Abstract
Methods that enabled the identification, detection, and enumeration of Bifidobacterium species by PCR targeting the transaldolase gene were tested. Bifidobacterial species isolated from the feces of human adults and babies were identified by PCR amplification of a 301-bp transaldolase gene sequence and comparison of the relative migrations of the DNA fragments in denaturing gradient gel electrophoresis (DGGE). Two subtypes of Bifidobacterium longum, five subtypes of Bifidobacterium adolescentis, and two subtypes of Bifidobacterium pseudocatenulatum could be differentiated using PCR-DGGE. Bifidobacterium angulatum and B. catenulatum type cultures could not be differentiated from each other. Bifidobacterial species were also detected directly in fecal samples by this combination of PCR and DGGE. The number of species detected was less than that detected by PCR using species-specific primers targeting 16S ribosomal DNA (rDNA). Real-time quantitative PCR targeting a 110-bp transaldolase gene sequence was used to enumerate bifidobacteria in fecal samples. Real-time quantitative PCR measurements of bifidobacteria in fecal samples from adults correlated well with results obtained by culture when either a 16S rDNA sequence or the transaldolase gene sequence was targeted. In the case of samples from infants, 16S rDNA-targeted PCR was superior to PCR targeting the transaldolase gene for the quantification of bifidobacterial populations.
Bifidobacterial species are common members of the gut microflora of humans, comprising up to 3% of the total fecal microflora of adults (11, 16, 28). They are more numerous in the infant gut, where they form up to 91% of the total microflora in breast-fed babies and up to 75% in formula-fed infants (14). Perhaps because of their prevalence in the feces of infants suckled at the breast, bifidobacteria are considered to be beneficial bacteria, and they are used in the preparation of probiotic products (13). The bifidobacteria are obligately anaerobic bacteria and require the provision of a reduced environment for cultivation in the laboratory. Selective media for the cultivation of bifidobacteria have been described, but bias may be introduced through the use of such media if they do not equally support the growth of all of the Bifidobacterium species encountered in human feces (2). The identification of bifidobacterial species is considered to be difficult when based on phenotypic characteristics of the bacteria (30). All of these culture-related factors limit the opportunities to conduct quality research about bifidobacterial populations in the gut.
The derivation of PCR primers that are species specific has greatly aided the detection and identification of these bacteria in microbial ecological studies (18). To date, species-specific PCR primers for bifidobacteria have targeted 16S rRNA gene sequences. Detection and identification of bifidobacteria by using these primers are, however, logistically demanding, since at least eight species are commonly encountered among the human gut microflora (3, 4, 20). The use of genus-specific PCR primers targeting the 16S rRNA gene, coupled with denaturing gradient gel electrophoresis (DGGE), has provided a screening method for the detection and identification of bifidobacterial species (26). 16S rRNA sequences are highly conserved among the bifidobacteria, and there are multiple copies of the 16S rRNA gene per chromosome (6, 12). These features might influence the outcome of nucleotide sequence-based identifications and quantitative PCR methods. We investigated the use of an alternative gene sequence for the identification, detection, and enumeration of bifidobacterial species by PCR. Bifidobacteria produce at least 14 types of transaldolase that can be differentiated by protein electrophoresis and by serology (29). This suggested to us that the amino acid sequences of these isoenzymes of transaldolase were different and that, in turn, the nucleotide base sequence of the transaldolase gene might be variable between species. In this paper, we demonstrate that this is indeed so and that PCR-DGGE targeting of the transaldolase gene enables Bifidobacterium species to be detected and differentiated. Additionally, we demonstrate that real-time, quantitative PCR targeting the transaldolase gene can be used to enumerate bifidobacteria in fecal samples collected from adult humans.
MATERIALS AND METHODS
Identification and detection of Bifidobacterium species by using species-specific primers.
The species-specific PCR method described previously (18, 19) was used to identify the strains among randomly selected bifidobacteria isolated on Rogosa SL agar (31) (Table 1) and from DNAs extracted from fecal samples that had been collected from healthy adults and infants.
TABLE 1.
Identification of bifidobacterial isolates by different molecular methods
| Isolate | Species (identified by species-specific PCR primers) | Species identification confirmed by 16S rDNA sequence | Identification by transaldolase (migration in DGGE) |
|---|---|---|---|
| A6a | B. longum | NDa | B. longum I |
| HB2 B-1 | B. longum | ND | B. longum I |
| NH1-B2 | B. longum | ND | B. longum I |
| GT10-1C | B. longum | B. longum | B. longum I |
| JBA1 B-1 | B. longum | ND | B. longum I |
| MH B-1 | B. longum | B. longum | B. longum II |
| GT9 T-1C | B. adolescentis | ND | B. adolescentis I |
| GT FOS1 | B. adolescentis | B. adolescentis | B. adolescentis I |
| GT FOS8 | B. adolescentis | B. adolescentis | B. adolescentis I |
| MH2 B-10 | B. adolescentis | ND | B. adolescentis I |
| MH2 B-2 | B. adolescentis | ND | B. adolescentis I |
| HB2 B-3 | B. adolescentis | ND | B. adolescentis I |
| A12 | B. adolescentis | B. adolescentis | B. adolescentis II |
| A15 | B. adolescentis | B. adolescentis | B. adolescentis III |
| JB1 B-1 | B. adolescentis | ND | B. adolescentis IV |
| KB1 | B. adolescentis | ND | B. adolescentis IV |
| ALM2 B-2 | B. adolescentis | ND | B. adolescentis IV |
| HB1 B-5 | B. adolescentis | B. adolescentis | B. adolescentis IV |
| JB4 T-1C | B. adolescentis | ND | B. adolescentis IV |
| G1 | B. adolescentis | B. adolescentis | B. adolescentis IV |
| NH2 B-5 | B. adolescentis | B. adolescentis | B. adolescentis V |
| G2 | B. catenulatum group | B. pseudocatenulatum | B. pseudocatenulatum I |
| NH1 B-4 | B. catenulatum group | B. pseudocatenulatum | B. pseudocatenulatum I |
| G4 | B. catenulatum group | B. pseudocatenulatum | B. pseudocatenulatum II |
| G3 | B. catenulatum group | B. pseudocatenulatum | B. pseudocatenulatum II |
| G3a | B. catenulatum group | B. pseudocatenulatum | B. pseudocatenulatum II |
| JB FOS5 | B. bifidum | ND | B. bifidum |
ND, not done.
Sequencing of 16S rRNA genes of bifidobacterial isolates.
16S rRNA genes were amplified by PCR using primers SacI-POmod and SalI-T7-PC5 together with the PCR program described previously by Rodtong and Tannock (24) (Table 2). Additional primers used to assist in sequencing were SmaI-T3-P3rev and 16Smidfor (Table 2). Sequencing was accomplished by the dideoxy chain termination method (25) using a 373A automated DNA sequencing system (PE Applied Biosystems, Foster City, Calif.). Computer-assisted alignment of sequences was carried out with the MegAlign module of the Lasergene software package (DNAStar Inc., Madison, Wis.). A search of the GenBank DNA database by using the BLAST algorithm (1) permitted identification of the isolates on the basis of highest score.
TABLE 2.
PCR primers used in this study
| Primer | Sequence |
|---|---|
| SacI-POmod | 5′-CCG AGC TCA ACA GAG TTT GAT CCT GGC TCA G-3′ |
| SalI-T7-PC5 | 5′-GGT CGA CCG TTA ATA CGA CTC ACT ATA GGG ATA CCT TGT TAC GAC TT-3′ |
| SmaI-T3-P3rev | 5′-GCC CGG GCG ATT AAC CCT CAC TAA AGG GAA TTA GAT ACC CTA GTA GTC C-3′ |
| 16Smidfor | 5′-GGC CGT TAC TGA CGC TGA G-3′ |
| 7716 | 5′-CGG CGT TTC CAT CTG GC-3′ |
| 7717 | 5′-GCG GAA GAT GTC GGT GG-3′ |
| ForTal | 5′-CGT CGC CTT CTT CTT CGT CTC-3′ |
| RevTal | 5′-CTT CTC CGG CAT GGT GTT GAC-3′ |
| g-Bifid-F | 5′-CTC CTG GAA ACG GGT GG-3′ |
| g-Bifid-R | 5′-GGT GTT CTT CCC GAT ATC TAC A-3′ |
| T7 | 5′-TAA TAC GAC TCA CTA TAG GGC-3′ |
| SP6 | 5′-ATT TAG GTG ACA CTA TAG AAT-3′ |
| TAQfor | 5′-GCG TCC GCT GTG GGC-3′ |
| TAQrev | 5′-CTT CTC CGG CAT GGT GTT G-3′ |
Determination of the transaldolase gene sequence of Bifidobacterium infantis ATCC 15702.
The transaldolase was purified from cell lysates (with a French press at a pressure of 20,000 lb/in2) of B. infantis ATCC 15702 by ammonium sulfate precipitation followed by ion-exchange chromatography (Q-Sepharose, Fast Flow; Pharmacia LKB Biotechnology). Transaldolase activity was measured as described by Tchola and Horeker (32), and protein concentrations were determined by the Bradford method (7). Protein fractions were further investigated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis with the Tris-Tricine discontinuous buffer system of Schagger and von Jagow (27). A protein fraction that yielded a single polypeptide in SDS-polyacrylamide gel electrophoresis was transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore) by electroblotting. The N-terminal amino acid sequence of the polypeptide was determined by the Protein Microchemistry Facility, University of Otago. The amino acid sequence was used to derive oligonucleotides that served as PCR primers that, together with other degenerate primers based on the type II transaldolase (15) obtained from other bacterial species deposited in GenBank, enabled part of the B. infantis gene to be amplified, cloned, and sequenced.
The plasmid vector pBluescript II KS(±) (Stratagene, La Jolla, Calif.) was digested within the multiple cloning site with the restriction enzymes EcoRI and XbaI (Roche, Mannheim, Germany) and was gel purified (QiaexII; Qiagen, Hilden, Germany). Bifidobacterial chromosomal DNA was extracted by the method of Delley and colleagues (9), except that the cells were incubated for 1 h at 37°C with a mixture of mutanolysin (120 μg/ml) and lysozyme (1 mg/ml). The chromosomal DNA was simultaneously digested with restriction enzymes as for the vector. A 246-bp PCR product (nucleotides 33 to 279; primers 7716 and 7717 [Table 2]) derived from the transaldolase sequence obtained by PCR using degenerate primers was used as a probe for Southern hybridizations of membrane-bound, digested chromosomal DNA. This showed that part of the gene was on an approximately 5-kb doubly digested DNA fragment. The digested vector (0.4 μg) was ligated (1 U of ligase [Roche]) with 4 μg of gel-purified chromosomal fragments 3 to 7 kb in size. After electroporation into competent cells of Escherichia coli XL-1 Blue (Stratagene) and typical blue-white selection on medium containing ampicillin (100 μg/ml), an alkaline lysis procedure (5) was used to extract the plasmid DNA. Identification of plasmids with the correct insert was carried out by dot blotting of plasmid DNA and Southern hybridization using the probe described above. Hybridization conditions were 65°C overnight with wash conditions of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS, twice at room temperature and once for 30 min at 65°C. A clone containing an insert of approximately 5 kb was sequenced by a combination of PCR, primer walking, and subcloning using AccI with sequencing primers internal to the plasmid (T7 and SP6 [Table 2]) (8).
Design of bifidobacterial transaldolase-specific PCR primers.
DNA was extracted from cultures of B. infantis ATCC 15697T, Bifidobacterium adolescentis DSM 20083T, Bifidobacterium longum ATCC 15707T, Bifidobacterium breve ATCC 15698T, Bifidobacterium bifidum DSM 20456T, Bifidobacterium catenulatum DSM 20103T, Bifidobacterium pseudocatenulatum ATCC 27919T, Bifidobacterium angulatum ATCC 27535T, and Bifidobacterium lactis DSM 10140T by the method of Walter et al. (33). Primer pairs derived on the basis of the B. infantis ATCC 15702 transaldolase sequence were used to amplify the gene from B. bifidum and B. breve DNAs (8). Comparison of these sequences revealed a region of DNA that was variable between bifidobacterial species and which was flanked by conserved regions (nucleotides 588 to 889 with reference to the B. infantis ATCC 15702 gene).
PCR primers that permitted the amplification of this 301-bp region from all of the species listed above were derived (ForTal and RevTal [Table 2]). The PCR mixture contained 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, a 200 μM concentration of each primer, 1 μg of DNA, and 2.5 U of Taq DNA polymerase (Roche). The amplification program was as follows: 94°C for 3 min; 10 cycles of 94°C for 30 s, 60°C for 20 s, and extension at 72°C for 20 s; repeated for 10 cycles but with an extension time of 25 s; repeated for 15 cycles but with an extension time of 30 s; and then a final extension time of 72°C for 5 min.
Differentiation of Bifidobacterium species by PCR-DGGE.
The 301-bp sequence of the transaldolase gene was amplified using ForTal and RevTal-GC (RevTal modified by the addition of a 40-bp GC clamp as described previously [33]) primers and the PCR program given above. DGGE was performed with a DCode apparatus (Bio-Rad, Hercules, Calif.), using a 6% polyacrylamide gel with a 52 to 65% gradient of 7.0 M urea and 40% (vol/vol) formamide that increased in the direction of electrophoresis. Electrophoresis was carried out in 1× TAE (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) buffer at 130 V and 60°C for about 4.5 h. The gel was stained with ethidium bromide solution (5 μg/ml) for 20 min and viewed by UV transillumination. This methodology was used to compare the relative migrations of 301-bp PCR products derived from pure cultures of the type strains and from pure cultures of bifidobacteria isolated from humans (Table 1).
Detection of bifidobacterial species in fecal samples by PCR-DGGE.
DNA was extracted from fecal samples obtained from 10 adults (single samples) and 2 infants (multiple samples) as described by Tannock et al. (31), using a FastDNA kit (BIO 101, Carlsbad, Calif.). The samples had been collected during studies of the fecal microflora of healthy humans and were included to provide a range of samples to test the efficacy of the transaldolase-based analytical methods. The feces had been stored at −20°C. Prior to storage, the bifidobacterial CFU per gram had been determined by diluting fecal homogenates and culturing aliquots of the dilutions on Rogosa SL agar for 3 days under anaerobic conditions, as described previously (31), for 7 of the adult samples and all 10 of the infant samples. DNAs extracted from the fecal samples were used as templates (100 ng) in PCRs with the ForTal and RevTal-GC primers, and the PCR amplicons were used in DGGE as described above.
Cloning of DNA fragments eluted from DGGE gels.
Transaldolase gene fragments were cut from the polyacrylamide gel using a sterile surgical scalpel after the stained gel had been photographed. The fragments were placed in Microfuge tubes, and DNA was eluted by the addition of 100 μl of diffusion buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA [pH 8.0], and 0.1% SDS) and incubation for 30 min at 50°C. After vortex mixing and brief centrifugation, the supernatant was transferred to a QIAquick gel extraction kit spin column (Qiagen), and DNA purification proceeded according to the manufacturer's instructions. The purified DNA was used as a template in a PCR using the ForTal and RevTal primers as described above. PCR products were cleaned using a QIAquick PCR purification kit and ligated with plasmid pGEM-T Easy (Promega, Madison, Wis.) according to the manufacturer's instructions. Cells of E. coli DH5α were electrotransformed (Bio-Rad Gene Pulser) with recombinant plasmids by a standard method (10). Selection of transformants was on MacConkey agar (red-clear colony screening) containing 100 μg of ampicillin per ml. Recombinant plasmids were purified from E. coli colonies (5), and the cloned DNA was amplified using primers T7 and SP6 (Table 2). The PCR program was 94°C for 3 min; 15 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 1 min; and 68°C for 7 min. The amplified DNA was sequenced after cleaning (QIAquick PCR purification kit) using the T7 and SP6 oligonucleotides as sequencing primers. In some instances, transaldolase DNA was cloned directly from PCR products into pGEM-T Easy.
Enumeration of bifidobacteria in fecal samples by real-time PCR targeting the 16S rRNA gene.
DNA purified from B. longum ATCC 15707T (for preparation of a standard curve) and fecal DNA samples were prepared in New Zealand as described above and transported to Japan. PCR was performed in a fluorescence temperature cycler (LightCycler; Roche). Amplification was performed in a 10-μl final volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 500 ng of bovine serum albumin per μl, a 200 μM concentration of each deoxynucleoside triphosphate, template DNA, a 1:30,000 dilution of SYBR Green I (Molecular Probes, Eugene, Oreg.), 11 ng of TaqStart antibody (Clontech, Palo Alto, Calif.) per μl, and 0.05 U of Taq DNA polymerase (Takara, Tokyo, Japan) per μl, using a 0.25 μM concentration of the Bifidobacterium group-specific primers g-Bifid-F and g-Bifid-R (Table 2). These primers have been validated for bifidobacterial specificity (T. Matsuki and K. Watanabe, unpublished data). The amplification program included 40 cycles of three steps each, comprised of heating at 20°C/s to 95°C with a 0-s hold, cooling at 20°C/s to 55°C with a 10-s hold, and heating at 20°C/s to 72°C with a 20-s hold. A fluorescent product was detected at the last step of each cycle. After amplification, a melting curve was obtained by heating at 20°C/s to 96°C, cooling at 20°C/s to 60°C, and slowly heating at 0.2°C/s to 96°C, with fluorescence collection at 0.2°C intervals. Melting curves were used to determine the specificity of the PCR (23). The lower limit of detection was approximately 106 bifidobacterial cells per g of feces.
Enumeration of bifidobacteria by using real-time PCR targeting the transaldolase gene.
A conserved 110-bp region (nucleotides 781 to 888) within the previously described 301-bp region of the transaldolase gene was targeted for use in the Taqman assay with the ABI Prism 7700 Sequence Detection System (PE Applied Biosystems). Primers TAQfor and TAQrev (Table 2) and the fluorogenic probe (5′-[6-carboxyfluoroscein]-TCC ACC GGC ACC AAG AAC GC-[6-carboxytetramethylrhodamine]-3′) for this system were derived using Primer Express (version 1.5) ABI Prism and guidelines provided by PE Biosystems. Amplification reaction mixtures contained 10 to 50 ng of DNA, 5 mM MgCl2, a 200 to 400 μM concentration of each deoxynucleoside triphosphate, a 300 nM concentration of each primer, 200 nM fluorogenic probe, 0.25 U of AmpErase uracyl N-glycosylase, and 2.5 U of AmpliTaq Gold DNA polymerase. The PCR program was 50°C for 2 min, 95°C for 10 min, and then 40 cycles of two-temperature PCR (95°C for 15 s and 58°C for 60 s). The cycle threshold values generated by real-time PCR from dilutions of DNA extracted from 3 × 108 CFU of B. longum ATCC 15707T cells were used to plot a standard curve from which the number of bifidobacteria represented by the target transaldolase sequences in DNA extracted from feces or cultures could be calculated. The lower limit of detection was approximately 105 bifidobacterial cells per g of feces.
Nucleotide sequence accession numbers.
Transaldolase gene sequences from type cultures of B. breve, B. bifidum, and B. infantis ATCC 15702 were deposited in GenBank (accession numbers AF417539, AF417538, and AF417540, respectively), as were the 301-bp variable region sequences targeted by primers ForTal and RevTal (GenBank accession numbers AF417529, AF417530, AF417531, AF417532, AF417533, AF417534, AF417535, AF417536, and AF417537).
RESULTS
Characterization of the transaldolase gene sequences from type cultures of bifidobacterial species.
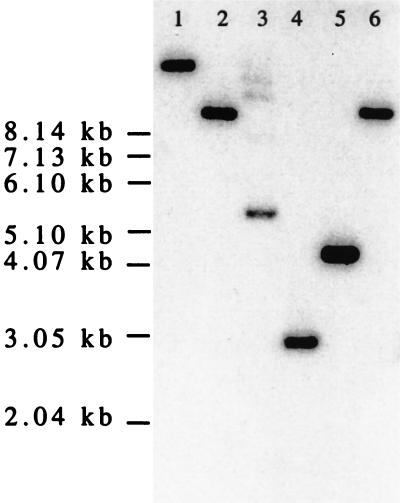
The transaldolase gene was present in chromosomal DNA of B. infantis ATCC 15702 as a single copy, because the 246-bp radiolabeled transaldolase gene probe hybridized with a single fragment in restriction endonuclease digests of chromosomal DNA (Fig. 1). Characterization of partial transaldolase gene sequences from other bifidobacterial species was based on the use of PCR primer pairs derived on the basis of the B. infantis ATCC 15702 sequence. The 301-bp variable region targeted by primers ForTal and RevTal is shown, for all nine type cultures that we tested, in Fig. 2.
FIG. 1.
Hybridization of 246-bp radiolabeled PCR product to restriction endonuclease digests of chromosomal DNAs purified from B. infantis ATCC 15702. Lane 1, BamHI digestion; lane 2, EcoRI digestion; lane 3, KpnI digestion; lane 4, PstI digestion; lane 5, SphI digestion; lane 6, XbaI digestion.
FIG. 2.
Alignment of 301-bp transaldolase gene sequence targeted in PCRs. PCR primer annealing sites are indicated in boldface (ForTal and RevTal) or boxed (TAQfor and TAQrev). The target for the fluorogenic probe used in real-time quantitative PCR is shaded. DSM 20083, B. adolescentis; ATCC 27535, B. angulatum; DSM 20456, B. bifidum; ATCC 15698, B. breve; DSM 20103, B. catenulatum; ATCC 15697, B. infantis; DSM 10140, B. lactis; ATCC 15707, B. longum; ATCC 27919, B. pseudocatenulatum.
Differentiation of bifidobacterial species by PCR-DGGE.
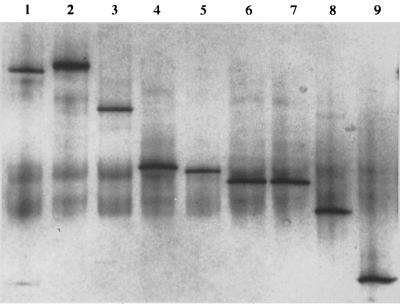
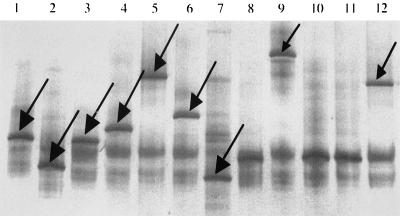
Differentiation of bifidobacterial species type cultures by relative migration distances of the 301-bp transaldolase gene fragments is demonstrated in Fig. 3. All of the species could be differentiated from one another, except that the transaldolase sequences from B. catenulatum and B. angulatum migrated the same distance in the DGGE gel. PCR amplification of transaldolase sequences from DNAs extracted from bifidobacteria cultured from fecal samples (Table 1) showed that variants of the transaldolase gene occurred among B. longum, B. adolescentis, and B. pseudocatenulatum. (Table 1; Fig. 4). It was concluded that variants occurred within these species because species-specific PCR identified the isolates as belonging to particular species, yet a range of intraspecies DNA fragment migration distances was observed. PCR artifacts (multiple, less intensely stained, diffuse DNA fragments in Fig. 4) that we have encountered previously (33) were present in the DGGE gels.
FIG. 3.
Migration of transaldolase DNA fragments generated from bifidobacterial type cultures in DGGE. Lane 1, B. breve ATCC 15698T; lane 2, B. lactis DSM 10140T; lane 3, B. pseudocatenulatum ATCC 27919T; lane 4, B. adolescentis DSM 20083T; lane 5, B. longum ATCC 15707T; lane 6, B. catenulatum DSM 20103T; lane 7, B. angulatum ATCC 27535; lane 8, B. bifidum DSM 20456T; lane 9, B. infantis ATCC 15697T.
FIG. 4.
Migration of transaldolase DNA fragments (arrows) in DGGE. Lane 1, B. longum I; lane 2, B. longum II; lane 3, B. adolescentis I, lane 4, B. adolescentis II, lane 5, B. adolescentis III; lane 6, B. adolescentis IV; lane 7, B. adolescentis V; lane 9, B. pseudocatenulatum II; lane 12, B. pseudocatenulatum I.
Detection of bifidobacterial species in fecal samples by PCR-DGGE.
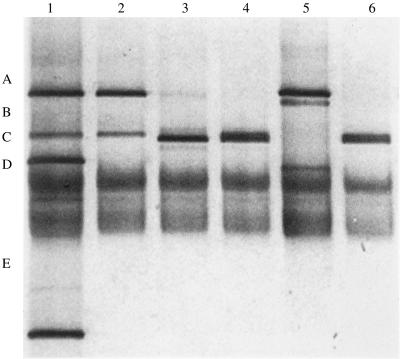
Comparison of results obtained from the PCR amplification of transaldolase gene sequences and DGGE or by the use of species-specific PCR primers showed that more species were detected in the fecal samples by the latter method (Table 3; Fig. 5). In some samples, transaldolase fragments generated from fecal DNA did not match those in the identification ladder (Fig. 5; Table 3). Excision and sequencing of the fragment provided an identification by comparison with sequences obtained from pure cultures, because they were at least 98% similar to that of a type culture.
TABLE 3.
Detection of Bifidobacterium species in fecal samples
| Subjecta | Species detected by:
|
|
|---|---|---|
| Species-specific primers | PCR-DGGE | |
| F1 | B. adolescentis, B. longum, B. catenulatum | B. adolescentis I, B. adolescentis IV |
| F2 | B. adolescentis, B. longum, B. catenulatum | B. longum I |
| F3 | B. bifidum | B. bifidum |
| F4 | B. adolescentis, B. longum, B. bifidum | B. adolescentis I, B. longum I |
| F5 | B. adolescentis, B. longum, B. bifidum, B. catenulatum | B. adolescentis I, B. longum I |
| F6 | B. adolescentis, B. longum, B. catenulatum | B. adolescentis I, B. longum I |
| F7 | B. adolescentis, B. longum, B. catenulatum | Not identifiedb |
| F8 | B. adolescentis, B. longum, B. bifidum, B. catenulatum | B. adolescentis IV, not identified |
| F9 | B. adolescentis, B. longum, B. infantis, B. catenulatum | B. adolescentis I, B. longum I |
| F10 | B. adolescentis, B. longum, B. infantis, B. catenulatum | B. longum I |
| B1 | No product | No product |
| B2 | B. bifidum | B. bifidum, not identified |
| B3 | B. bifidum | B. bifidum |
| B4 | B. bifidum | B. bifidum |
| B5 | B. bifidum | B. bifidum |
| B6 | B. bifidum, B. longum | B. bifidum |
| B7 | B. bifidum, B. longum | B. bifidum |
| B8 | B. bifidum, B. longum, B. catenulatum | B. bifidum |
| B9 | No product | B. infantis, B. longum, not identified |
| B10 | B. bifidum | B. bifidum |
F, adult; B, baby.
Not identified, a satisfactory sequence could not be obtained.
FIG. 5.
Examples of transaldolase profiles from adult fecal samples in DGGE gel. Lane 1, identification ladder (A, B. adolescentis subtype IV; B, B. adolescentis subtype I; C, B. catenulatum; D, B. bifidum; E, B. infantis); lane 2, F1; lane 3, F2; lane 4, F5; lane 5, F8; lane 6, F9.
Comparison of real-time quantitative PCR methods.
Comparison of the bifidobacterial CFU per gram obtained by culture and real-time PCR showed that there was good correlation (Pearson linear correlation test) whether fecal samples from adults were examined by PCR targeting the transaldolase gene sequence or 16S ribosomal DNA (rDNA) (Table 4). Means and standard errors (SE) were not different whether the bifidobacteria were enumerated by PCR targeting the transaldolase gene or 16S rDNA (for transaldolase PCR, mean = 9.0 and SE = 0.1; for 16S rDNA PCR, mean = 8.7 and SE = 0.1 [Mann-Whitney U test, P = 0.0753]). In the case of fecal samples from infants, however, results obtained by 16S rDNA-targeted PCR correlated better with culture results than did those of transaldolase-targeted PCR (Table 4). This was probably because real-time PCR conditions did not favor amplification of the transaldolase gene sequence from B. bifidum, which was the species that predominated in the infant samples (Table 3). The number of B. bifidum cells in pure culture determined by transaldolase-based PCR was lower than that detected by culture (Table 5).
TABLE 4.
Correlation between bifidobacterial populations detected in fecal samples by culture and real-time PCR.
| Comparison (n) | Linear correlation (r) |
|---|---|
| Adult feces 16S rDNA-based PCR and transaldolase- based PCR (7) | 0.818 |
| Adult feces culture and transaldolase-based PCR (7) | 0.896 |
| Adult feces culture and 16S rDNA-based PCR (7) | 0.729 |
| Infant feces culture and transaldolase-based PCR (10) | 0.660 |
| Infant feces culture and 16S rDNA-based PCR (10) | 0.942 |
TABLE 5.
Comparison of bifidobacterial populations in brain heart infusion broth (duplicate cultures) determined by culture and real-time PCR targeting the transaldolase gene
| Strain | Log10 bifidobacteria/ml determined by:
|
|
|---|---|---|
| Culture | Transaldolase-based PCR | |
| B. infantis ATCC 15697T | 9.1, 9.2 | 8.7, 9.1 |
| B. longum ATCC 15707T | 8.7, 9.0 | 8.9, 9.0 |
| B. breve ATCC 15698T | 9.1, 9.3 | 8.6, 9.1 |
| B. catenulatum DSM 20103T | 8.5, 8.7 | 8.3, 8.4 |
| B. adolescentis DSM 20083T | 8.5, 8.5 | 8.5, 8.7 |
| B. bifidum DSM 20456T | 7.9, 8.8 | 5.8, 6.1 |
DISCUSSION
Nucleotide base sequence-targeted methods of analysis provide considerable advantages over culture-based approaches for the analysis of complex bacterial communities. Both cultivable and noncultivable populations can be detected, and international cooperative studies can be initiated because freshly collected samples can be frozen and sent to analytical laboratories in other locations. The provision of laboratory conditions for the culture of fastidious anaerobic bacteria is unnecessary. Moreover, nucleotide base sequences provide tangible evidence of the identity of a bacterial isolate compared to the sometimes subjective interpretation of phenotypic observations. PCR coupled with DGGE provides a valuable tool in microbial ecological studies because the composition of the bacterial community can be screened in numerous samples with a minimal expenditure of resources and time (34). Real-time quantitative PCR for the enumeration of specific groups of bacteria in complex communities holds much promise because, relative to culture-based techniques, it removes bias related to selective culture media. Doubtless other biases are introduced, because PCRs are known to amplify DNA sequences from mixed populations with different efficiencies (22). Real-time PCR assays can be performed rapidly (about 1 h 50 min per run).
Nucleotide base sequence-based analytical methods in microbial ecology target mainly the V regions of 16 rRNA genes. It is common for multiple copies of the rRNA operon to be present on the bacterial chromosome (6, 17), which might affect the accuracy of real-time quantitative PCR (multiple 16S target sites). Additionally, there is heterogeneity with regard to 16S rRNA sequences even within a bacterial cell that might affect the binding of a probe to target DNA (21). We reasoned, therefore, that an alternative target sequence might be preferable for the enumeration, detection, and identification of bifidobacteria. The transaldolase gene was worthy of consideration because it is common to bifidobacteria, was present as a single copy on the genome of B. infantis ATCC 15702, and contained conserved sequences that were suitable targets for PCR primers and a real-time quantitative PCR probe. A BLAST search of GenBank showed that the primers would not anneal with the sequences of transaldolase genes of other bacterial genera, and only sequences resembling those obtained from bifidobacterial cultures were detected in DGGE gels in which PCR amplicons from fecal DNA had been tested.
We tested the ability of PCR-DGGE to identify bifidobacterial species isolated from human feces. Our “gold standards” for comparison were identification using species-specific PCR primers (18, 19) and 16S rDNA sequence. We observed that the majority of type cultures of bifidobacterial species reported to occur in human feces could be differentiated by transaldolase-specific PCR-DGGE. Intraspecies heterogeneity in the transaldolase target sequence resulted in the recognition of two subtypes of B. longum, two subtypes of B. pseudocatenulatum, and five subtypes of B. adolescentis among the fecal isolates. Screening the identity of bifidobacterial isolates was simpler with PCR-DGGE than with species-specific primers because only a single PCR with Tal primers was required. Thus, PCR-DGGE targeting the transaldolase gene will prove to be a useful method for bifidobacterial identification in microbial ecological studies. It has the advantage over 16S rDNA-based differentiation of species (18, 19, 26) that B. catenulatum and B. pseudocatenulatum can be differentiated from each other. From our observations, B. pseudocatenulatum is relatively common in human feces (Table 3). Moreover, we were able to recognize subgroups within B. longum, B. adolescentis, and B. pseudocatenulatum. This may be useful information in tracking the fate of particular cultures of bifidobacteria in future probiotic and prebiotic studies. Transaldolase-specific PCR-DGGE did not differentiate B. catenulatum from B. angulatum, however, although these species can be separated on the basis of 16S rDNA sequences (26).
PCR-DGGE detection of bifidobacterial species in feces was not as sensitive as species-specific PCR. We speculate that only bifidobacterial species whose cells were the most numerous were detected in feces by PCR targeting the transaldolase gene and DGGE. For comparison, we detected species in some of the fecal samples by using primers targeting the 16S rRNA gene and DGGE (26), and although intensely staining DNA fragments matching the migration of the B. adolescentis markers were detected in the DGGE gel, other faintly staining fragments did not match the migration distances of fragments generated from type cultures (observations not shown). Thus, PCR-DGGE can be used to detect bifidobacterial populations in fecal samples, but some of the constituent species detected by species-specific PCR will be missed.
Real-time quantitative PCR results correlated adequately with enumeration of bifidobacteria by culture from adult feces. Culture-based enumeration was a satisfactory gold standard because we have demonstrated previously that results using our culture method are the same as those obtained by fluorescent in situ hybridization (31). Results obtained from the enumeration of bifidobacteria in infant feces by using transaldolase and Taqman did not correlate well with culture results. This was probably because under Taqman conditions, the transaldolase target region did not amplify well from B. bifidum. This was the species present in the infant samples. B. bifidum has been detected in the feces of adult humans occasionally (20). If the cells of this species were numerous in an adult sample, PCR targeting the transaldolase gene would give an inaccurate result. Quantitative PCR targeting the 16S rDNA provided results that correlated adequately with culture results from both adult and infant feces.
We conclude that PCR-DGGE targeting the transaldolase gene provides a useful method for the identification of bifidobacterial isolates from human feces. Although culture of the bacteria is necessary, DGGE of a single PCR amplicon per isolate can provide an identification. It can probably also be used to screen fecal samples for the presence of predominant bifidobacterial species, but this requires confirmation by a study in which individual bifidobacterial species in the fecal samples would be enumerated. Both transaldolase- and 16S rDNA-targeted quantitative PCR methods can be used to enumerate bifidobacteria in feces obtained from adult humans. These methodologies promise to enhance the scope and accuracy of microbial ecological studies of the gut microflora.
Acknowledgments
The support of the University of Otago Grants Committee is gratefully acknowledged. T.R. was supported by an MEC grant (Estancias de Investigadores Espanoles en Centros Extranjeros) and Project AGL2000-07270C03-02.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Beerens, H. 1990. An elective and selective isolation medium for Bifidobacterium spp. Lett. Appl. Microbiol. 11:155-157. [Google Scholar]
- 3.Biavati, B., P. Castagnoli, F. Crociani, and L. D. Trovatelli. 1984. Species of the Bifidobacterium in the feces of infants. Microbiologica 7:341-345. [PubMed] [Google Scholar]
- 4.Biavati, B., P. Castagnoli, and L. D. Trovatelli. 1986. Species of the genus Bifidobacterium in the feces of human adults. Microbiologica 9:39-45. [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourget, N., J.-M. Simonet, and B. Decaris. 1993. Analysis of the genome of the five Bifidobacterium breve strains: plasmid content, pulsed-field gel electrophoresis genome size estimation and rrn loci number. FEMS Microbiol. Lett. 110:11-20. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Burton, J. P. 2000. The development of molecular methods for the identification and enumeration of bifidobacteria. Ph.D. thesis. University of Otago, Dunedin, New Zealand.
- 9.Delley, M., B. Mollet, and H. Hottinger. 1990. DNA probe for Lactobacillus delbrueckii. Appl. Environ. Microbiol. 56:1967-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High-efficiency transformation of E. coli by high-voltage electroporation. Nucleic Acids Res. 16:6127-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham, R., A. J. Duncan, and K. H. Wilson. 1993. Ribosomal DNA sequences of bifidobacteria: implications for sequence-based identification of the human colonic flora. Microb. Ecol. Health Dis. 6:23-27. [Google Scholar]
- 13.Goldin, B. R., and S. L. Gorbach. 1992. Probiotics for humans, p. 355-376. In R. Fuller (ed.), Probiotics. The scientific basis. Chapman and Hall, London, United Kingdom.
- 14.Harmsen, H. J. M., A. C. M. Wildeboer, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 15.Kohler, U., R. Verff, and H. Brinkmann. 1996. Transaldolase genes from the cyanobacteria Anabaena variabilis and Synechocystis sp. PCC 6803: comparison with other eubacterial and eukaryotic homologues. Plant Mol. Biol. 30:213-218. [DOI] [PubMed] [Google Scholar]
- 16.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig, W., and H.-P. Klenk. 2001. Overview: a phylogenetic backbone and taxonomic framework for procaryotic systematics, p. 49-65. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.)., Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 18.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuki, T., K. Watanabe, R. Tanaka, and H. Oyaizu. 1998. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species- and group-specific primers. FEMS Microbiol. Lett. 167:113-121. [DOI] [PubMed] [Google Scholar]
- 20.Mitsuoka, T. 1992. The human gastrointestinal tract, p. 69-114. In B. J. B. Wood (ed.), The lactic acid bacteria, vol. 1. Elsevier Applied Science, London, United Kingdom. [Google Scholar]
- 21.Nubel, U., B. Engelen, A. Felske, J. Snaidr, A. Weishuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reysenbach, A.-L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1998. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245:154-160. [DOI] [PubMed] [Google Scholar]
- 24.Rodtong, S., and G. W. Tannock. 1993. Differentiation of Lactobacillus strains by ribotyping. Appl. Environ. Microbiol. 59:3480-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Saarela, and W. M. De Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 28.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within the human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sgorbati, B., B. Biavati, and D. Palenzona. 1995. The genus Bifidobacterium, p. 279-306. In B. J. B. Wood and W. H. Holzapfel (ed.), The lactic acid bacteria, vol. 2. The genera of lactic acid bacteria. Blackie Academic and Professional, London, United Kingdom.
- 30.Tannock, G. W. 1999. Identification of lactobacilli and bifidobacteria, p. 45-56. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, United Kingdom.
- 31.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tchola, O., and B. L. Horecker. 1966. Transaldolase. Methods Enzymol. 9:499-506. [Google Scholar]
- 33.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoetendal, E., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]