Abstract
Experimentally inoculated sheep and cattle were used as models of natural ruminant infection to investigate the pattern of Escherichia coli O157:H7 shedding and gastrointestinal tract (GIT) location. Eighteen forage-fed cattle were orally inoculated with E. coli O157:H7, and fecal samples were cultured for the bacteria. Three distinct patterns of shedding were observed: 1 month, 1 week, and 2 months or more. Similar patterns were confirmed among 29 forage-fed sheep and four cannulated steers. To identify the GIT location of E. coli O157:H7, sheep were sacrificed at weekly intervals postinoculation and tissue and digesta cultures were taken from the rumen, abomasum, duodenum, lower ileum, cecum, ascending colon, descending colon, and rectum. E. coli O157:H7 was most prevalent in the lower GIT digesta, specifically the cecum, colon, and feces. The bacteria were only inconsistently cultured from tissue samples and only during the first week postinoculation. These results were supported in studies of four Angus steers with cannulae inserted into both the rumen and duodenum. After the steers were inoculated, ruminal, duodenal, and fecal samples were cultured periodically over the course of the infection. The predominant location of E. coli O157:H7 persistence was the lower GIT. E. coli O157:H7 was rarely cultured from the rumen or duodenum after the first week postinoculation, and this did not predict if animals went on to shed the bacteria for 1 week or 1 month. These findings suggest the colon as the site for E. coli O157:H7 persistence and proliferation in mature ruminant animals.
For many years it has been known that healthy ruminants transiently harbor the human pathogen Escherichia coli O157:H7 in their gastrointestinal tract; however, the conditions that lead to its acquisition, persistence, and clearance from that site are not clearly understood (18, 30). Elucidating the relationship between cattle and E. coli O157:H7 may impact the development of interventions to curb its presence in ruminants and thereby reduce the incidence of human infections with this pathogen.
The hallmark of human disease with E. coli O157:H7 is a hemorrhagic colitis that is most often self-limiting (2, 18). However, among reported outbreaks, as many as 25% of infected individuals have been hospitalized, 6% have developed life-threatening sequelae, the hemolytic uremic syndrome, and 1% have died from their infection. For children infections are even more severe, with 5 to 10% progressing to hemolytic uremic syndrome and a 3 to 5% mortality rate (2, 6, 18). E. coli O157:H7 is transmitted by ingestion of contaminated bovine food products, contaminated drinking water or fruit juice, contact with contaminated recreation water or culture-positive animals, and in rare cases, human-to-human transmission via the fecal-oral route (2, 3, 18, 24).
Although healthy cattle (5, 9, 15, 21, 37), sheep (9, 26, 28), deer (16, 34), and goats (9, 32) have been shown to transiently harbor E. coli O157:H7 naturally, cattle are the main source of human infections (2, 17, 18). Studies show a nearly ubiquitous distribution of E. coli O157:H7 in cattle (31). Longitudinal studies of cattle show that up to 75% of dairy herds and 63% of feedlots have culture-positive animals (12, 19, 20, 22). The prevalence of E. coli O157:H7 within a single herd can be as high as 10%, and the overall prevalence of E. coli O157 in the feces of beef cattle is estimated at 1 to 3% (19, 20, 22, 36, 37) but has even been recently reported at upwards of 28% (14, 17). However, Meyer-Broseta et al. suggest that these prevalence estimates may be low for several reasons: too few herds have been tested, too few animals within the herds have been tested, winter studies may misclassify herds as negative due to seasonal variation, and small fecal samples and low sensitivity of culturing methods may miss the bacteria (31).
Sanderson et al. found the average duration that calves shed E. coli O157 to be 30 days (35). However, this average and others like it are calculated from animals with very diverse E. coli O157:H7 shedding patterns. Some animals clear E. coli O157:H7 from the gastrointestinal tract (GIT) in a few days, while others remain culture positive for up to 1 year or more (4, 22, 39). The factors that contribute to the persistence of E. coli O157:H7 in ruminants are mostly unknown but likely include the ability of the bacteria to colonize a particular location within the GIT. Few experiments have been reported investigating the site of persistence in ruminants, and the few that exist contain conflicting conclusions. For example, Brown et al. reports that the forestomachs are the primary sites of E. coli O157:H7 localization and proliferation, while Dean-Nystrom et al. and Buchko et al. suggest that the rectum and the cecum are the primary sites of E. coli O157:H7 colonization (7, 8, 13).
We previously reported that fast intestinal cell proliferation in the cecum and the distal colon correlates with rapid clearance of E. coli O157 from the bovine GIT (30). To complement this finding, here we compared the GIT site of E. coli O157 and the duration that animals remain culture positive. In this study, we used experimentally inoculated sheep and cattle as models of natural ruminant infection. We investigated (i) the pattern of E. coli O157 shedding, (ii) the GIT location of E. coli O157 in sheep over the course of an infection, and (iii) the GIT location of E. coli O157 in cattle harboring the bacteria for various times.
(Part of this research was presented in the 1998 Proceedings of the 31st Annual Convention of the American Association of Bovine Practitioners [Spokane, Wash.]).
MATERIALS AND METHODS
Experimental animals.
Animals were held in quarantined areas in which feed, water, bedding, manure, and waste was not in contact with nonexperimental animals and all personnel and animal handlers followed strict aseptic safety precautions.
(i) Cattle.
Healthy 1-year-old Holstein steers, 1-year-old Charolais/Hereford heifers, and 3- to 5-year-old crossbred beef steers that were at least 50% Black Angus (referred to throughout as Angus) were used. Weights ranged from 360 to 450 kg for the Holsteins and Charolais/Herefords and from 500 to 640 kg for the Black Angus steers. All animals were housed individually, identified by ear tags, had water ad libitum, and were fed twice daily throughout the studies. Cattle were adapted to a forage-based diet for at least 3 weeks before oral inoculation with E. coli O157:H7. The Charolais/Hereford heifers (designated CH1 through CH4) were fed a diet containing the following: 71.1% alfalfa-grass silage, 11.3% grass hay, 6.9% barley, 6.9% corn, 3.0% soybean meal, 0.4% limestone, 0.2% dicalcium phosphate, and 0.2% trace mineralized salt. Eight Holsteins (designated H1 through H8) were fed alfalfa hay, and six Holsteins (designated H9 through H14) were fed grass hay. Four Angus (designated A1 through A4) were fed a diet containing 82.5% triticale silage, 7.1% triticale hay, 7.4% barley, 2.9% dried wheat, and 0.1% calcium carbonate.
The Angus steers were surgically fitted with both ruminal and duodenal cannula a minimum of 1 year prior to the start of this study. The surgeries were performed at Washington State University (Pullman, Wash.) under the supervision of a licensed veterinarian with specialized training in large animal surgery. The University of Idaho Animal Care and Use Committee approved the surgical protocol. The ruminal cannulation was performed by exteriorizing the dorsal rumen through the muscle of the body wall and sutured to the incision in the hide. After formation of an adhesion to the hide, a 10-cm rumen cannula (Ankom Technology, Fairport, N.Y.) was placed in the opening. A closed, T-type intestinal cannula (Ankom Technology) was placed approximately 30 cm distal to the pyloric sphincter to allow access to the duodenum.
(ii) Sheep.
All sheep were adapted to feed and housing conditions for at least 3 weeks prior to experimentation. Twenty-four healthy sheep (12 ewes and 12 rams), 6 to 7 months old, of cross-bred Columbia/Panama/Dorset/Heavy muscle variety were used. The sheep were housed as a group in a cement floor pen and fed pelleted alfalfa twice daily, with free access to drinking water. No supplements were included in their diets. One fatality occurred in the course of the experiment, due to causes unrelated to the study. Postinoculation, sheep were sacrificed (4 per day) from day 7 through day 43 at weekly intervals. Animals were euthanatized with sodium pentobarbital.
In addition, five 1 year-old, healthy ewes, of Columbia/Panama breeds, that remained culture-positive for more than 1 month in a previous study (29) were housed in raised, grated-floor pens, had free access to drinking water, and were fed twice daily. Two sheep were fed 100% grass-hay (forage-based), and three were fed 50% corn-50% pelleted alfalfa (grain-based). On day 53 postinoculation, all five sheep were euthanatized with sodium pentobarbitol. For all sheep, fecal, digesta, and tissue samples were collected and cultured as described below.
Bacteria.
E. coli O157:H7 ATCC 43894 or 43895 bacteria were used. Both strains are isolates from human outbreaks of disease and both express Shiga toxin types 1 and 2. Also, an E. coli O157:H7 resistant to naladixic acid (Nalr) was used as the inoculum for three sheep (strain provided by T. E. Besser, Washington State University, Pullman, Wash.) (29). Inocula were prepared as previously described (29). Briefly, bacteria were grown overnight in Luria-Bertani (LB) broth at 37°C, with aeration, for 18 h. The bacterial cells were harvested by centrifugation for 20 min at 2,600 × g 4°C, and the pellets were resuspended at appropriate concentrations of sterile saline (0.15 M NaCl) or LB broth. Viable cell counts were determined by plate count on LB agar. Bacteria were administered in a 10-ml total volume using three mechanisms: orally to sheep, via a gastric tube followed by a 3-liter fresh water wash to cattle, or by direct placement into the rumen of the cannulated steers.
Samples.
All samples were held on ice and immediately transferred to the laboratory for culture. Samples were collected prior to bacterial inoculation (predose) and thereafter as indicated in Results (see Tables 1 to 4).
TABLE 1.
Fecal E. coli O157 in forage-fed cattlea
| Subject | No. of E. coli O157:H7 CFU/g of feces on day(s) postinoculationc:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1-3 | 5-7 | 8-10 | 12-14 | 15-17 | 18-19 | 21-22 | 24-26 | 28-29 | 31-33 | 35-38 | |
| Charolais/Herefordb | ||||||||||||
| CH1 | 0 | 2.5 × 103 | 4.1 × 105 | 5.0 × 101 | 4.0 × 102 | E+d | E+ | 0 | E+ | E+ | 0 | 0 |
| CH2 | 0 | 2.0 × 103 | 5.0 × 103 | E+ | 0 | 0 | 0 | 0 | 0 | 0 | NDe | |
| CH3 | 0 | 1.5 × 103 | 2.9 × 105 | E+ | E+ | E+ | E+ | E+ | E+ | E+ | 0 | 0 |
| CH4 | 0 | 7.9 × 103 | 5.0 × 101 | E+ | 0 | 0 | 0 | 0 | 0 | 0 | ND | |
| Holsteinf | ||||||||||||
| H1 | 0 | 1.7 × 105 | 2.0 × 104 | E+ | E+ | E+ | ND | E+ | 0 | E+ | 0 | 0 |
| H2 | 0 | 2.0 × 104 | 1.0 × 103 | 0 | 0 | 0 | ND | 0 | 0 | 0 | 0 | 0 |
| H3 | 0 | E+ | 0 | 2.0 × 104 | E+ | E+ | ND | E+ | 0 | 0 | 0 | 0 |
| H4 | 0 | 3.5 × 104 | 1.9 × 105 | 0 | 0 | 0 | ND | 2.5 × 103 | 5.0 × 104 | E+ | E+ | 0 |
| H5 | 0 | E+ | E+ | E+ | E+ | E+ | ND | E+ | E+ | E+ | E+ | E+ |
| H6 | 0 | 1.0 × 102 | E+ | E+ | E+ | E+ | ND | 0 | E+ | E+ | 0 | E+h |
| H7 | 9.0 × 103g | 2.0 × 103 | E+ | E+ | E+ | 1.0 × 103 | ND | E+ | E+ | E+ | E+ | E+i |
| H8 | E+g | E+ | 2.5 × 104 | 0 | 4.5 × 104 | E+ | ND | 2.0 × 104 | E+ | E+ | E+ | 1.0 × 103j |
| H9 | 0 | E+ | 5.0 × 103 | 5.0 × 103 | 5.0 × 102 | E+ | E+ | E+ | E+ | ND | E+ | ND |
| H10 | 0 | E+ | 2.5 × 103 | 1.5 × 105 | 5.0 × 102 | 1.5 × 103 | 0 | 0 | E+ | ND | 1.4 × 104 | ND |
| H11 | 0 | 2.0 × 103 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 | ND |
| H12 | 0 | E+ | 1.0 × 104 | 1.0 × 104 | E+ | E+ | E+ | E+ | E+ | ND | E+ | ND |
| H13 | 0 | 0 | E+ | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 | ND |
| H14 | 0 | 5.0 × 102 | 1.0 × 104 | 1.4 × 103 | E+ | E+ | E+ | E+ | E+ | ND | E+ | ND |
Cattle were inoculated with E. coli O157:H7 ATCC 43894 using a gastric tube placed directly into the rumen. CH1-4 and H1-8 received 4 × 1010 CFU of E. coli O157:H7. H9-14 received 3 × 108 CFU of E. coli O157:H7. Fecal samples were cultured on one of the days indicated.
One-year old Charolais/Hereford heifers fed silage (see Materials and Methods).
Results from nonenrichment culture.
E+, fecal sample culture positive for E. coli O157 after enrichment culture but not by nonenrichment culture technique.
ND, not determined.
One-year-old Holstein steers. H1 to H8 were fed alfalfa, and H9 to H14 were fed grass hay (see Materials and Methods).
Animals H7 and H8 were culture positive for E. coli O157 before experimental inoculation.
Animal H6 remained E. coli O157 positive until day 69 by enrichment culture.
Animal H7 remained E. coli O157 positive until day 56 by enrichment culture.
Animal H8 remained E. coli O157 positive until day 69 by nonenrichment culture.
TABLE 4.
E. coli O157 in GIT compartments of reinoculated steers
| No. of day postinoculationa | No. of E. coli O157 CFU/g of digesta fromb:
|
|||||
|---|---|---|---|---|---|---|
| Rumen
|
Duodenum
|
Feces
|
||||
| A1b | A2 | A1 | A2 | A1 | A2 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0.25 | 1.0 × 103 | 1.0 × 102 | 1.0 × 103 | E+ | 0 | 5.0 × 103 |
| 1 | E+c | 1.0 × 102 | E+ | 1.0 × 102 | 1.6 × 104 | 5.0 × 104 |
| 2 | E+ | E+ | E+ | E+ | 1.0 × 103 | 5.0 × 102 |
| 3 | E+ | E+ | 0 | E+ | 2.0 × 102 | 1.0 × 103 |
| 6 | 0 | E+ | E+ | 0 | 1.0 × 101 | 5.0 × 101 |
| 9 | 0 | 0 | 0 | 0 | 0 | 1.5 × 102 |
| 13 | 0 | E+ | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | E+ | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | E+ | 0 |
| 30 | 0 | 0 | 0 | 0 | E+ | 0 |
| 34 | 0 | 0 | 0 | 0 | 0 | E+ |
| 41 | 0 | 0 | 0 | 0 | 0 | 0 |
Two steers received 2 × 109 E. coli O157:H7 ATCC 43895 directly into the rumen 132 days after the first inoculation (see Table 3).
A1, A2, etc. identify animals from which samples were taken.
E+, digesta sample culture positive for E. coli O157 after enrichment culture only.
(i) Sheep.
Fecal, digesta, and tissue samples (10 g each) were aseptically collected from all 29 sheep tested. GIT locations included the rumen, abomasum, duodenum, lower ileum, cecum, ascending colon, and descending colon. Digesta was removed from each GIT location and cultured for E. coli O157. Ten grams of GIT tissue was prepared for bacterial culture from a 4- to 5-inch sample of GIT tissues removed from each location. Tissue was aseptically opened with a scalpel, rinsed with sterile saline to remove visible digesta, and cultured for E. coli O157. Feed was withheld for 24 h from three animals prior to sacrifice at 43 days postinoculation (see Table 2). Also, a rectal tissue sample was collected on that day.
TABLE 2.
E. coli O157 in the GIT compartments and tissues of sheep
| GIT sites | Resultsa at day postinoculation:
|
|||||
|---|---|---|---|---|---|---|
| 7 | 15 | 22 | 28 | 35 | 43b | |
| Digesta and feces | ||||||
| Rumen | 0c | 1/4 | 0 | 0 | 0 | 0 |
| Abomasum | 0 | 0 | 0 | 0 | 0 | 0 |
| Duodenum | 0 | 0 | 0 | 0 | 0 | 0 |
| Lower Ileum | 2/4 | 0 | 0 | 0 | 0 | 0 |
| Cecum | 2/4 | 2/4 | 0 | 0 | 0 | 0 |
| Ascending colon | 1/4 | 2/4 | 0 | 0 | 0 | 0 |
| Descending colon | 0 | 0 | 0 | 0 | 0 | 1/3 |
| Feces | 4/4 | 3/4 | 2/4 | 2/4 | 2/4 | 1/3 |
| Tissues | ||||||
| Rumen | 1/4 | 0 | 0 | 0 | 0 | 0 |
| Abomasum | 2/4 | 0 | 0 | 0 | 0 | 0 |
| Duodenum | 2/4 | 0 | 0 | 0 | 0 | 0 |
| Lower ileum | 2/4 | 0 | 0 | 0 | 0 | 0 |
| Cecum | 2/4 | 0 | 0 | 0 | 0 | 0 |
| Ascending colon | 2/4 | 0 | 0 | 0 | 0 | 0 |
| Descending colon | 0 | 0 | 0 | 0 | 0 | 0 |
| Rectum | NDd | ND | ND | ND | ND | 1/3 |
No. of culture-positive animals/no. of animals tested.
Feed was withheld for 24 h prior to sampling.
0, all samples cultured negative for E. coli O157.
ND, not determined.
(ii). Cattle.
Fecal samples (10 g) were collected aseptically from all 22 cattle tested. In addition, samples of digesta were taken from the rumen and duodenum of four cannulated steers. Fecal and ruminal digesta samples were obtained aseptically using a sterile-gloved hand; duodenal samples were obtained by inserting a sterile shunt into the cannula and allowing peristalsis to move duodenal digesta into a sterile tube.
Sample E. coli O157 culture and analysis.
Samples were cultured for E. coli O157 by nonenrichment and enrichment procedures as previously described (27). Briefly, initial suspensions were made with 10-g samples diluted 1:5 in Trypticase soy broth (BBL/Becton Dickinson, Detroit, Mich.) supplemented with cefexime (50 μg/liter; Lederle Laboratories, Pearl River, N.Y.) (generously provided by D. D. Hancock, Washington State University), potassium tellurite (2.5 mg/liter; Sigma Chemical Co., St. Louis, Mo.), and vancomycin (40 mg/liter; Sigma) (referred to hereafter as TSB-CTV) (24). Nonenrichment cultures were prepared by performing serial dilutions in sterile saline (0.15 M NaCl) and plating onto sorbitol MacConkey agar supplemented with 4-methylunbelliferyl-β-d-glucuronide (MUG), cefexime, potassium tellurite, and vancomycin (SMAC-CTVM). Plates were incubated at 37°C overnight and observed for colonies not fermenting sorbitol (white colonies) and not hydrolyzing MUG (no florescence at 363 nm). Presumptive E. coli O157:H7 colonies were confirmed by latex agglutination (Pro-Lab Diagnostics, Toronto, Canada). Positive cultures from this technique result in a quantitative measure of E. coli O157:H7 bacteria in CFU/gram. Negative cultures were further analyzed by selective enrichment that entailed incubating the TSB-CTV/sample flasks with aeration at 37°C for 18 h, plating onto SMAC-CTVM without vancomycin (SMAC-CTM), and confirming presumptive E. coli O157 colonies as described above. This technique results in a positive or negative O157 culture, referred to as either E+ or 0.
The Nalr strain of E. coli O157 was identified by simultaneously subculturing presumptive E. coli O157 colonies on LB agar with and without naladixic acid (20 μg/ml; Sigma). Sample pH values were measured using a Φ 45 pH meter (Beckmann Instruments, Fullerton, Calif).
E. coli O157 fingerprint by pulsed-field gel electrophoresis (PFGE) of chromosomal DNA.
Genomic DNA from E. coli O157 isolates, cultured from steers H7 and H8 (see Table 1), were prepared as previously described (1). The agarose-embedded DNA was digested with 10 U of XbaI/plug (GibcoBRL) at 37°C overnight. PFGE was performed in a CHEF-DR II unit (Bio-Rad Laboratories, Hercules, Calif.) using 1% PFGE grade agarose-tris borate buffer gels (Boehringer Mannheim, Indianapolis, Ind.). The DNA was electrophoresed for 20 h at a constant voltage of 200 V (6 V/cm), pulse time of 5 to 50 s, an electric field angle of 120o, and a temperature of 15°C, before being stained with ethidium bromide. Resulting patterns were analyzed on a DNA ProScan ProRFLP program (DNA Proscan, Inc., Nashville, Tenn.), and the number and size of the DNA fragments were used as criteria for categorizing distinct patterns.
E. coli O157:H7 in vitro survival in digesta.
In vitro analysis of E. coli O157:H7 survival in ruminal and duodenal fluid was performed by inoculating E. coli O157:H7 ATCC 43895 into 5 ml of fresh ruminal or duodenal fluid, incubating at 37°C without shaking, and culturing for the bacteria at 0, 1, and 6 h postinoculation by the nonenrichment method described above. Preinoculation culturing was also performed to ensure E. coli O157 culture-negative status prior to inoculation.
Statistical analysis.
The SAS statistical package was used to compare data for significance with the Student's t test, analysis of variance (ANOVA), and repeated measures ANOVA. To analyze the data for possible differences between GIT locations or time and shedding status, for each animal the proportion of sampling times at which the animal was positive was recorded separately from rumen, duodenum, and fecal samples for days 1 to 7 postinoculation and for days 10 to 34 postinoculation. These proportions were arcsin-square root transformed (a standard method to improve variance homogeneity) prior to data analysis to help meet model assumptions (25). Repeated measures ANOVA was used to test for differences in transformed proportions due to GIT location, time, or GIT and time interaction (25).
RESULTS
All animals were healthy and free from disease at the onset of and throughout each study, even when they were actively shedding E. coli O157:H7. As in natural E. coli O157:H7 infections in ruminants, the experimental infections resulted in shedding of high-titer amounts of the bacteria in the feces, followed by declining titers and clearance of the bacteria over time.
Inoculation of cattle with E. coli O157:H7 resulted in three patterns of shedding.
Charolais/Hereford heifers and Holstein steers were inoculated with a single oral dose of E. coli O157:H7, and the results of fecal culture are shown in Table 1. We observed three distinct patterns of E. coli O157 shedding: animals that shed for about 1 week; animals that shed for about 1 month; and animals that shed for 2 months or more. The most common pattern we observed, among the 18 animals, was fecal shedding of E. coli O157 for about 1 month (9 of the 18 animals shed for 29 to 33 days). These animals shed E. coli O157 with titers of bacteria detectable by nonenrichment culture for 7 to 10 days followed by either consistent or intermittent enrichment culture-positive results for an average of 30.5 days. Three animals (H2, CH2, and CH4) were E. coli O157 culture positive for only 1 week (7 or 8 days), and four animals (H5, H6, H7, and H8) shed E. coli O157 for more than 2 months. Similar to those animals that shed for 1 month, these short-term and long-term shedders usually showed high titers during the first week postinoculation and thereafter were culture negative or positive with low titers (predominantly E+). Interestingly, the titers of fecal E. coli O157 during the first week were not significantly different (log transformed titers; P = 0.29, from ANOVA) between animals that went on to clear the bacteria rapidly or remained culture positive for 69 days. Also, there was no significant difference between the mean no. of E. coli O157 CFU/g of feces cultured from animals dosed with 108 CFU of O157/ml and animals dosed with 1010 CFU of O157/ml (P = 0.29).
Two animals (H7 and H8) were culture positive for E. coli O157 prior to experimental inoculation. Both of these animals shed for an extended period of time, though neither showed an unusually low or high number of E. coli O157:H7 bacteria in their feces nor did they show an unusual shedding pattern. As determined by analysis of E. coli O157 isolates by PFGE of chromosomal DNA, the strains shed by these animals postinoculation were the inoculum strain and not the original wild-type strain (data not shown).
We considered two animals (H11 and H13) not to have been colonized by E. coli O157 since they were fecal culture positive only once after inoculation.
To confirm the suggestion that cattle shed E. coli O157 in three distinct patterns, all cattle described in this paper (total of 24 animals represented in Tables 1, 3, and 4) were analyzed to determine the last day they were E. coli O157 culture positive. The distribution showed eight animals with values between 3 and 10 days (about 1 week), 13 animals with values between 22 and 38 days (about 1 month), and 3 animals with values between 56 and 69 days (more than 2 months). This pattern, with three sets of values that are separated from each other by either 12 or 18 days, suggests the existence of three distinct shedding patterns and was reinforced by density estimates and cluster analysis (data not shown).
TABLE 3.
E. coli O157 in GIT compartments of steers
| Day postinoculationa | No. of E. coli O157 CFU/g of digesta fromd:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rumen
|
Duodenum
|
Feces
|
||||||||||
| A1 | A2 | A3 | A4 | A1 | A2 | A3 | A4 | A1 | A2 | A3 | A4 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0.04 (1 h) | 3.2 × 104 | 4.6 × 104 | 1.3 × 105 | 3.9 × 104 | E+ | E+ | E+ | 1.9 × 103 | ND | ND | ND | ND |
| 0.08 (2 h) | 3.9 × 104 | 6.3 × 104 | 8.6 × 104 | 1.3 × 104 | E+ | 1.0 × 102 | 1.3 × 103 | 0 | ND | ND | ND | ND |
| 0.17 (4 h) | 3.1 × 103 | 2.4 × 103 | 2.5 × 104 | 7.5 × 104 | E+ | E+ | 5.0 × 103 | 5.0 × 101 | ND | ND | ND | ND |
| 0.25 (6 h) | 2.4 × 103 | 3.0 × 103 | 2.2 × 104 | 1.0 × 103 | E+ | E+ | 6.9 × 103 | E+ | E+ | 0 | E+ | 7.5 × 103 |
| 1 | 4.3 × 103 | 3.7 × 103 | 3.5 × 103 | 3.7 × 103 | E+ | E+ | E+ | 4.0 × 102 | 3.0 × 104 | 1.9 × 104 | 1.6 × 105 | 1.1 × 104 |
| 2 | E+b | E+ | 5.0 × 101 | E+ | E+ | E+ | E+ | 0 | 2.5 × 102 | 3.6 × 103 | 1.3 × 104 | 2.5 × 103 |
| 3 | E+ | E+ | 0 | E+ | 0 | E+ | E+ | 0 | 7.0 × 103 | 4.7 × 103 | 3.1 × 103 | 4.6 × 103 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | E+ | 0 | 3.7 × 103 | E+ | 2.0 × 102 | 2.0 × 102 |
| 10 | E+ | E+ | E+ | E+ | 0 | 0 | NDc | 0 | E+ | 0 | E+ | E+ |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | E+ | 0 |
| 16 | 0 | E+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | E+ | 0 | 0 | 0 |
| 24 | ND | 0 | ND | ND | ND | ND | ND | ND | E+ | ND | 0 | ND |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | E+ | 0 | E+ | 0 |
Angus steers with rumenal and duodenal cannulas were given a single dose of 2×1011 CFU E. coli O157:H7 ATCC 43894 directly into the rumen.
E+, digesta sample culture positive for E. coli O157 after enrichment culture only.
ND, not determined.
A1, A2, etc. identify animals from which samples were taken.
E. coli O157 persisted in the lower GIT of sheep.
To identify the GIT location of E. coli O157:H7 in animals that shed for various times, sheep were used. Twenty-four sheep, fed a pelleted alfalfa diet, were inoculated orally with 109 CFU of E. coli O157 ATCC 43894, serially sacrificed on a weekly interval, and fecal, tissue, and digesta samples along the GIT were cultured for the bacteria. The results are shown in Table 2. Prior to inoculation, no E. coli O157 was isolated from any of the fecal samples collected. Two days postinoculation, all sheep tested positive for E. coli O157. Seven days postinoculation, E. coli O157 was isolated from the feces of all four sacrificed animals, but it was not found in the ruminal, abomasal, or duodenal digesta and was only found in the lower ileum and lower GIT digesta of one or two animals. Similar to the digesta culture results during this first week, E. coli O157 was cultured from two of four tissue samples from the lower GIT locations. However, E. coli O157 was cultured from tissue samples in the rumen, abomasum, and duodenum of some (one or two) of the sacrificed sheep. No lesions were observed in any tissue sample.
By day 15 postinoculation, the number of E. coli O157:H7-positive animals decreased to three of the four sheep sacrificed. The bacteria were isolated from the feces and digesta, but not the tissues, collected from these animals. In fact, no subsequent tissue samples were culture positive for E. coli O157 and no lesions were observed. The digesta samples positive for E. coli O157:H7 were primarily restricted to the lower GIT, cecum, and ascending colon. Although one animal had cleared E. coli O157 by day 15, the remaining sheep were still fecal culture positive for the bacteria (data not shown). Titers of fecal E. coli O157 ranged from 104 CFU/g to levels detectable by enrichment culture only (less than 10 CFU/g). However, on days 22 and 35, E. coli O157 was not isolated from any digesta or tissue samples, even when half the animals sacrificed on those days had the bacterium in their feces. By day 42, two of the three remaining sheep had cleared E. coli O157 and one animal was positive only by enrichment culture (data not shown). To enhance shedding of E. coli O157 by an abrupt dietary change, feed was withheld for 24 h prior to sacrifice. In the one E. coli O157-positive animal, fecal shedding increased to 5.5 × 102 CFU E. coli O157/g of feces. The bacterium was also isolated from the descending colonic digesta and rectal tissue of this animal.
To further investigate the GIT location of E. coli O157:H7 in animals that harbored the bacteria for at least one month, five sheep that were consistently culture positive for 41 days were sacrificed on day 53 postinoculation. Fecal, digesta, and tissue samples were cultured. Two sheep had been fed forage and dosed with 1010 CFU of E. coli O157 ATCC 43894; three had been fed grain and dosed with 1010 CFU of Nalr E. coli O157. Postnecropsy, E. coli O157 was isolated from the feces of both forage-fed sheep and from the feces and the cecal digesta of one grain-fed sheep (data not shown). Digesta from all other GIT locations were culture negative. E. coli O157 was not isolated from any of tissue samples, and no tissue lesions were observed (data not shown). Two grain-fed sheep were culture negative the day of sacrifice.
Thus, in both sheep experiments, most animals shed E. coli O157 bacteria for about 1 month, but the bacteria were rarely cultured from upper GIT locations. In 10 animals that harbored E. coli O157 for more than 2 weeks, the bacteria were easily cultured from feces, but surprisingly were not found in any other GIT tissue or digesta samples. No differences in the E. coli O157 colonization pattern were observed based on diet or inoculum strain in this study.
E. coli O157 persisted in the lower GIT of cattle.
To confirm our sheep-related findings with cattle, four dually cannulated steers were inoculated in the rumen with a single bolus of E. coli O157:H7, and the results of digesta and fecal cultures are shown in Table 3. The pattern of fecal shedding of E. coli O157 by these cannulated steers was similar to the 1-week and 1-month patterns seen previously in the cattle (Table 1) and sheep (Table 2). Fecal samples from animals A2 and A4 were culture positive until days 7 and 10, while fecal samples from animals A1 and A3 were culture positive until the study ended at 34 days. Thus, among these animals, there were two patterns of E. coli O157 shedding, about 1 week and 1 month. Also similar to the data in Table 1, the initial titers of E. coli O157 in the feces were detectable by nonenrichment culture for 7 to 10 days, followed by either culture-negative status or intermittent enrichment culture-positive status.
From day 1 postinoculation through the end of the study, the most prevalent location of E. coli O157 was the lower GIT. We cultured E. coli O157 from the feces in three of four animals at 6 h postinoculation, indicating rapid travel of the inoculum through the GIT. In all animals at 6 h, E. coli O157 was cultured from all three locations, though titers in the feces were higher than titers in either the rumen or the duodenum. In addition, after 24 hours, the titers of E. coli O157 bacteria remaining in the rumen were low (≤50 CFU/g of digesta) and did not persist. Likewise, E. coli O157 did not persist in the duodenum and was not cultured by nonenrichment from this site in any animal longer than 24 h after inoculation. Also, the number of E. coli O157 CFU/g cultured from the duodenum was markedly lower than titers from the rumen and colon. By day three (72 h), only two of the four animals remained enrichment culture positive in the duodenum. Repeated-measures ANOVA showed that there was a significant difference among the three GIT locations for the proportion of samples that tested positive (P = 0.031). Individual tests showed that the duodenum and feces were significantly different, with a P value of 0.026. The duration that animals harbored E. coli O157 in the rumen and duodenum did not predict the persistence of E. coli O157 in the feces. Also, the titers of E. coli O157 bacteria in the rumen and duodenum during the first week postinoculation showed no difference between animals that shed for 1 week and those that shed for 1 month (P = 0.55).
Prior exposure to E. coli O157 did not effect the GIT location or shedding pattern.
Two steers (A1 and A2) were reinoculated with a lower (109) bolus of E. coli O157:H7 132 days after the initial inoculation to determine if prior exposure to the bacterium changed the quantity of O157 shed in the feces or its clearance from the rumen or duodenum. Two steers were chosen for reinoculation. Steer A1 previously shed E. coli O157 bacteria in its feces intermittently for 34 days, and steer A2 shed for 7 days. Upon reinoculation, the feces of both steers was E. coli O157 culture positive on or after day 30, and both harbored O157 in the rumen for less than 2 weeks and in the duodenum for 3 and 6 days, respectively (Table 4). This suggests that prior exposure to the bacteria did not prevent infection and/or affect the duration of fecal shedding. Again, E. coli O157 persisted in the lower GIT and not the rumen or the duodenum. Also, there was no significant difference between the numbers of E. coli O157 CFU/g shed in the feces by steers after the first and second inoculations (P = 0.93).
The location of positive E. coli O157 cultures from all cannulated steers were compiled and are shown in Table 5. After the initial days postinoculation, positive cultures were primarily from the feces. The bacteria rapidly dissipated from the rumen so that they were never cultured from that compartment after 16 days. They disappeared from the duodenal compartment even more rapidly. In fact, recovery of E. coli O157 from duodenal fluid declined within 48 h postinoculation until no sample was positive after 1 week, even in animals that shed the bacteria 34 days postinoculation. Combining the data from sheep and cattle, showed 17 animals in which E. coli O157 persisted for more than 1 week. E. coli O157 was cultured only from the lower GIT and not from the forestomach for 14 of these animals, while the bacteria were cultured from both the lower GIT and the forestomach (rumen) in three animals. Therefore, the proportion of animals in which E. coli O157 was only in the colon was 82% (95% confidence interval of 64 to 100%). This clearly suggests that ruminant animals that shed E. coli O157 bacteria for more than 1 week are colonized by the bacteria in the colon and not the rumen.
TABLE 5.
Compilation of the E. coli O157:H7 culture status in the GIT compartments of steers over the course of an infection
E. coli O157 survived in ruminal and duodenal fluid in vitro
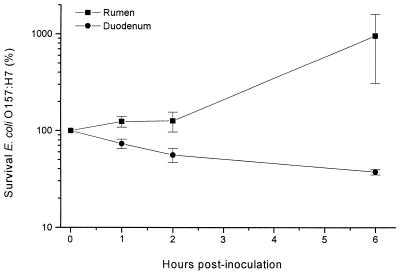
Since our in vivo findings indicated that the upper GIT was not the site of E. coli O157 persistence (Tables 2, 3, 4, and 5), the survival of this organism in ruminal and duodenal fluid was examined in vitro. To determine the variability of the GIT environment in forage-fed cannulated steer, we measured the mean pH values of digesta samples at various times. At hourly intervals, fluids were collected aseptically from two animals on two separate days. Rumen fluid was extracted from digesta particulate upon sampling. pH values were determined immediately after transport to the laboratory. The mean pH of the ruminal fluid and feces was neutral, ranging from 7.43 to 6.39 throughout the day. The mean pH of duodenal fluid was acidic, ranging from 4.84 to 4.19. Since little variation in pH was measured, ruminal and duodenal fluids were obtained from the cannulated steers at just one time (before feeding), inoculated in vitro with E. coli O157:H7, and monitored for bacterial survival. E. coli O157:H7 survived in both ruminal and duodenal fluid over the 6-h analysis (Fig. 1). However, the number of E. coli O157:H7 bacteria increased by about 10-fold in ruminal fluid and decreased by about 5-fold in duodenal fluid. Similar increases in the titers of E. coli O157 in the rumen of cannulated animals were not measured (Tables 3 and 4). This may be due to several factors, including digesta passage from the rumen, added volume to the rumen from feed and water consumption, and the competitive effects of the strict anaerobic microbiota in the rumen that did not survive or thrive under the conditions of this in vitro assay.
FIG. 1.
In vitro survival of E. coli O157:H7 in rumen or duodenal fluid. Fluids were collected aseptically from cannulated Angus steers, and rumen fluid was extracted from digesta particulate upon sampling. E. coli O157:H7 bacteria were added to GIT fluids at time zero. At various times, samples were analyzed for titers of E. coli O157 by dilution spread plate culture. Data are expressed as a percentage of the initial inoculum (± standard error) and were calculated from three independent experiments.
DISCUSSION
The most significant finding in this ruminant study was that E. coli O157 persisted in the lower GIT and was not harbored for long periods of time in the stomach (rumen, reticulum, omasum, and abomasum) or duodenum. In addition, the titers and location of E. coli O157 along the GIT early in an infection (first week postinoculation) did not predict the duration that animals harbored the bacteria. The infrequent ability to culture E. coli O157 from the rumen or the duodenum was not supported by in vitro studies of the bacterial survival in fluids from those compartments. Finally, three patterns of E. coli O157 shedding were observed among ruminants that were infected with a single bolus of E. coli O157:H7: animals that shed for less than 1 week, animals that shed for about 1 month, and animals that shed for longer than 2 months.
Ruminants that are naturally or experimentally exposed to E. coli O157:H7 appear to have identical infections. Adult animals harboring these bacteria are not sick and shed the bacteria in the feces in titers ranging from 105 CFU/g of feces to levels detectable only by sensitive selective enrichment culture procedures. The course of an infection is not different upon secondary oral exposure to the bacteria. Cohorts within a herd or an experiment vary in the rate at which they clear E. coli O157:H7 from the GIT. The fecal shedding measured in this study included animals that cleared the bacteria immediately (uninfected), after about 1 week, after about 1 month, or after more than 2 months post inoculation. This variation was observed in sheep and cattle regardless of breed, age, inoculum level, inoculum strain, or type of forage fed. The reason(s) for these differences in persistence are likely complex and may include animal immunity, physiological and biochemical GIT conditions, competing microbial flora, or the GIT location of infection.
This study identified the site of E. coli O157:H7 in the GIT of experimentally infected ruminants. Sheep were inoculated with the bacteria and serially sacrificed at weekly intervals to culture for the bacteria in GIT tissues and digesta. The recovery of E. coli O157 from the GIT always correlated with a positive fecal test. Early after inoculation (7 days) was the only time E. coli O157 was recovered from tissue samples along the GIT. In general, E. coli O157:H7 appeared to colonize the middle to lower GIT (lower ileum, cecum, and colon) of sheep during the first 2 weeks of infection. Subsequently, E. coli O157 was isolated only from the feces. The additional culturing of the organism from the cecum, descending colon, and rectum of some animals may indicate a dispersed presence of E. coli O157:H7 in the lower GIT, especially in animals that shed the bacteria for about 1 month (5 to 7 weeks). After feed was withheld for 1 day, increased excretion of E. coli O157 bacteria was induced in one sheep, and the bacteria were isolated from the descending colonic digesta and rectal tissue. This observation is consistent with our earlier report that feed disruptions enhance shedding of E. coli O157:H7 bacteria (27). In several of the sheep, we were unable to isolate E. coli O157 from either the digesta or tissue samples although the feces were culture positive. This may have been due to the presence of E. coli O157:H7 in low numbers not detectable by our culture protocol, our oversight in not sampling the appropriate GIT site, or the inability of our culture protocol to detect colonization of the tissue.
The relationship between sheep and E. coli O157:H7 is similar to that between cattle and this human pathogen. We and others have used sheep successfully to predict interactions between cattle and E. coli O157:H7 (9, 10, 26, 28). Our findings in sheep were confirmed in cattle fitted with dual cannulae so that digesta from the rumen and the duodenum could be accessed through the course of an infection. Animals were fed a forage-based diet because all previous experiments using this infection model in cattle and sheep show that forage-fed animals shed E. coli O157 longer than grain-fed cohorts (23, 27-29). Our goal was to compare the GIT location of the bacteria among animals that cleared the infection early and those that harbored the bacteria long term. To this end, inoculated steers cleared E. coli O157:H7 bacteria after 1 week (A2 and A4; Table 3), while others shed the bacteria for 1 month, albeit intermittently (A3, A4; Table 3) (A1; Table 4).
Like the results with sheep, after the first week, the recovery of E. coli O157 was primarily from the feces, presumably representative of the colonic digesta. During this time, recovery of the bacteria from the rumen and duodenum almost always correlated with positive fecal tests. The initial large bolus of bacteria moved rapidly through the GIT and was found within 1 h in the duodenum and 6 h postinoculation in the feces (Table 3). The low number of times that the bacteria were cultured from the rumen and not the duodenum or feces suggests that after the initial inoculum, E. coli O157:H7 bacteria in the rumen do not survive passage downstream (Table 5, days 10 to 16). This distribution of E. coli O157 in the GIT was the same in animals reinoculated with the bacteria. The phenomenon that oral exposure to E. coli O157:H7 does not result in protective immunity has previously been reported by us and others (11, 29).
The finding that E. coli O157:H7 resides primarily in the hindgut in both sheep and cattle is consistent with the distribution of generic E. coli along the bovine GIT. Although E. coli is found in the rumen and the upper GIT, both its titer and its proportion relative to total bacterial flora increase in the lower GIT (11). Also, our findings confirm the work of two other groups and most closely parallels that of Buchko et al., who found E. coli O157:H7 was rapidly eliminated from the rumen but persisted in the hindgut (cecum and colon) of 1-year-old steers fed three different grain diets (8). Earlier work by Dean-Nystrom et al. also suggests a hindgut location for E. coli O157:H7 in cattle that shed the organism long term (13). This is based on observations in a weaned calf model of EHEC infections that attaching and effacing lesions were found only in the rectum and cecum. In contrast, other researchers have found the primary site of E. coli O157:H7 to be the rumen and report that factors influencing the proliferation of E. coli O157:H7 in the forestomachs determine the magnitude of fecal shedding (7, 38). In this study, the E. coli O157:H7 titers and location in the rumen and the duodenum were not different among animals that went on to shed the bacteria for 1 week and those that shed for 1 month. The reason for both discrepancies may be differences between calves and adult cattle. The studies demonstrating persistence of E. coli O157:H7 in the rumen were done with 8- to 11-week old calves that had been weaned at 4 weeks old. These young animals may not have acquired the full component of ruminal microbiota and therefore may not have been fully ruminant, as was the case for the older animal in this and other studies (8, 13). The complex ruminal environment in an adult animal may limit the survival of E. coli O157:H7 (33), while the flora in a very young animal whose rumen is still developing may not.
The finding that E. coli O157 resides primarily in the hindgut also complements our earlier work showing that rapid colonic and cecal epithelial cell proliferation correlates with clearance of E. coli O157. It is hard to imagine that hindgut epithelial cell division would influence clearance of bacteria if they were originating with the rumen.
The striking fact that we were unable to culture E. coli O157 from the duodenal fluid so early after inoculation could be because the bacteria do not survive in that environment. However, here we showed by in vitro analysis that very few E. coli O157:H7 bacteria were killed in duodenal fluid from these steers. The lack of E. coli O157 in duodenal digesta may be explained by the possibility that the titers of E. coli O157:H7 were so low that it was not detected by our sensitive culture protocol (previously shown to detect as few as 1 CFU/10 g of feces [27]). However, it is most likely that E. coli O157:H7 does not pass from the rumen to the lower GIT because it is killed in the abomasum (gastric stomach). In forage-fed animals, E. coli O157 might be rapidly killed in the abomasum. Whether the abrupt disappearance of E. coli O157 from the upper GIT is unique to forage-fed animals was not tested here, but the work of Buchko et al. suggests that results with grain-fed animals will be similar. Studies are planned in our laboratory to compare these results to experiments with grain-fed animals. In any case, the persistence of E. coli O157:H7 depended on the ability of the bacteria to survive in the hindgut and not the rumen. Therefore, the site of E. coli O157:H7 persistence in the adult ruminant GIT is likely the colon.
Acknowledgments
This work was supported, in part, by the Idaho Agriculture Experiment Station, U.S. Department of Agriculture NRICGP grants 92-04350, 95-37201-1979, and 99-35201-8539, Public Health Service grant AI33981, and NO1-HD-0-3309 from the National Institutes of Health, and by grants from the United Dairymen of Idaho and the Idaho Beef Council.
We thank T. E. Besser for providing the NalR E. coli O157:H7 strain and D. D. Hancock for providing the cefixime. We acknowledge the technical assistance provided by H. Sheng, S. Wynia, K. R. Moore, J. G. Andrae, C. G. Doggett, J. V. Anderson, B. J. Sanford, U. M. Nance, K. A. Cloud, P. R. Austin, and T. Austin. We thank W. E. Carpenter for invaluable assistance in cattle handling and feeding. We also acknowledge D. Falk, S. Maki, and the University of Idaho farm personnel for support services.
L. J. Grauke and I. T. Kudva made similar contributions to this study.
REFERENCES
- 1.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besser, R. E., P. M. Griffin, and L. Slutsker. 1999. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu. Rev. Med. 50:355-367. [DOI] [PubMed] [Google Scholar]
- 3.Besser, R. E., S. M. Lett, J. T. Weber, M. P. Doyle, T. J. Barrett, J. G. Wells, and P. M. Griffin. 1993. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh-pressed apple cider. JAMA 269:2217-2220. [PubMed] [Google Scholar]
- 4.Besser, T. E., D. D. Hancock, L. C. Pritchett, E. M. McRae, D. H. Rice, and P. I. Tarr. 1997. Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. J. Infect. Dis. 175:726-729. [DOI] [PubMed] [Google Scholar]
- 5.Borczyk, A. A., M. A. Karmali, H. Lior, and L. M. Duncan. 1987. Bovine reservoir for verotoxin-producing Escherichia coli O157:H7. Lancet i:98. [DOI] [PubMed] [Google Scholar]
- 6.Boyce, T. G., D. L. Swerdlow, and P. M. Griffin. 1995. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N. Engl. J. Med 333:364-368. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. A., B. G. Harmon, T. Zhao, and M. P. Doyle. 1997. Experimental Escherichia coli O157:H7 carriage in calves. Appl. Environ. Microbiol. 63:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchko, S. J., R. A. Holley, W. O. Olson, V. P. Gannon, and D. M. Veira. 2000. The effect of different grain diets on fecal shedding of Escherichia coli O157:H7 by steers. J. Food Prot. 63:1467-1474. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, P. A. 2000. Sources of Escherichia coli O157 and experiences over the past 15 years in Sheffield, UK. J. Appl. Microbiol. 88(Suppl.):51S-60S. [DOI] [PubMed] [Google Scholar]
- 10.Cornick, N. A., S. L. Booher, T. A. Casey, and H. W. Moon. 2000. Persistent colonization of sheep by Escherichia coli O157:H7 and other E. coli pathotypes. Appl. Environ. Microbiol. 66:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cray, W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dargatz. 1997. Factors associated with the presence of Escherichia coli O157 in feces of feedlot cattle. J. Food Prot. 60:466-470. [DOI] [PubMed] [Google Scholar]
- 13.Dean-Nystrom, E. A., B. T. Bosworth, and H. W. Moon. 1999. Pathogenesis of Escherichia coli O157:H7 in weaned calves. Adv. Exp. Med. Biol. 473:173-177. [DOI] [PubMed] [Google Scholar]
- 14.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, J. R., T. Zhao, M. P. Doyle, M. R. Goldberg, C. A. Brown, C. T. Sewell, D. M. Kavanaugh, and C. D. Bauman. 2001. Experimental and field studies of Escherichia coli O157:H7 in white-tailed deer. Appl. Environ. Microbiol. 67:1218-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gansheroff, L. J., and A. D. O'Brien. 2000. Escherichia coli O157:H7 in beef cattle presented for slaughter in the U.S.: higher prevalence rates than previously estimated. Proc. Natl. Acad. Sci. USA 97:2959-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 19.Hancock, D. D. 1997. Effects of farm manure-handling practices on Escherichia coli O157 prevalence in cattle. J. Food Protect. 60:363-366. [DOI] [PubMed] [Google Scholar]
- 20.Hancock, D. D. 1997. Epidemiology of Escherichia coli O157 in feedlot cattle. J. Food Prot. 60:462-465. [DOI] [PubMed] [Google Scholar]
- 21.Hancock, D. D., T. E. Besser, M. L. Kinsel, P. I. Tarr, D. H. Rice, and M. G. Paros. 1994. The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington state. Epidemiol. Infect. 113:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock, D. D., T. E. Besser, D. H. Rice, D. E. Herriott, and P. I. Tarr. 1997. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 118:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hovde, C. J., P. R. Austin, K. A. Cloud, C. J. Williams, and C. W. Hunt. 1999. Effect of cattle diet on Escherichia coli O157:H7 acid resistance. Appl. Environ. Microbiol. 65:3233-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaper, J. B., and A. D. O'Brien (ed.). 1998. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains, p. 1-81. ASM Press, Washington, D.C.
- 25.Kirk, R. E. 1995. Experimental design: procedures for the behavioral sciences. Brooks/Cole Publishing Company, Pacific Grove, Calif.
- 26.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1997. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli serotypes isolated from sheep. J. Clin. Microbiol. 35:892-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1995. Effect of diet on the shedding of Escherichia coli O157:H7 in a sheep model. Appl. Environ. Microbiol. 61:1363-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1996. Escherichia coli O157:H7 in microbial flora of sheep. J. Clin. Microbiol. 34:431-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudva, I. T., C. W. Hunt, C. J. Williams, U. M. Nance, and C. J. Hovde. 1997. Evaluation of dietary influences on Escherichia coli O157:H7 shedding by sheep. Appl. Environ. Microbiol. 63:3878-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnuson, B. A., M. Davis, S. Hubele, P. R. Austin, I. T. Kudva, C. J. Williams, C. W. Hunt, and C. J. Hovde. 2000. Ruminant gastrointestinal cell proliferation and clearance of Escherichia coli O157:H7. Infect. Immun. 68:3808-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer-Broseta, S., S. N. Bastian, P. D. Arne, O. Cerf, and M. Sanaa. 2001. Review of epidemiological surveys on the prevalence of contamination of healthy cattle with Escherichia coli serogroup O157:H7. Int. J. Hyg. Environ. Health 203:347-361. [DOI] [PubMed] [Google Scholar]
- 32.Pritchard, G. C., G. A. Willshaw, J. R. Bailey, T. Carson, and T. Cheasty. 2000. Verocytotoxin-producing Escherichia coli 0157 on a farm open to the public: outbreak investigation and longitudinal bacteriological study. Vet. Rec. 147:259-264. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen, M. A., W. C. Cray, Jr., T. A. Casey, and S. C. Whipp. 1993. Rumen contents as a reservoir of enterohemorrhagic Escherichia coli. FEMS Microbiol. Lett. 114:79-84. [DOI] [PubMed] [Google Scholar]
- 34.Rice, D. H., D. D. Hancock, and T. E. Besser. 1995. Verotoxigenic E coli O157 colonisation of wild deer and range cattle. Vet. Rec. 137:524. [DOI] [PubMed] [Google Scholar]
- 35.Sanderson, M. W., T. E. Besser, J. M. Gay, C. C. Gay, and D. D. Hancock. 1999. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Vet. Microbiol. 69:199-205. [DOI] [PubMed] [Google Scholar]
- 36.Sargeant, J. M., J. R. Gillespie, R. D. Oberst, R. K. Phebus, D. R. Hyatt, L. K. Bohra, and J. C. Galland. 2000. Results of a longitudinal study of the prevalence of Escherichia coli O157:H7 on cow-calf farms. Am. J. Vet. Res. 61:1375-1379. [DOI] [PubMed] [Google Scholar]
- 37.Wells, J. G., L. D. Shipman, K. D. Greene, E. G. Sowers, J. H. Green, D. N. Cameron, F. P. Downes, M. L. Martin, P. M. Griffin, S. M. Ostroff, et al. 1991. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J. Clin. Microbiol. 29:985-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, T., M. P. Doyle, B. G. Harmon, C. A. Brown, P. O. Mueller, and A. H. Parks. 1998. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J. Clin. Microbiol. 36:641-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]