable elements with considerable heterogeneity in gene composition and arrangement (7, 12).
Only a limited number of genes have previously been shown to be involved in the adherence of STEC to host cells, and especially the initial attachment step of adherence is poorly understood (45). In this study, one fragment of interest, S-22, presented 87% amino acid similarity to the O#154 putative fimbrial usher protein of O157:H7 strain EDL933 and 66% similarity to that of S. enterica serovar Typhimurium. Whether this fragment is part of a new functional fimbrial operon and could have a role in the initial adherence step of colonization must be investigated. By colony blot hybridization, the fragment seems to be present in four of the six hemolytic-uremic syndrome-associated pathogenic strains.
Comparison of our results with the analysis of the O157:H7 EDL933 genome supports the idea that genomes of the pathogenic STEC strains are particularly subject to recombinational evolution. Of 29 sequences obtained, 19 presented similarities to the O157:H7 EDL933 genome but were absent from the O6:H10 genomes. Among them, 12 fragments were recovered, some of which were present in several copies on the CH014 chromosome, that had similarities to the sequences of 7 of the 18 multigenic bacteriophage-related regions recently described in O157:H7 (32).
It is possible that such prophages contain new, unidentified virulence factors. The fact that stx2 genes appear to be located in the same regions of the CH014 chromosome suggests that some of them could correspond to stx-encoding phages. Unkmeir and Schmidt (46) have recently shown that crucial differences exist in the stx-flanking regions of different Stx-converting bacteriophages. Although the phages are similar in morphology, have similar modes of replication, and are similar in genomic structure, they may be unrelated at the nucleotide level (17, 21, 47). They might excise and reintegrate at different sites on the bacterial chromosome and could be a source of genetic heterogeneity in STEC isolates (23). This is also supported by our finding that strain CH014 contains sequences that originally stem from different phages. The role of stx2-encoding bacteriophages as a source of genetic diversity in closely related STEC strains needs to be further investigated. Furthermore, hybridization of XbaI-digested CH014 genomic DNA with an stx2-specific probe showed a pattern of three bands, indicating the presence of at least three copies of the stx2 gene in strain CH014 versus one, shown by a pattern of one band, in strains NV183 and NV110. Such a difference in the number of stx2 copies could explain the pathogenicity of CH014.
In summary, this study shows that the subtractive hybridization technique is an efficient way to generate DNA fragments highly enriched in unique sequences. Most of the recovered fragments represented previously uncharacterized phage- or plasmid-borne sequences. Thus, the utility of the procedure is demonstrated by the generation of new and potentially important information on the genetic organization of STEC. The genomic subtraction method is not labor intensive and is applicable to the genomes of STEC strains. Our findings may facilitate the understanding of the evolution and the virulence properties of STEC and reveal a surprising level of differences between pathogenic O91:H21 strain CH014 and O6:H10 strains commonly found in cattle and food products in the same geographic area. Many similarities have been shown between the genome of CH014 and several mobile elements distributed on the genome of O157:H7 strain EDL933. The data suggest that pathogenic STEC strains have been submitted to extensive lateral gene transfer. Most differences in overall gene content are attributable to horizontal transfer and offer a wealth of candidate genes that may be part of new virulence determinants. We suggest that STEC strains comprise a heterogeneous set of pathogens that share specific phage- and plasmid-borne genes. However, the role of these various sequences in STEC pathogenicity warrants further investigation. Furthermore, additional comparisons of the genomes of other STEC strains are necessary before the complex relationships between STEC strains can be understood.
Acknowledgments
We thank C. P. Vivares, from the Laboratoire Parasitologie Moléculaire et Cellulaire, UMR CNRS 6023, Université Blaise Pascal, for providing the PFGE apparatus and for advice on PFGE experiments and Frédéric Carvalho for technical assistance with genomic subtraction.
REFERENCES
- 1.Bäumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjelloun-Touimi, Z., P. J. Sansonetti, and C. Parsot. 1995. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol. Microbiol. 17:123-135. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., D. Geier, H. Steinruck, S. Zimmermann, and F. Scheutz. 1993. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J. Clin. Microbiol. 31:2483-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L., D. Geier, S. Zimmermann, S. Aleksic, H. A. Gillepsie, and T. S. Whittam. 1997. Epidemiological relatedness and clonal types of natural populations of Escherichia coli strains producing Shiga toxins in separate populations of cattle and sheep. Appl. Environ. Microbiol. 63:2175-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjourson, A. J., and J. E. Cooper. 1988. Isolation of Rhizobium loti strain-specific DNA sequences by subtraction hybridization. Appl. Environ. Microbiol. 54:2852-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco, M., J. E. Blanco, J. Blanco, A. Mora, C. Prado, M. P. Alonso, M. Mourino, C. Madrid, C. Balsalobre, and A. Juarez. 1997. Distribution and characterization of faecal verotoxin-producing Escherichia coli (VTEC) isolated from healthy cattle. Vet. Microbiol. 54:309-319. [DOI] [PubMed] [Google Scholar]
- 7.Boerlin, P., S. Chen, J. K. Colbourne, R. Johnson, S. DeGrandis, and C. Gyles. 1998. Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in Shiga toxin-producing E. coli. Infect. Immun. 66:2553-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böhm, H., and H. Karch. 1992. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnet, R., B. Souweine, G. Gauthier, C. Rich, V. Livrelli, J. Sirot, B. Joly, and C. Forestier. 1998. Non-O157 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J. Clin. Microbiol. 36:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, P. K., and R. Curtiss, I. I. I. 1996. Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 93:11149-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 12.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 13.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. d'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 14.Burland, V., Y. Shao, N. T. Perna, G. Plunkett III, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cebula, T. A., W. L. Payne, and P. Feng. 1995. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33:248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darrasse, A., A. Kotoujansky, and Y. Bertheau. 1994. Isolation by genomic subtraction of DNA probes specific for Erwinia carotovora subsp. atroseptica. Appl. Environ. Microbiol. 60:298-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datz, M., C. Janetzki-Mittmann, S. Franke, F. Gunzer, H. Schmidt, and H. Karch. 1996. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl. Environ. Microbiol. 62:791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, P., K. A. Lampel, H. Karch, and T. S. Whittam. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750-1753. [DOI] [PubMed] [Google Scholar]
- 21.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 23.Iyoda, S., K. Tamura, K. Itoh, H. Izumiya, N. Ueno, K. Nagata, M. Togo, J. Terajima, and H. Watanabe. 2000. Inducible stx2 phages are lysogenized in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patients. FEMS Microbiol. Lett. 191:7-10. [DOI] [PubMed] [Google Scholar]
- 24.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic-uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 26.Keskimäki, M., M. Saari, T. Heiskanen, and A. Siitonen. 1998. Shiga toxin-producing Escherichia coli in Finland from 1990 through 1997: prevalence and characteristics of isolates. J. Clin. Microbiol. 36:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuflu, K. M., and D. A. Cuppels. 1997. Development of a diagnostic DNA probe for xanthomonads causing bacterial spot of peppers and tomatoes. Appl. Environ. Microbiol. 63:4462-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1999. Promiscuous origin of a chimeric sequence in the Escherichia coli O157:H7 genome. J. Bacteriol. 181:7614-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maas, R. 1983. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid 10:296-298. [DOI] [PubMed] [Google Scholar]
- 30.Makino, K., K. Ishii, T. Yasunaga, M. Hattori, K. Yokoyama, C. H. Yutsudo, Y. Kubota, Y. Yamaichi, T. Iida, K. Yamamoto, T. Honda, C. G. Han, E. Ohtsubo, M. Kasamatsu, T. Hayashi, S. Kuhara, and H. Shinagawa. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5:1-9. [DOI] [PubMed] [Google Scholar]
- 31.Nishizawa, T., M. Asayama, K. Fujii, K. Harada, and M. Shirai. 1999. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 126:520-529. [DOI] [PubMed] [Google Scholar]
- 32.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 33.Picard-Pasquier, N., M. Ouagued, B. Picard, P. Goullet, and R. Krishnamoorthy. 1989. A simple, sensitive method of analyzing bacterial ribosomal DNA polymorphism. Electrophoresis 10:186-189. [DOI] [PubMed] [Google Scholar]
- 34.Piérard, D., L. VanDamme, L. Moriau, D. Stevens, and S. Lauwers. 1997. Virulence factors of verocytotoxin-producing Escherichia coli isolated from raw meats. Appl. Environ. Microbiol. 63:4585-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pradel, N., K. Boukhors, Y. Bertin, C. Forestier, C. Martin, and V. Livrelli. 2001. Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients, cattle, and food samples in central France. Appl. Environ. Microbiol. 67:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pradel, N., V. Livrelli, C. DeChamps, J.-B. Palcoux, A. Reynaud, F. Scheutz, J. Sirot, B. Joly, and C. Forestier. 2000. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J. Clin. Microbiol. 38:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pupo, G. M., D. K. R. Karolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramachandran, V., M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2001. The common ovine Shiga toxin 2-containing Escherichia coli serotypes and human isolates of the same serotypes possess a Stx2d toxin type. J. Clin. Microbiol. 39:1932-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 41.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey, V. J. DiRita, and M. Bagdasarian. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, H., C. Geitz, P. I. Tarr, M. Frosch, and H. Karch. 1999. Non-O157:H7 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J. Infect. Dis. 179:115-123. [DOI] [PubMed] [Google Scholar]
- 44.Straus, D., and F. M. Ausubel. 1990. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc. Natl. Acad. Sci. USA 87:1889-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatsuno, I., H. Kimura, A. Okutani, K. Kanamaru, H. Abe, S. Nagai, K. Makino, H. Shinagawa, M. Yoshida, K. Sato, J. Nakamoto, T. Tobe, and C. Sasakawa. 2000. Isolation and characterization of mini-Tn 5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect. Immun. 68:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, P. L., D. W. K. Acheson, and M. K. Waldor. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wells, J. G., L. D. Shipman, K. D. Greene, E. G. Sowers, J. H. Green, et al. 1991. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J. Clin. Microbiol. 29:985-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Ørskov, I. Ørskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wieler, L. H., T. K. McDaniel, T. S. Whittam, and J. B. Kaper. 1997. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol. Lett. 156:49-53. [DOI] [PubMed] [Google Scholar]